The Complex World of Emaraviruses—Challenges, Insights, and Prospects
Abstract
1. Introduction
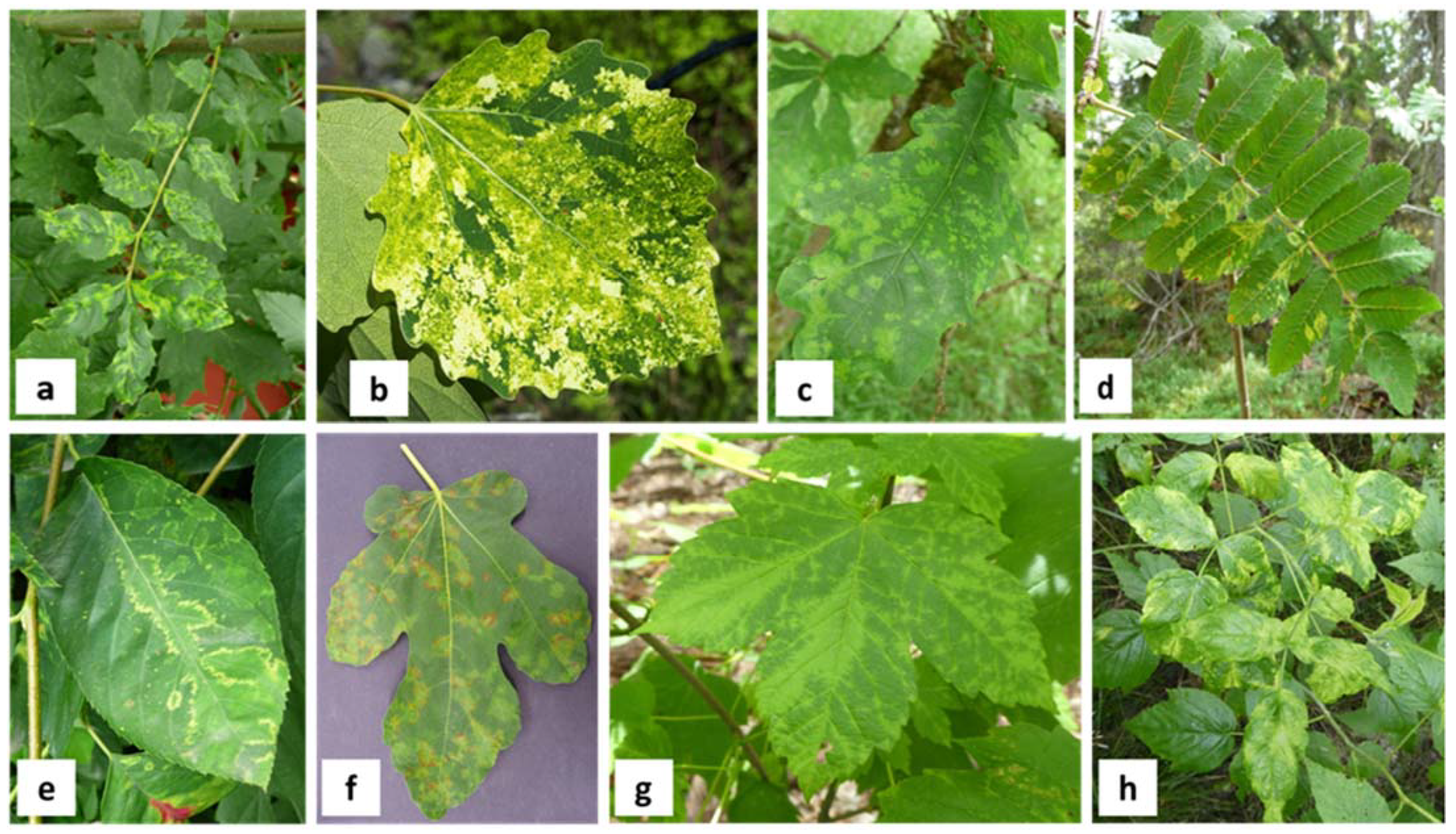
2. Symptomatology
3. Transmission and Host Range
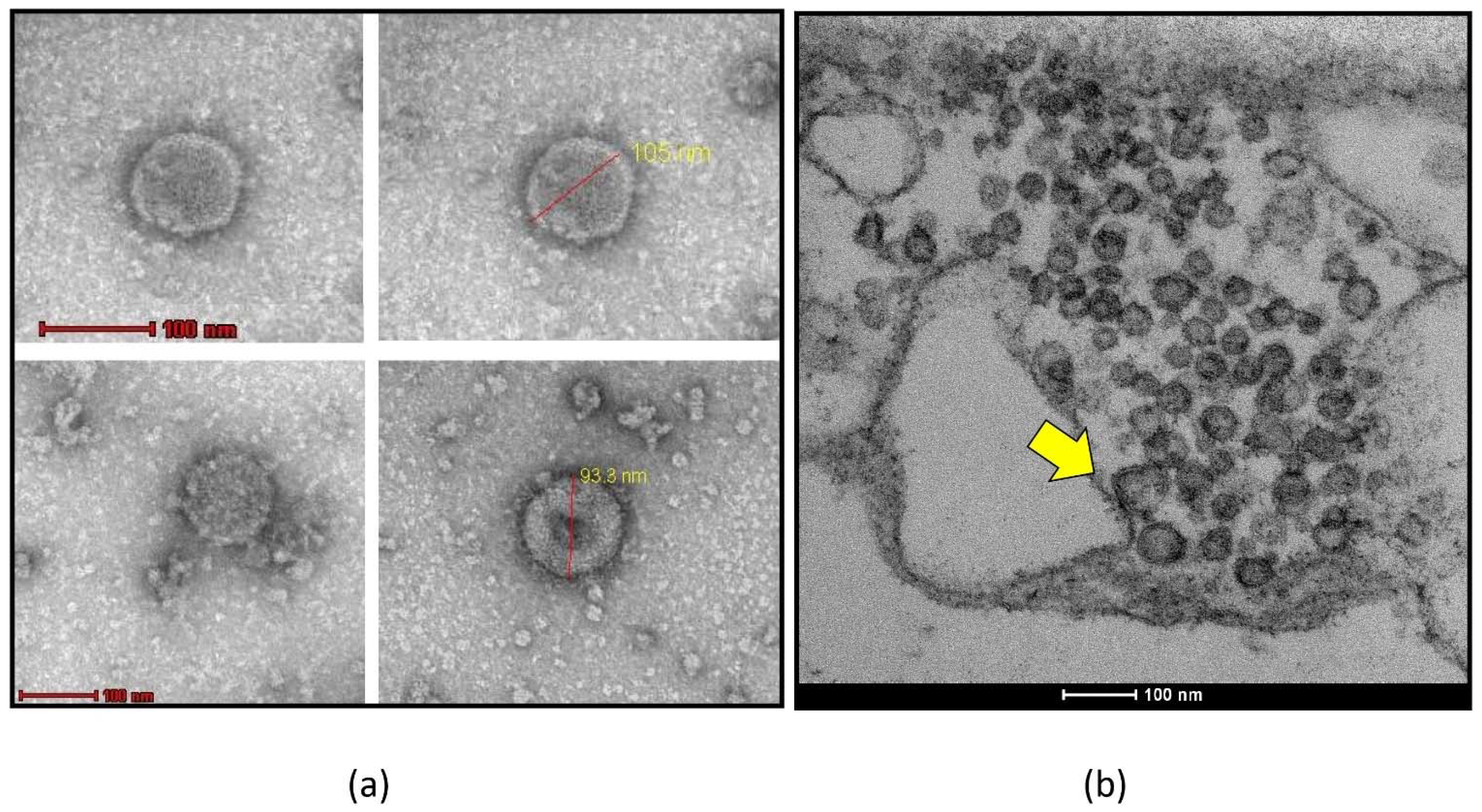
3.1. Transmission
3.2. Host Range
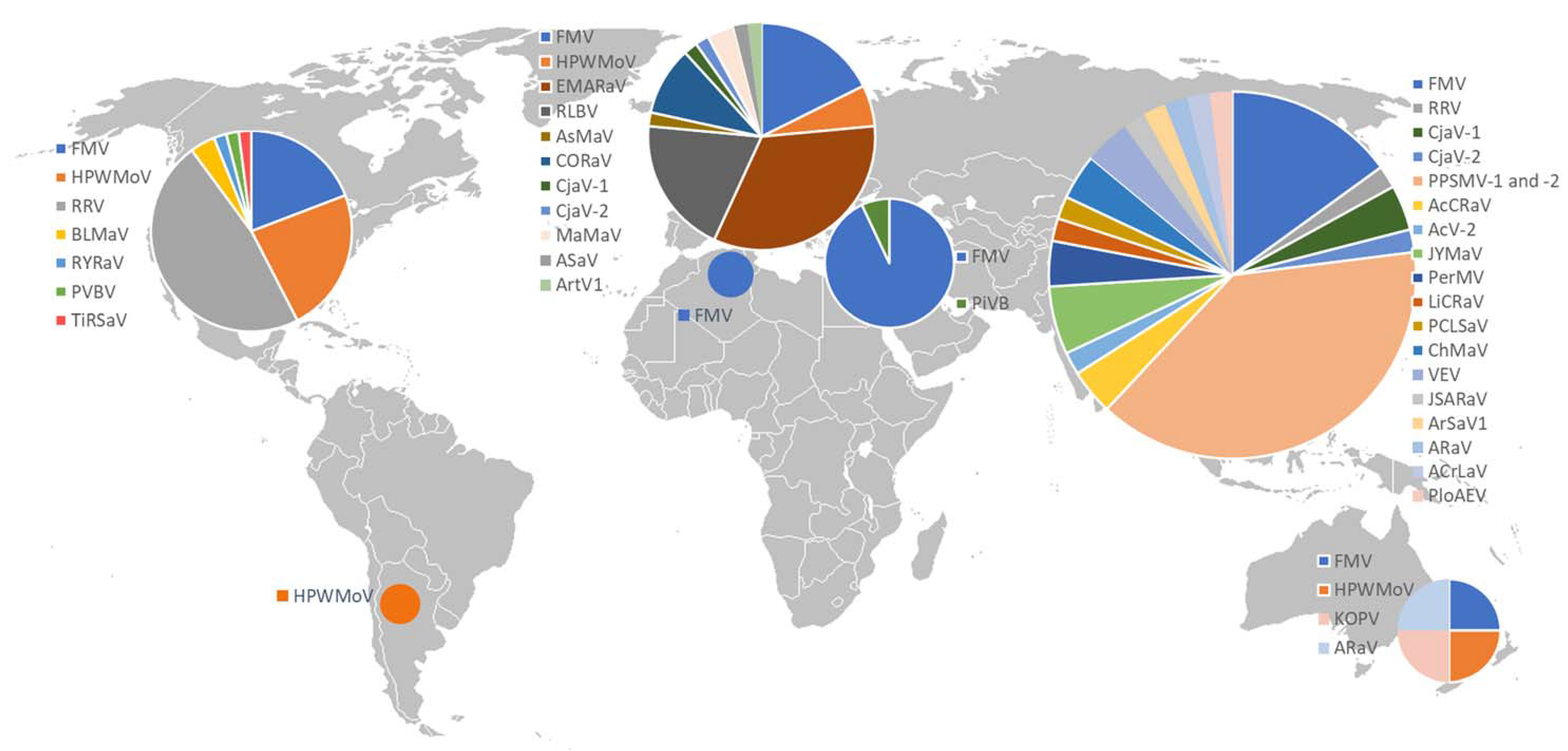
4. Geographic Occurrence
- In North America, endemic emaraviruses have been reported in large areas of the US and Canada. HPWMoV was reported from several US states comprising the Great Plains region [10,99,100]. RRV, initially reported from the Eastern USA, has also been detected in 36 US states including middle and western states and in East Canada [101,102]. Outside North America, RRV has so far only been reported in India [103]. In the Eastern United States, BLMaV was detected [36]. RYRaV, palo verde broom virus (PVBV), and TiRSaV have been reported in spatially limited regions.
- In Europe, RLBV has been reported from several northern countries [104], western countries [28], and the Balkan regions [105,106]. In recent years, a constantly growing number of emaraviruses affecting important deciduous tree species have been detected. Since its first description in German rowan trees in 2005 [6], EMARaV has been reported in several European countries including Austria, the Czech Republic, Sweden, Finland, Norway, and the United Kingdom [107]. Novel emaraviruses affecting oak, sycamore, ash, and aspen trees in forests and urban landscapes were detected in several Northern and Central European countries [17,19,20,21].
- In Asia, particularly in the southern and eastern region, a variety of emaraviruses affecting important commercial fruits and ornamental crops have been reported. In India, pigeonpea-infecting emaraviruses PPSMV-1 and PPSMV-2 are a major threat in most pigeonpea-producing regions where sterility mosaic disease is endemic. The disease was also reported in other parts of South-East Asia including Bangladesh, Nepal, Thailand, Myanmar, and Sri Lanka [108]. In China, AcCRaV and Actinidia virus 2 (AcV-2), both infecting kiwifruits, have been found in several provinces [35,39]. Further emaraviruses including JYMaV [40], PerMV [109], LiCRaV [47], Camellia chlorotic ringspot virus 1 (CaCRSV-1), and Camellia chlorotic ringspot virus 2 (CaCRSV-2) [44], PCLSaV [48], chrysanthemum mosaic-associated virus (ChMaV) [50,110], Japanese star anise ringspot-associated virus (JSARaV) [57], Vitis emaravirus (VEV) [53,54], alfalfa ringspot-associated virus (ARaV) [52], Ailanthus crinkle leaf-associated emaravirus (ACrLaV) [60], and PloAEV that infect important commercial fruits, ornamental crops, and trees have been described in China and Japan.
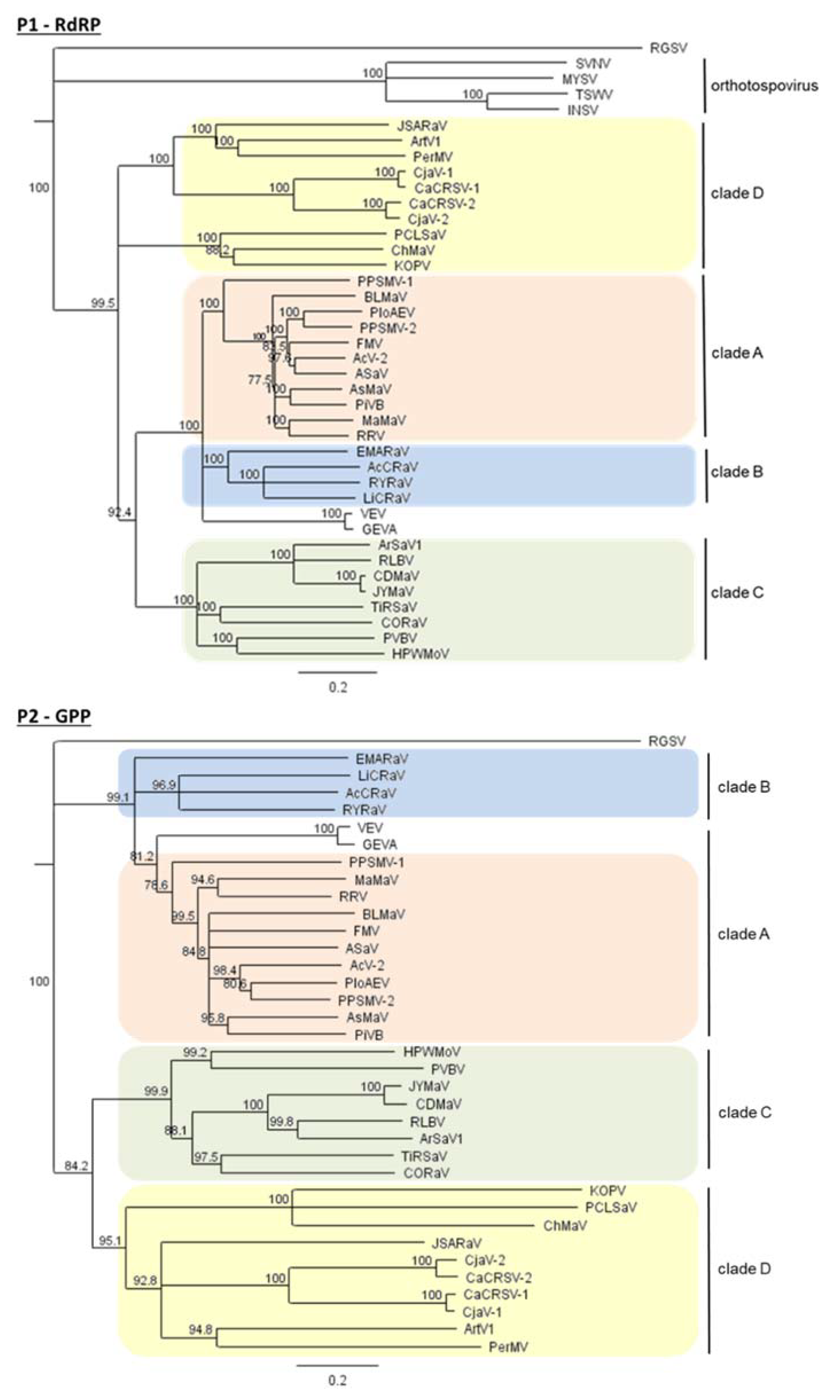
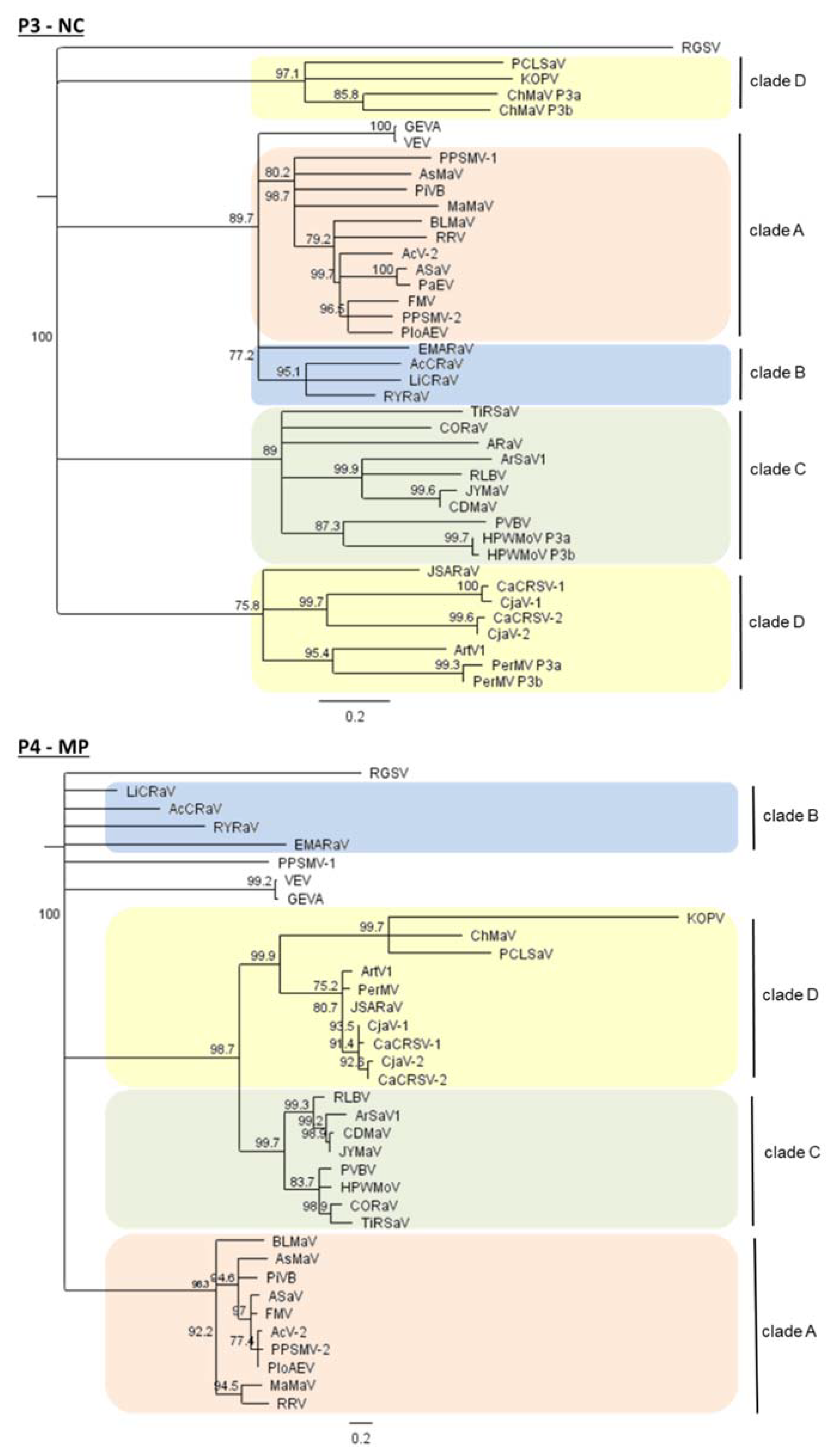
5. The Four Main Phylogenetic Clades of Emaraviruses
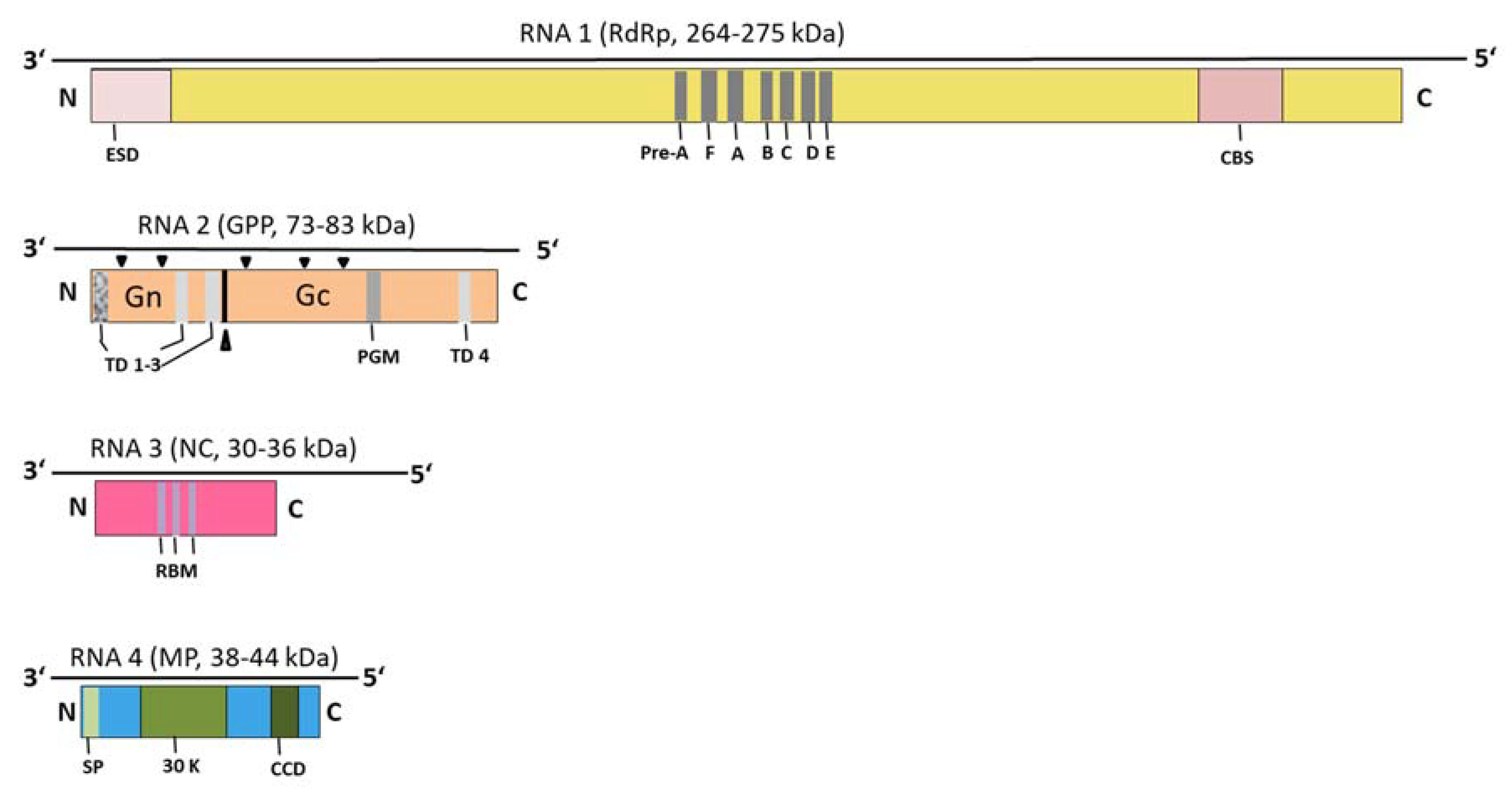
6. Genome Organization and Genetic Diversity
6.1. The Core Genome of Emaraviruses
6.2. Most Emaraviruses Encode Additional (Non-Core) Genome Segments
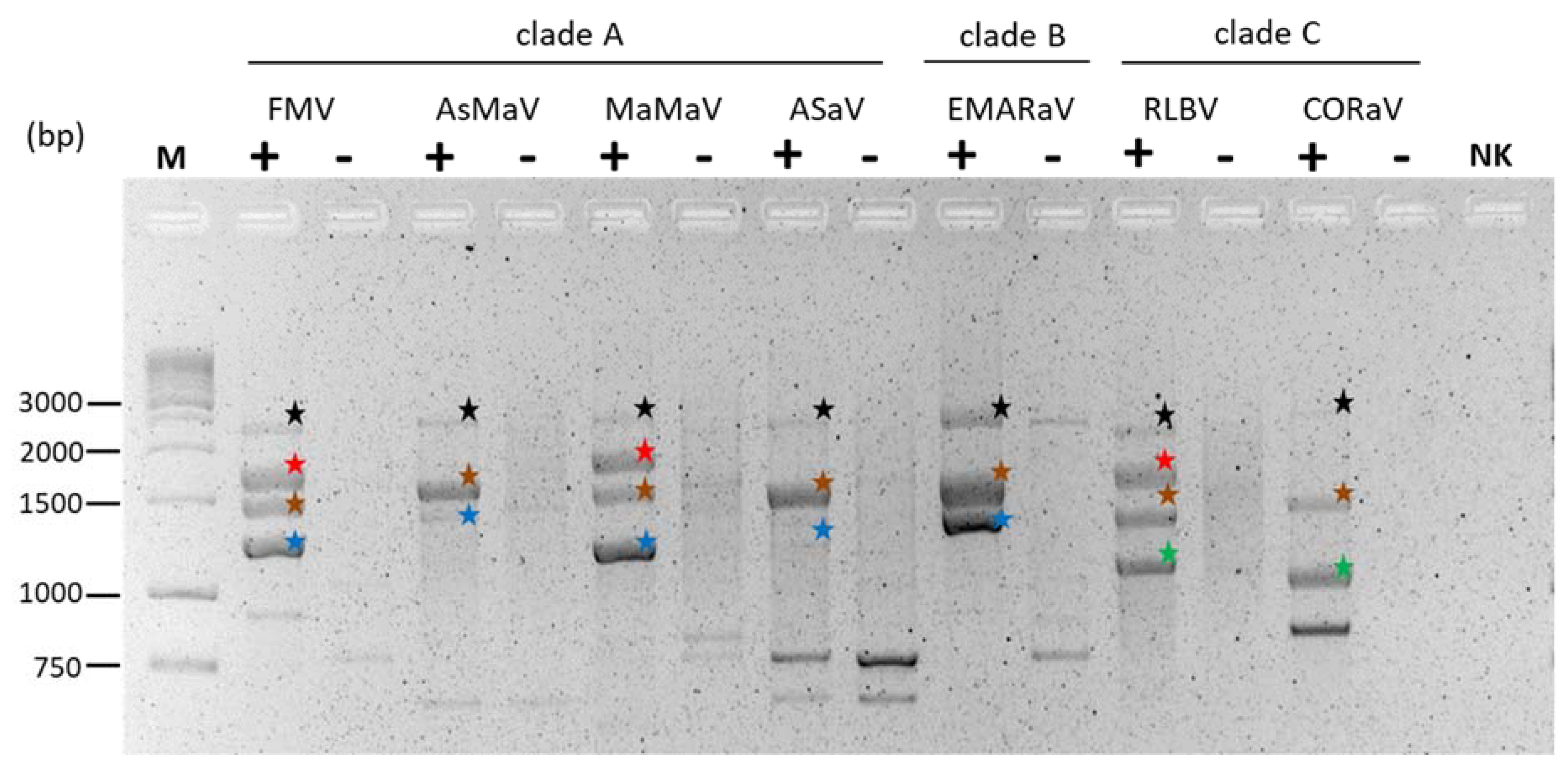
6.2.1. Full-Length RT–PCR as a Useful Approach to Detect and Characterize Emaraviral Genome Segments
6.2.2. Proteins Encoded by Non-Core Genome Segments Can Be Clustered into Three Main Groups
- (1)
- A protein of 460–500 aa, named P55 (in orange in Figure 8), found in some members of clades A, C, and D;
- (2)
- A protein of 170–250 aa, named “ABC” (in green in Figure 8), found in almost all members of clades A, B, and C;
- (3)
- A homolog of the sadwavirus Glu2–Pro glutamic protease (in yellow in Figure 8), found in some members of clades C and D.
6.2.3. Some Emaraviruses in Clades A, C, D Encode a Protein of ~55 kDa, P55, Containing a Highly Conserved C-Terminal Domain
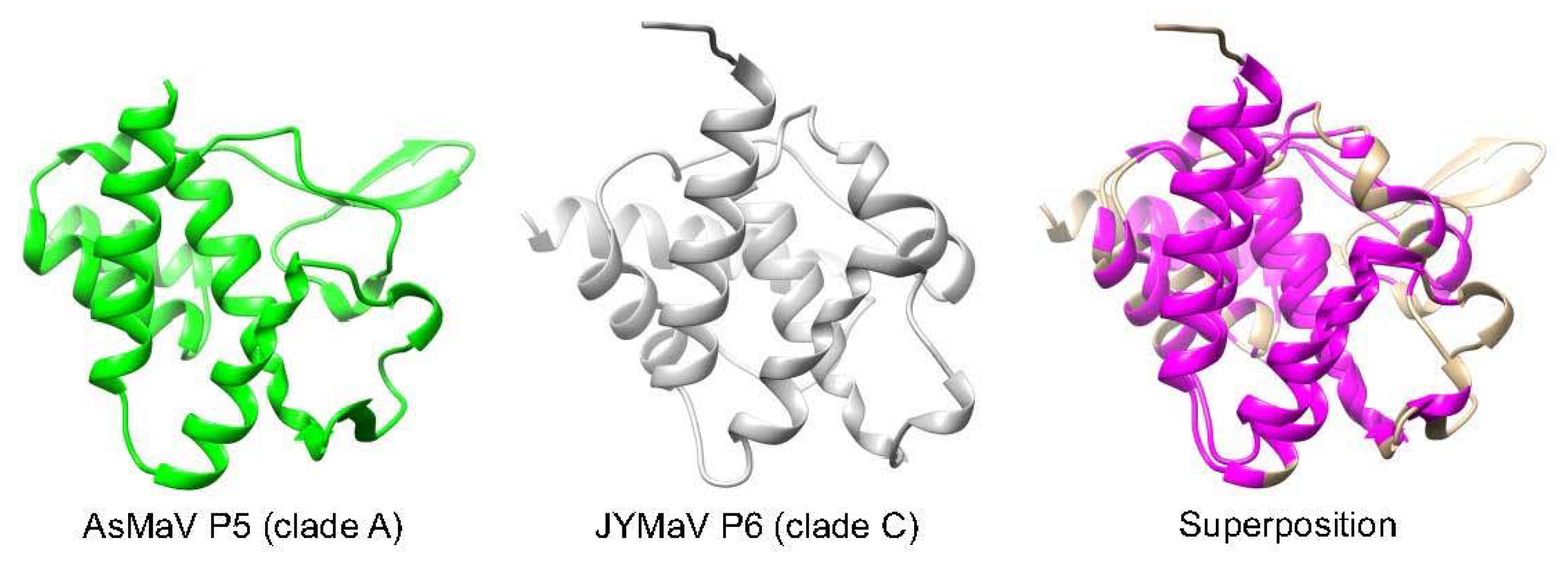
6.2.4. Almost All Emaraviruses in Clades A, B, and C Encode a Homologous “ABC” Protein of 18–27 kD, Involved in Pathogenicity
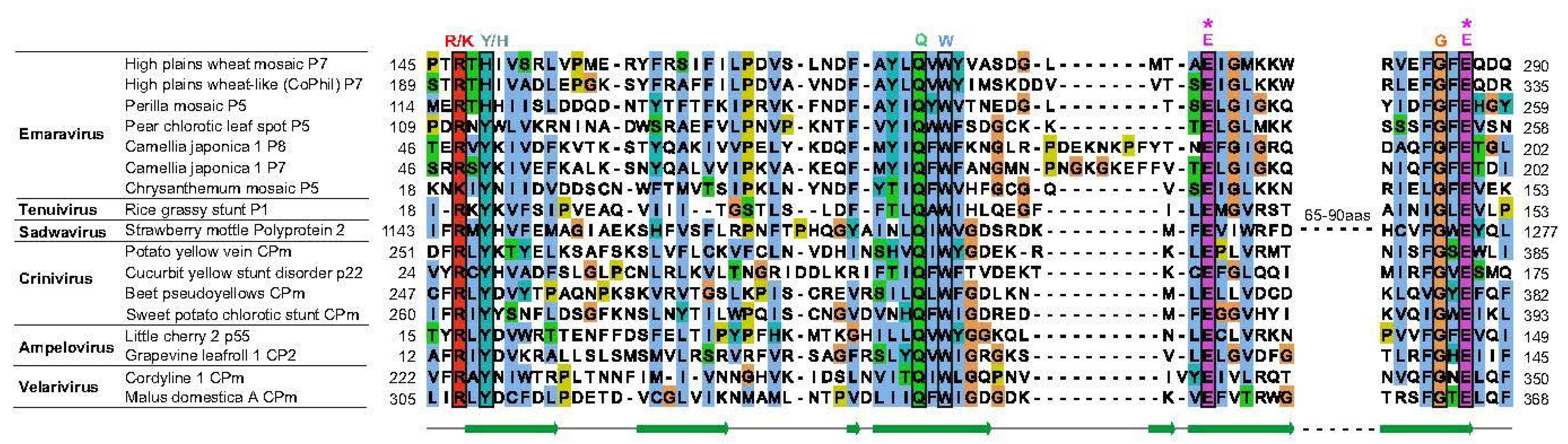
6.2.5. Some Emaraviruses from Clades C and D Encode a Homolog of the Sadwavirus Glu2–Pro Glutamic Protease
6.2.6. Species of Clade D Contain Non-Core Segments That Encode a Wider Variety of Proteins than in Other Clades
6.3. Incomplete Genome Information and Harmonization of Proteins Names
6.4. Terminal Ends of Genome Segments Are Conserved across Emaraviruses, with Variations in a Subgroup of Clade D
6.5. Emaraviruses Show Significant Intraspecific Genetic Variability
7. A Proposal: Dividing the Current Genus Emaravirus into At Least Two Genera
8. Established and Putative Members of the Genus
8.1. Established Emaraviruses
8.1.1. Pigeonpea Sterility Mosaic Virus 2
8.1.2. Blackberry Leaf Mottle-Associated Virus
8.1.3. Actinidia-Infecting Emaraviruses
8.1.4. Redbud Yellow Ringspot-Associated Virus
8.1.5. Pistacia Virus B
8.1.6. Palo Verde Broom Virus
8.1.7. Jujube-Infecting Emaraviruses
8.1.8. Ti Ringspot-Associated Virus
8.1.9. Perilla Mosaic Virus
8.1.10. Aspen Mosaic-Associated Virus
8.1.11. Lilac Chlorotic Ringspot-Associated Virus
8.1.12. Pear Chlorotic Leaf Spot-Associated Virus
8.1.13. Camellia-Infecting Emaraviruses
8.1.14. Common Oak Ringspot-Associated Virus
8.1.15. Maple Mottle-Associated Virus
8.1.16. Chrysanthemum Mosaic-Associated Virus
8.2. Putative Emaraviruses
8.2.1. Alfalfa Ringspot-Associated Virus
8.2.2. Ash Shoestring-Associated Virus
8.2.3. Karaka Okahu Purepure Virus
8.2.4. Emaraviruses in Grapevine
8.2.5. Japanese Star Anise Ringspot-Associated Virus
8.2.6. Arceuthobium Sichuanense-Associated Virus 1
8.2.7. Artemisia Fimovirus 1
8.2.8. Ailanthus Crinkle Leaf-Associated Emaravirus
8.2.9. Pueraria Lobata-Associated Emaravirus
8.2.10. Woolly Burdock Yellow Vein Virus
8.2.11. Yunnan Emara-like Virus
- partial sequence of RNA 1 of Illicium anisatum ringspot-associated virus
- sequence for two different RNA 1 similar to established emaraviruses, identified in wild potato species in South Africa [64]
9. Diagnosis and Control Strategies
9.1. Diagnosis Needs
9.1.1. Worldwide Dissemination
9.1.2. Detrimental Effect on Plant Health
9.1.3. Latent Infections
9.1.4. Incomplete Knowledge of Transmission Modes
9.1.5. Limited Availability of Serological Detection Methods
| Species | Detection Assay | Reference(s) |
|---|---|---|
| Genus-specific | RT–PCR targeting RNA 1 pre-motif A, motif A, and motif C RT–PCR targeting RNA 1 motifs F, A, and B | [110,158] |
| EMARaV | Species-specific RT–PCR, EBIA, qRT–PCR, Dot blot hybridization | [7,14,22,23,69,79,167] |
| FMV | Species-specific RT–PCR, RT–LAMP, electrochemical immunosensor, Western Blot, Dot immuno-binding and DAS–ELISA | [25,125,147,168,169,170] |
| RRV | ELISA, immuno dip-stick, IC–RT–PCR, multiplex RT–PCR, quantitative RT–PCR, RT–RPA, LAMP | [26,90,171,172,173,174,175,176] |
| RLBV | Species-specific RT–PCR, Dot-blot hybridization | [28,29,149] |
| PPSMV-1 | Species-specific RT–PCR, DAS–ELISA, DIBA, ELISA | [11,12,31,121,177] |
| PPSMV-2 | RT–PCR, ELISA | [31,32,121] |
| HPWMoV | Species-specific RT–PCR, ELISA (AGDIA), Northern Blot | [33,178] |
| RYRaV | Species-specific RT–PCR | [34] |
| AcCRaV | Species-specific RT–PCR, Multiplex RT–PCR | [35,179] |
| BLMaV | Species-specific RT–PCR, qRT–PCR | [36,180] |
| PiVB | Species-specific RT–PCR | [37] |
| PVBV | Species-specific RT–PCR | [38] |
| AcV-2 | Species-specific RT–PCR | [39] |
| JYMaV | Species-specific RT–PCR targeting RNA 3-RNA 6, virus protein detection with antibody by Western Blot | [40,41,153] |
| TiRSaV | RT–PCR using genus-specific primers and species-specific primers targeting RNA 1 | [42] |
| CjaV-1 and -2 | Species-specific qRT–PCR, RT–PCR targeting RNA 1, RNA 5 and RNA 6 | [43,45] |
| PerMV | Species-specific RT–PCR, immunoblot with specific antibody raised against peptide of viral P3 protein | [46] |
| AsMaV | Species-specific RT–PCR | [17] |
| LiCRaV | Species-specific RT–PCR | [47] |
| PCLSaV | Species-specific RT–PCR | [48] |
| CORaV | Species-specific RT–PCR | [18,19] |
| MaMaV | Species-specific RT–PCR | [20] |
| ChMaV | Species-specific RT–PCR | [50,110] |
| ARaV | HTS-based detection | [51] |
| VEV | Species-specific RT–PCR | [53] |
| ASaV | Species-specific RT–PCR | [21] |
| KOPV | Species-specific RT–PCR | [56] |
| JSARaV | Species-specific RT–PCR | [57] |
| ArSaV1 | HTS-based detection only | [58] |
| ArtV1 | HTS-based detection only | [59] |
| ACrLaV | HTS-based detection only | [60] |
| PloAEV | Species-specific RT–PCR | [61] |
9.2. Control Strategies
10. Research Requirements for Emaraviruses
- (1)
- Mapping all genome segments for each emaravirus;
- (2)
- Determining the functions of proteins encoded by non-core genome segments;
- (3)
- Understanding emaravirus–plant interactions;
- (4)
- Breeding resistant plants;
- (5)
- Clarifying transmission modes;
- (6)
- Investigating the evolutionary link between host range, transmission, and biology.
- Mapping all genome segments. The bioinformatic analysis presented here (Section 6.2) strongly suggests that some “non-core” genome segments might have been overlooked in some emaraviruses. One of our first priorities should be to map them and to ensure that no genome segment is overlooked when a new emaravirus is discovered. Our description of non-core segments typically found in some clades will help to achieve this aim.
- Determining the function of proteins encoded by non-core segments. The development of reverse genetic tools, recently introduced for RRV, has opened new possibilities for exploring the roles of emaravirus proteins [119]. This reverse-genetics approach can also be used to determine the function(s) of proteins encoded by non-core segments. The homologies we have identified and the conserved domains shared between many accessory proteins should help to accelerate this process, by suggesting functions common to homologs.
- Understanding emaravirus–plant interactions. The newly developed reverse genetic tools should also help us map molecular interactions between emaraviruses and their hosts or vectors [119]. For example, Pang et al. [93] observed different symptoms between Arabidopsis plants experimentally infected with Agrobacterium-delivered RRV cDNA clones and between naturally infected roses or infected N. benthamiana.
- Breeding resistant plants. Reverse genetics will also accelerate progress in breeding research to identify resistant host genotypes. Such research is essential for emaraviruses that cause strong damage to affected plants, but was until now painstakingly slow.
- Clarifying transmission modes. As long as the modes of transmission of emaraviruses are not fully characterized, no comprehensive strategy can be developed to combat them. Additional factors that contribute to virus spread cannot be ruled out at that time. They would tremendously influence the way control approaches can be best implemented. In particular, determining whether emaraviruses can be transmitted through water and soil would be especially interesting for long-living woody hosts in which the role of viruses on the overall health status is harder to elucidate [18].
- Investigating the evolutionary link between host range, transmission, and biology.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elbeaino, T.; Digiaro, M.; Mielke-Ehret, N.; Muehlbach, H.P.; Martelli, G.P.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Fimoviridae. J. Gen. Virol. 2018, 99, 1478–1479. [Google Scholar] [CrossRef] [PubMed]
- Baur, E. Über infektiöse Chlorosen bei Ligustrum Laburnum, Fraxinus, Sorbus und Ptelea. Ber. Der Dtsch. Bot. Gesellschaft. 1907, 25, 410–413. [Google Scholar]
- Kegler, H. Das Ringfleckenmosaik Der Eberesche (Sorbus Aucuparia L.). J. Phytopathol. 1960, 37, 214–216. [Google Scholar] [CrossRef]
- Büttner, C.; Führling, M. Studies on Virus Infection of Diseased Quercus Robur (L) from Forest Stands in Northern Germany. Ann. For. Sci. 1996, 53, 383–388. [Google Scholar] [CrossRef]
- Ebrahim-Nesbat, F.; Izadpanah, K. Viruslike Particles Associated with Ringfleck Mosaic of Mountain Ash and a Mosaic Disease of Raspberry in the Bavarian Forest. Eur. J. For. Pathol. 1992, 22, 1–10. [Google Scholar] [CrossRef]
- Benthack, W.; Mielke, N.; Büttner, C.; Mühlbach, H.P. Double-Stranded RNA Pattern and Partial Sequence Data Indicate Plant Virus Infection Associated with the Ringspot Disease of European Mountain Ash (Sorbus Aucuparia L.). Arch. Virol. 2005, 150, 37–52. [Google Scholar] [CrossRef]
- Mielke, N.; Muehlbach, H.P. A Novel, Multipartite, Negative-Strand RNA Virus Is Associated with the Ringspot Disease of European Mountain Ash (Sorbus Aucuparia L.). J. Gen. Virol. 2007, 88, 1337–1346. [Google Scholar] [CrossRef]
- Mielke-Ehret, N.; Mühlbach, H.P. Emaravirus: Anovel Genus of Multipartite, Negative Strand RNA Plant Viruses. Viruses 2012, 4, 1515–1536. [Google Scholar] [CrossRef]
- Elbeaino, T.; Digiaro, M.; Alabdullah, A.; De Stradis, A.; Minafra, A.; Mielke, N.; Castellano, M.A.; Martelli, G.P. A Multipartite Single-Stranded Negative-Sense RNA Virus Is the Putative Agent of Fig Mosaic Disease. J. Gen. Virol. 2009, 90, 1281–1288. [Google Scholar] [CrossRef]
- Jensen, S.G.; Lane, L.C.; Seifers, D.L. A New Disease of Maize and Wheat in the High Plains. Plant Dis. 1996, 80, 1387–1390. [Google Scholar] [CrossRef]
- Patil, B.L.; Kumar, P.L. Pigeonpea Sterility Mosaic Virus: A Legume-Infecting Emaravirus from South Asia. Mol. Plant Pathol. 2015, 16, 775–786. [Google Scholar] [CrossRef]
- Elbeaino, T.; Digiaro, M.; Uppala, M.; Sudini, H. Deep Sequencing of Pigeonpea Sterility Mosaic Virus Discloses Five RNA Segments Related to Emaraviruses. Virus Res. 2014, 188, 27–31. [Google Scholar] [CrossRef]
- King, A.M.Q.; Lefkowitz, E.; Adams, M.J.; Carstens, E.B. Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses; Academic Press: London, UK, 2012; pp. 767–769. [Google Scholar]
- von Bargen, S.; Dieckmann, H.L.; Candresse, T.; Mühlbach, H.P.; Roßbach, J.; Büttner, C. Determination of the Complete Genome Sequence of European Mountain Ash Ringspot-Associated Emaravirus from Sorbus Intermedia Reveals Two Additional Genome Segments. Arch. Virol. 2019, 164, 1937–1941. [Google Scholar] [CrossRef]
- von Bargen, S.; Bandte, M.; Al Kubrusli, R.; Jalkanen, R.; Büttner, C. First Report of European Mountain Ash Ringspot-Associated Virus in Karpatiosorbus × Hybrida in Finland. New Dis. Rep. 2020, 42, 1. [Google Scholar]
- von Bargen, S.; Tischendorf, M.; Büttner, C. First Report of European Mountain Ash Ringspot-associated Virus in Serviceberry (Amelanchier spp.) in Germany. New Dis. Rep. 2018, 37, 19. [Google Scholar] [CrossRef]
- von Bargen, S.; Al Kubrusli, R.; Gaskin, T.; Fürl, S.; Hüttner, F.; Blystad, D.R.; Karlin, D.G.; Jalkanen, R.; Büttner, C. Characterisation of a Novel Emaravirus Identified in Mosaic-Diseased Eurasian Aspen (Populus Tremula). Ann. Appl. Biol. 2020, 176, 210–222. [Google Scholar] [CrossRef]
- Bandte, M.; Rehanek, M.; Leder, B.; von Bargen, S.; Büttner, C. Identification of an Emaravirus in a Common Oak (Quercus Robur L.) Conservation Seed Orchard in Germany: Implications for Oak Health. Forests 2020, 11, 1174. [Google Scholar] [CrossRef]
- Rehanek, M.; von Bargen, S.; Bandte, M.; Karlin, D.G.; Büttner, C. A Novel Emaravirus Comprising Five RNA Segments Is Associated with Ringspot Disease in Oak. Arch. Virol. 2021, 166, 987–990. [Google Scholar] [CrossRef]
- Rumbou, A.; Candresse, T.; von Bargen, S.; Büttner, C. Next-Generation Sequencing Reveals a Novel Emaravirus in Diseased Maple Trees from a German Urban Forest. Front. Microbiol. 2021, 11, 621179. [Google Scholar] [CrossRef]
- Gaskin, T.R.; Tischendorf, M.; Günther, I.; Rehanek, M.; Büttner, C.; von Bargen, S. Characterization of a Novel Emaravirus Affecting Ash Species (Fraxinus spp.) in Europe. Forests 2021, 12, 1574. [Google Scholar] [CrossRef]
- Führling, M.; Büttner, C. Transmission Experiments of Viruses to Woody Seedlings (Quercus Robur and Sorbus Aucuparia) by Grafting and Mechanical Inoculation. Eur. J. For. Path. 1995, 25, 129–135. [Google Scholar]
- Grimová, L.; Marek, M.; Konrady, M.; Ryšánek, P. Newly Identified Host Range of European Mountain Ash Ringspot-Associated Virus (EMARaV) and Its Distribution in the Czech Republic. For. Pathol. 2015, 45, 177–189. [Google Scholar] [CrossRef]
- Walia, J.J.; Salem, N.M.; Falk, B.W. Partial Sequence and Survey Analysis Identify a Multipartite, Negative-Sense RNA Virus Associated with Fig Mosaic. Plant Dis. 2009, 93, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Maejima, K.; Komatsu, K.; Kitazawa, Y.; Hashimoto, M.; Takata, D.; Yamaji, Y.; Namba, S. Identification and Characterization of Two Novel Genomic RNA Segments of Fig Mosaic Virus, RNA5 and RNA6. J. Gen. Virol. 2012, 93, 1612–1619. [Google Scholar] [CrossRef] [PubMed]
- Laney, A.G.; Keller, K.E.; Martin, R.R.; Tzanetakis, I.E. A Discovery 70 Years in the Making: Characterization of the Rose Rosette Virus. J. Gen. Virol. 2011, 92, 1727–1732. [Google Scholar] [CrossRef]
- Di Bello, P.L.; Ho, T.; Tzanetakis, I.E. The Evolution of Emaraviruses Is Becoming More Complex: Seven Segments Identified in the Causal Agent of Rose Rosette Disease. Virus Res. 2015, 210, 241–244. [Google Scholar] [CrossRef]
- McGavin, W.J.; Mitchell, C.; Cock, P.J.A.; Wright, K.M.; MacFarlane, S.A. Raspberry Leaf Blotch Virus, a Putative New Member of the Genus Emaravirus, Encodes a Novel Genomic RNA. J. Gen. Virol. 2012, 93, 430–437. [Google Scholar] [CrossRef]
- Lu, Y.; McGavin, W.; Cock, P.J.A.; Schnettler, E.; Yan, F.; Chen, J.; MacFarlane, S. Newly Identified RNAs of Raspberry Leaf Blotch Virus Encoding a Related Group of Proteins. J. Gen. Virol. 2015, 96, 3432–3439. [Google Scholar] [CrossRef]
- Kumar, P.L.; Jones, A.T.; Sreenivasulu, P.; Reddy, D.V.R. Breakthrough in the Identification of the Causal Virus of Pigeonpea Sterility Mosaic Disease. J. Mycol. Plant Pathol. 2000, 30, 249. [Google Scholar]
- Kumar, P.L.; Jones, A.T.; Reddy, D.V.R. A Novel Mite-Transmitted Virus with a Divided RNA Genome Closely Associated with Pigeonpea Sterility Mosaic Disease. Phytopathology 2003, 93, 71–81. [Google Scholar] [CrossRef]
- Elbeaino, T.; Digiaro, M.; Uppala, M.; Sudini, H. Deep sequencing of dsRNAs recovered from mosaic-diseased pigeonpea reveals the presence of a novel emaravirus: Pigeonpea sterility mosaic virus 2. Arch. Virol. 2015, 160, 2019–2029. [Google Scholar] [CrossRef]
- Tatineni, S.; McMechan, A.J.; Wosula, E.N.; Wegulo, S.N.; Graybosch, R.A.; French, R.; Hein, G.L. An Eriophyid Mite-Transmitted Plant Virus Contains Eight Genomic RNA Segments with Unusual Heterogeneity in the Nucleocapsid Protein. J. Virol. 2014, 88, 11834–11845. [Google Scholar] [CrossRef]
- Di Bello, P.L.; Laney, A.G.; Druciarek, T.; Ho, T.; Gergerich, R.C.; Keller, K.E.; Martin, R.R.; Tzanetakis, I.E. A Novel Emaravirus Is Associated with Redbud Yellow Ringspot Disease. Virus Res. 2016, 222, 41–47. [Google Scholar] [CrossRef]
- Zheng, Y.; Navarro, B.; Wang, G.; Wang, Y.; Yang, Z.; Xu, W.; Zhu, C.; Wang, L.; Di Serio, F.; Hong, N. Actinidia Chlorotic Ringspot-Associated Virus: A Novel Emaravirus Infecting Kiwifruit Plants. Mol. Plant Pathol. 2017, 18, 569–581. [Google Scholar] [CrossRef]
- Hassan, M.; Di Bello, P.L.; Keller, K.E.; Martin, R.R.; Sabanadzovic, S.; Tzanetakis, I.E. A New, Widespread Emaravirus Discovered in Blackberry. Virus Res. 2017, 235, 1–5. [Google Scholar] [CrossRef]
- Buzkan, N.; Chiumenti, M.; Massart, S.; Sarpkaya, K.; Karadağ, S.; Minafra, A. A New Emaravirus Discovered in Pistacia from Turkey. Virus Res. 2019, 263, 159–163. [Google Scholar] [CrossRef]
- Ilyas, M.; Avelar, S.; Schuch, U.K.; Brown, J.K. First Report of an Emaravirus Associated with Witches’ Broom Disease and Eriophyid Mite Infestations of the Blue Palo Verde Tree in Arizona. Plant Dis. 2018, 102, 1863. [Google Scholar] [CrossRef]
- Wang, Y.; Zhai, L.; Wen, S.; Yang, Z.; Wang, G.; Hong, N. Molecular Characterization of a Novel Emaravrius Infecting Actinidia Spp. in China. Virus Res. 2020, 275, 197736. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, S.; Han, T.; Fu, J.; Di Serio, F.; Cao, M. Identification and Characterization of a Novel Emaravirus Associated with Jujube (Ziziphus JujubaMill.) Yellow Mottle Disease. Front. Microbiol. 2019, 10, 1417. [Google Scholar] [CrossRef]
- Liu, D.; Bai, J.; Fang, R.; Zhang, L. Full Genome Sequence of Chinese Date Mosaic-Associated Virus, a New Member of the Genus Emaravirus. Acta Microbiol. Sin. 2020, 60, 397–405. [Google Scholar] [CrossRef]
- Olmedo-Velarde, A.; Park, A.C.; Sugano, J.; Uchida, J.Y.; Kawate, M.; Borth, W.B.; Hu, J.S.; Melzer, M.J. Characterization of Ti Ringspot-Associated Virus, a Novel Emaravirus Associated with an Emerging Ringspot Disease of Cordyline Fruticosa. Plant Dis. 2019, 103, 2345–2352. [Google Scholar] [CrossRef]
- Peracchio, C.; Forgia, M.; Chiapello, M.; Vallino, M.; Turina, M.; Ciuffo, M. A Complex Virome Including Two Distinct Emaraviruses Associated with Virus-like Symptoms in Camellia Japonica. Virus Res. 2020, 286, 197964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, L.; Ma, L.; Tian, X.; Li, R.; Zhou, C.; Cao, M. Virome of Camellia Japonica: Discovery of and Molecular Characterization of New Viruses of Different Taxa in Camellias. Front. Microbiol. 2020, 11, 945. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, J.; Cheng, X. First Report of Camellia Japonica Associated Emaravirus 1 Associated with Camellia Leaf Ringspot and Flower Color-Breaking Disease in China. Plant Dis. 2020, 104, 3271. [Google Scholar] [CrossRef]
- Kubota, K.; Usugi, T.; Tomitaka, Y.; Shimomoto, Y.; Takeuchi, S.; Kadono, F.; Yanagisawa, H.; Chiaki, Y.; Tsuda, S. Perilla Mosaic Virus Is a Highly Divergent Emaravirus Transmitted by Shevtchenkella sp. (Acari: Eriophyidae). Phytopathology 2020, 110, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, Y.; Cao, M.; Cheng, Q.; Wu, J.; Hu, T. Identification of a Novel Emaravirus Infecting Lilac through Next-Generation Sequencing. J. Integr. Agric. 2020, 19, 2064–2071. [Google Scholar] [CrossRef]
- Liu, H.; Wang, G.; Yang, Z.; Wang, Y.; Zhang, Z.; Li, L.; Waqas, M.; Hong, N.; Liu, H.; Wang, G.; et al. Identification and Characterization of a Pear Chiorotic Leaf Spot-Associated Virus, a Novel Emaravirus Associated with a Severe Disease of Pear Trees in China. Plant Dis. 2020, 104, 2786–2798. [Google Scholar] [CrossRef]
- Führling, M.; Büttner, C. Nachweis von Tobamo-Viren in Bergahorn (Acer Pseudoplatanus L.) Mit Scheckung Und Blattdeformation. Forstw. Cbl. 1998, 117, 92–97. [Google Scholar] [CrossRef]
- Kubota, K.; Yanagisawa, H.; Chiaki, Y.; Yamasaki, J.; Horikawa, H.; Tsunekawa, K.; Morita, Y.; Kadono, F. Complete Nucleotide Sequence of Chrysanthemum Mosaic-Associated Virus, a Novel Emaravirus Infecting Chrysanthemum. Arch. Virol. 2021, 166, 1241–1245. [Google Scholar] [CrossRef]
- Samarfard, S.; McTaggart, A.R.; Sharman, M.; Bejerman, N.E.; Dietzgen, R.G. Viromes of Ten Alfalfa Plants in Australia Reveal Diverse Known Viruses and a Novel RNA Virus. Pathogens 2020, 9, 214. [Google Scholar] [CrossRef]
- Li, J.; Shang, Q.; Liu, Y.; Dai, W.; Li, X.; Wei, S.; Hu, G.; Mcneill, M.R.; Ban, L. Occurrence, Distribution, and Transmission of Alfalfa Viruses in China. Viruses 2022, 14, 1519. [Google Scholar] [CrossRef]
- Nabeshima, T.; Abe, J. High-Throughput Sequencing Indicates Novel Varicosavirus, Emaravirus, and Deltapartitivirus Infections in Vitis Coignetiae. Viruses 2021, 13, 827. [Google Scholar] [CrossRef]
- Fan, X.; Li, C.; Zhang, Z.; Ren, F.; Hu, G.; Shen, H.; Zhang, B.; Dong, Y. Identification and Characterization of a Novel Emaravirus From Grapevine Showing Chlorotic Mottling Symptoms. Front. Microbiol. 2021, 12, 694601. [Google Scholar] [CrossRef]
- Gaafar, Y.Z.A.; Herz, K.; Hartrick, J.; Fletcher, J.; Blouin, A.G.; MacDiarmid, R.; Ziebell, H. Investigating the Pea Virome in Germany—Old Friends and New Players in the Field(S). Front. Microbiol. 2020, 11, 583242. [Google Scholar] [CrossRef]
- Rabbidge, L.O.; Blouin, A.G.; Chooi, K.M.; Higgins, C.M.; Macdiarmid, R.M. Characterisation and Distribution of Karaka Ōkahu Purepure Virus—A Novel Emaravirus Likely to Be Endemic to New Zealand. Viruses 2021, 13, 1611. [Google Scholar] [CrossRef]
- Shimomoto, Y.; Okada, T.; Ikeda, K.; Tatara, A.; Hasegawa, Y.; Yanagisawa, H.; Takeyama, S.; Hayashi, K.; Yano, K.; Morita, Y.; et al. Japanese Star Anise Ringspot-Associated Virus Is a Distinct Emaravirus Transmitted by the Eriophyid Mite (the Family Diptilomiopidae). J. Gen. Plant Pathol. 2022, 88, 69–80. [Google Scholar] [CrossRef]
- Sidharthan, V.K.; Chaturvedi, K.K.; Baranwal, V.K. Diverse RNA Viruses in a Parasitic Flowering Plant (Spruce Dwarf Mistletoe) Revealed through RNA-Seq Data Mining. J. Gen. Plant Pathol. 2022, 88, 138–144. [Google Scholar] [CrossRef]
- Rivarez, M.P.S.; Pecman, A.; Bačnik, K.; Carvalho Frreira, O.M.; Vučurović, A.; Seljak, G.; Mehle, N.; Gutierrrez-Aguirre, I.; Ravnikar, M.; Kutnjak, D. In-depth study of tomato and weed viromes reveals undiscovered plant virus diversity in an agroecosystem. bioRxiv 2022. [Google Scholar] [CrossRef]
- An, W.; Li, C.; Zhang, S.; Yu, M.; Cao, M.; Yang, C. A Putative New Emaravirus Isolated from Ailanthus Altissima (Mill.) Swingle with Severe Crinkle Symptoms in China. Arch. Virol. 2022, 167, 2403–2405. [Google Scholar] [CrossRef]
- Liang, X.; Mei, S.; Yu, H.; Zhang, S.; Wu, J.; Cao, M. Mixed Infection of an Emaravirus, a Crinivirus, and a Begomovirus in Pueraria Lobata (Willd) Ohwi. Front. Microbiol. 2021, 13, 926724. [Google Scholar] [CrossRef]
- Bi, Y.; Tugume, A.K.; Valkonen, J.P.T. Small-RNA Deep Sequencing Reveals Arctium Tomentosum as a Natural Host of Alstroemeria Virus X and a New Putative Emaravirus. PLoS ONE 2012, 7, e42758. [Google Scholar] [CrossRef]
- Chen, Y.M.; Sadiq, S.; Tian, J.H.; Chen, X.; Lin, X.D.; Shen, J.J.; Chen, H.; Hao, Z.Y.; Wille, M.; Zhou, Z.C.; et al. RNA Viromes from Terrestrial Sites across China Expand Environmental Viral Diversity. Nat. Microbiol. 2022, 7, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Mahlanza, T.; Pierneef, R.E.; Makwarela, L.; Roberts, R.; van der Merwe, M. Metagenomic Analysis for Detection and Discovery of Plant Viruses in Wild Solanum spp. in South Africa. Plant Pathol. 2022, 71, 1633–1644. [Google Scholar] [CrossRef]
- Mitra, H. Report of the Imperial Mycologist; Scientific Reports of the Agricultural Research Institute, Pusa; Superintendent of Government Printing: Calcutta, India, 1931.
- Condit, I.J.; Horne, W. A Mosaic of the Fig in California. Phytopathology 1933, 23, 887–896. [Google Scholar]
- Kumar, P.L.; Jones, A.T.; Waliyar, F. Biology, Etiology and Management of Pigeonpea Sterility Mosaic Disease. Annu. Rev. Plant Pathol. 2004, 3, 1–24. [Google Scholar]
- Allington, W.B.; Staples, R.; Viehmeyer, G. Transmission of Rose Rosette by the Eriophyid Mite, Phyllocoptes Fructiphilus. J. Econ. Entomol. 1968, 61, 1137–1140. [Google Scholar] [CrossRef]
- Kallinen, A.K.; Lindberg, I.L.; Tugume, A.K.; Valkonen, J.P.T. Detection, Distribution, and Genetic Variability of European Mountain Ash Ringspot-Associated Virus. Phytopathology 2009, 99, 344–352. [Google Scholar] [CrossRef]
- Martin, R.R.; MacFarlane, S.; Sabanadzovic, S.; Quito, D.; Poudel, B.; Tzanetakis, I.E. Viruses and Virus Diseases of Rubus. Plant Dis. 2013, 97, 168–182. [Google Scholar] [CrossRef]
- Di Bello, L.P. Understanding the Causal Agent of Rose Rosette Disease. Master’s Thesis, University of Arkansas, Fayetteville, AR, USA, 2015; pp. 1–76. [Google Scholar]
- Jamous, R.M.; Abu Zaitoun, S.Y.; Mallah, O.B.; Shtaya, M.; Elbeaino, T.; Ali-Shtayeh, M.S. Detection and Phylogenetic Analysis of Viruses Linked with Fig Mosaic Disease in Seventeen Fig Cultivars in Palestine. Plant Pathol. J. 2020, 36, 267–279. [Google Scholar] [CrossRef]
- Elbeaino, T.; Digiaro, M.; De Stradis, A.; Martelli, G.P. Partial Characterisation of a Closterovirus Associated with a Chlorotic Mottling of Fig. J. Plant Pathol. 2006, 88, 187–192. [Google Scholar]
- Elbeaino, T.; Digiaro, M.; De Stradis, A.; Martelli, G.P. Identification of a Second Member of the Family Closteroviridae in Mosaic-Diseased Figs. J. Plant Pathol. 2007, 89, 119–124. [Google Scholar]
- Elbeaino, T.; Digiaro, M.; Heinoun, K.; de Stradis, A.; Martelli, G.P. Fig Mild Mottle-Associated Virus, a Novel Closterovirus Infecting Fig. J. Plant Pathol. 2010, 92, 165–172. [Google Scholar]
- Elbeaino, T.; Kubaa, R.A.; Digiaro, M.; Minafra, A.; Martelli, G.P. The Complete Nucleotide Sequence and Genome Organization of Fig Cryptic Virus, a Novel Bipartite DsRNA Virus Infecting Fig, Widely Distributed in the Mediterranean Basin. Virus Genes 2011, 42, 415–421. [Google Scholar] [CrossRef]
- Preising, S.; Borges, D.F.; de Queiroz Ambrósio, M.M.; da Silva, W.L. A Fig Deal: A Global Look at Fig Mosaic Disease and Its Putative Associates. Plant Dis. 2021, 105, 727–738. [Google Scholar] [CrossRef]
- de Lillo, E.; Pozzebon, A.; Valenzano, D.; Duso, C. An Intimate Relationship Between Eriophyoid Mites and Their Host Plants—A Review. Front. Plant Sci. 2018, 9, 1786. [Google Scholar] [CrossRef]
- Mielke-Ehret, N.; Thoma, J.; Schlatermund, N.; Mühlbach, H.P. Detection of European Mountain Ash Ringspot-Associated Virus-Specific RNA and Protein P3 in the Pear Leaf Blister Mite Phytoptus Pyri (Eriophyidae). Arch. Virol. 2010, 155, 987–991. [Google Scholar] [CrossRef]
- Flock, R.A.; Wallace, J.M. Transmission of Fig Mosaic by the Eriophyid Mite Aceria Ficus. Phytopathology 1955, 45, 52–54. [Google Scholar]
- Seth, M.L. Transmission of Pigeon-Pea Sterility by an Eriophyid Mite. Indian Phytopathol. 1962, 15, 225–227. [Google Scholar]
- Kulkarni, N.K.; Kumar, P.L.; Muniyappa, V.; Jones, A.T.; Reddy, D.V.R. Transmission of Pigeon Pea Sterility Mosaic Virus by the Eriophyid Mite, Aceria Cajani (Acari: Arthropoda). Plant Dis. 2002, 86, 1297–1302. [Google Scholar] [CrossRef] [PubMed]
- Seifers, D.L.; Harvey, T.L.; Martin, T.J.; Jensen, S.G. Identification of the Wheat Curl Mite as the Vector of the High Plains Virus of Corn and Wheat. Plant Dis. 1997, 81, 1161–1166. [Google Scholar] [CrossRef]
- Gordon, S.C.; Taylor, C.E. Some Aspects of the Biology of the Raspberry Leaf and Bud Mite (Phyllocoptes (Eriophyes) Gracilis Nal.) Eriophyidae in Scotland. J. Hortic. Sci. 1976, 51, 501–508. [Google Scholar] [CrossRef]
- Lindquist, E.; Oldfield, G. Evolution of Eriophyoid Mites in Relation to Their Host Plants. In Eriophyoid Mites—Their Biology, Natural Enemies and Control; Lindquist, E., Sabelis, M., Bruin, J., Eds.; Elsevier Science Publishing: Amsterdam, The Netherlands, 1996; pp. 277–300. [Google Scholar]
- Proeseler, G. Transmission of Fig Mosaic Virus by the Gall Mite Aceria Ficus (Cotte). Zentralblatt für Bakteriol. Parasitenkunde Infekt. Hyg. 1969, 123, 288–292. [Google Scholar]
- Gray, S.M.; Banerjee, N. Mechanisms of Arthropod Transmission of Plant and Animal Viruses. Microbiol. Mol. Biol. Rev. 1999, 63, 128–148. [Google Scholar] [CrossRef] [PubMed]
- Alfieri, S.A.J. Fig Mosaic. Plant Pathol. Circ. 1967, 64, 1–2. Available online: https://www.fdacs.gov/content/download/11071/file/pp64.pdf (accessed on 18 October 2022).
- Büttner, C.; Koenig, R. Plant Viruses in Irrigation Water. In Biology, Detection, and Management of Plant Pathogens in Irrigation Water; Hong, C., Moorman, G.W., Wohanka, W., Büttner, C., Eds.; APS Print: St. Paul, ML, USA, 2014; ISBN 978-0-89054-491-4. [Google Scholar]
- Babu, B.; Washburn, B.K.; Miller, S.H.; Poduch, K.; Sarigul, T.; Knox, G.W.; Ochoa-Corona, F.M.; Paret, M.L. A Rapid Assay for Detection of Rose Rosette Virus Using Reverse Transcription-Recombinase Polymerase Amplification Using Multiple Gene Targets. J. Virol. Methods 2017, 240, 78–84. [Google Scholar] [CrossRef]
- Bandte, M.; von Bargen, S.; Landgraf, M.; Rybak, M.; Büttner, C. NNeue Erkenntnisse zu Viruserkrankungen an Bäumen Und Konsequenzen Für Die Baumpflege, 26th ed.; Haymarket Verlag: Braunschweig, Germany, 2022; ISBN 978-3-87815-279-8. [Google Scholar]
- Herath, V.; Romay, G.; Urrutia, C.D.; Verchot, J. Family Level Phylogenies Reveal Relationships of Plant Viruses within the Order Bunyavirales. Viruses 2020, 12, 1010. [Google Scholar] [CrossRef]
- Pang, M.; Gayral, M.; Lyle, K.; Shires, M.K.; Ong, K.; Byrne, D.; Verchot, J. Infectious DNA Clone Technology and Inoculation Strategy for Rose Rosette Virus That Includes All Seven Segments of the Negative-Strand RNA Genome. bioRxiv 2019, 712000. Available online: https://www.biorxiv.org/content/10.1101/712000v1 (accessed on 23 July 2019).
- Seifers, D.L.; Harvey, T.L.; Martin, T.J.; Jensen, S.G. A Partial Host Range of the High Plains Virus of Corn and Wheat. Plant Dis. 1998, 82, 875–879. [Google Scholar] [CrossRef]
- Elbeaino, T.; Marais, A.; Faure, C.; Trioano, E.; Candresse, T.; Parrella, G. High-Throughput Sequencing Reveals Cyclamen Persicum Mill. As a Natural Host for Fig Mosaic Virus. Viruses 2018, 10, 684. [Google Scholar] [CrossRef]
- Chirkov, S.; Tsygankova, S.; Rastorguev, S.; Mitrofanova, I.; Chelombit, S.; Boulygina, E.; Slobodova, N.; Sharko, F. First Report of Fig Mosaic Virus on Fig in Russia. Plant Dis. 2021, 105, 1–8. [Google Scholar] [CrossRef]
- Snihur, H.; Pozhylov, I.; Budzanivska, I.; Shevchenko, O. First Report of High Plains Wheat Mosaic Virus on Different Hosts in Ukraine. J. Plant Pathol. 2020, 102, 545–546. [Google Scholar] [CrossRef]
- Coutts, B.A.; Cox, B.A.; Thomas, G.J.; Jones, R.A.C. First Report of Wheat Mosaic Virus Infecting Wheat in Western Australia. Plant Dis. 2014, 98, 285. [Google Scholar] [CrossRef]
- Burrows, M.; Franc, G.; Rush, C.; Blunt, T.; Ito, D.; Kinzer, K.; Olson, J.; O’Mara, J.; Price, J.; Tande, C.; et al. Occurrence of Viruses in Wheat in the Great Plains Region, 2008. Plant Health Prog. 2009, 10, 1–7. [Google Scholar] [CrossRef]
- Stewart, L.R.; Paul, P.A.; Qu, F.; Redinbaugh, M.G.; Miao, H.; Todd, J.; Jones, M. Wheat Mosaic Virus (WMoV), the Causal Agent of High Plains Disease, Is Present in Ohio Wheat Fields. Plant Dis. 2013, 97, 1125. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Karlik, J.; Diaz-Lara, A.; Ong, K.; Mollov, D.; Haviland, D.; Golino, D. First Report of Rose Rosette Virus Associated with Rose Rosette Disease Affecting Roses in California. Plant Dis. 2019, 103, 380. [Google Scholar] [CrossRef]
- EPPO Global Database. Datasheet Rose Rosette Virus. Available online: https://www.cabi.org/isc/datasheet/47834#REF-DDB-140979 (accessed on 15 August 2022).
- Chakraborty, P.; Das, S.; Saha, B.; Karmakar, A.; Saha, D.; Saha, A. Rose Rosette Virus: An Emerging Pathogen of Garden Roses in India. Australas. Plant Pathol. 2017, 46, 223–226. [Google Scholar] [CrossRef]
- Bi, Y.; Artola, K.; Kurokura, T.; Hytönen, T.; Valkonen, J.P.T. First Report of Raspberry Leaf Blotch Virus in Raspberries in Finland. Plant Dis. 2012, 96, 1231. [Google Scholar] [CrossRef]
- Mavrič Pleško, I.; Viršček Marn, M.; Lazarova, S.; Peneva, V.; Širca, S.; Urek, G. First Detection of Raspberry Leaf Blotch Virus in Red Raspberry in Bulgaria. J. Plant Pathol. 2014, 96, 437. [Google Scholar] [CrossRef]
- Zindović, J.; Viršček Marn, M.; Mavrič Pleško, I. First Report of Raspberry Leaf Blotch Virus in Red Raspberry in Montenegro. J. Plant Pathol. 2015, 97, 398. [Google Scholar] [CrossRef]
- Roßbach, J.; Dieckmann, H.L.; Büttner, T.; Mühlbach, H.P.; Von Bargen, S.; Büttner, C. Genetic Variability and Phylogeny of European Mountain Ash Ringspot-Associated Virus RNA3 and RNA4. Forests 2015, 6, 4072–4087. [Google Scholar] [CrossRef]
- CABI/EPPO. Pigeonpea Sterility Mosaic Virus. [Distribution Map]. Available online: https://www.cabi.org/isc/datasheet/41746#toDistributionMaps (accessed on 16 August 2022).
- Kubota, K. Emergence of Emaraviruses, the Eriophyoid Mite-Transmitted Viruses in Plants. Uirusu 2017, 67, 37–48. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kubota, K.; Chiaki, Y.; Yanagisawa, H.; Yamasaki, J.; Horikawa, H.; Tsunekawa, K.; Morita, Y. Novel Degenerate Primer Sets for the Detection and Identification of Emaraviruses Reveal New Chrysanthemum Species. J. Virol. Methods 2021, 288, 113992. [Google Scholar] [CrossRef] [PubMed]
- Alemandri, V.; Mattio, M.F.; Rodriguez, S.M.; Truol, G. Geographical Distribution and First Molecular Detection of an Emaravirus, High Plains Wheat Mosaic Virus, in Argentina. Eur. J. Plant Pathol. 2017, 149, 743–750. [Google Scholar] [CrossRef]
- Mahmoud, S.Y.; Zeidan, E.S.H.; Fayez, K.A.; Shipat, R. Occurrence of Fig Mosaic Virus in Egypt. J. Agric. Technol. 2014, 10, 439–447. [Google Scholar]
- El Air, M.; Mahfoudhi, N.; Elbeaino, T.; Dhouibi, M.H.; Digiaro, M. Occurrence of Fig Mosaic Virus in Tunisian Fig Orchards. J. Plant Pathol. 2012, 94, S4.89. [Google Scholar] [CrossRef]
- Elçi, E.; Serçe, Ç.U.; Çaǧlayan, K. Phylogenetic Analysis of Partial Sequences from Fig Mosaic Virus Isolates in Turkey. Phytoparasitica 2013, 41, 263–270. [Google Scholar] [CrossRef]
- Alimoradian, M.; Rakhshandehroo, F.; Shams-Bakhsh, M. Prevalence and Phylogenetic Analysis of Fig Mosaic Virus and Fig Badnavirus-1 in Iran. J. Plant Prot. Res. 2016, 56, 122–128. [Google Scholar] [CrossRef][Green Version]
- Alhudaib, K. Incidence of Fig Leaf Mottle-Associated Virus and Fig Mosaic Virus in Eastern Province of Saudi Arabia. Int. J. Virol. 2011, 8, 128–132. [Google Scholar] [CrossRef]
- Minafra, A.; Chiumenti, M.; Martelli, G.P. Fig Tree Viruses in New Zealand. J. Plant Pathol. 2012, 94, S4.104. [Google Scholar] [CrossRef]
- Gergerich, R.C.; Kim, K.S. A Description of the Casual Agent of Rose Rosette Disease. Arkansas Farm Res. 1983, 32, 7. [Google Scholar]
- Verchot, J.; Herath, V.; Urrutia, C.D.; Gayral, M.; Lyle, K.; Shires, M.K.; Ong, K.; Byrne, D. Development of a Reverse Genetic System for Studying Rose Rosette Virus in Whole Plants. Mol. Plant-Microbe Interact. 2020, 33, 1209–1221. [Google Scholar] [CrossRef]
- Amroun, A.; Priet, S.; de Lamballerie, X.; Quérat, G. Bunyaviridae RdRps: Structure, Motifs, and RNA Synthesis Machinery. Crit. Rev. Microbiol. 2017, 43, 753–778. [Google Scholar] [CrossRef]
- Kumar, S.; Subbarao, B.; Hallan, V. Molecular Characterization of Emaraviruses Associated with Pigeonpea Sterility Mosaic Disease. Sci. Rep. 2017, 7, 1183. [Google Scholar] [CrossRef]
- Olschewski, S.; Cusack, S.; Rosenthal, M. The Cap-Snatching Mechanism of Bunyaviruses. Trends Microbiol. 2020, 28, 293–303. [Google Scholar] [CrossRef]
- Walia, J.J.; Falk, B.W. Fig Mosaic Virus MRNAs Show Generation by Cap-Snatching. Virology 2012, 426, 162–166. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Elbeaino, T.; Digiaro, M.; Martelli, G.P. Complete Nucleotide Sequence of Four RNA Segments of Fig Mosaic Virus. Arch. Virol. 2009, 154, 1719–1727. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Perrakis, A.; Sixma, T.K. AI Revolutions in Biology. EMBO Rep. 2021, 22, e54046. [Google Scholar] [CrossRef]
- Marx, V. Method of the Year: Protein Structure Prediction. Nat. Methods 2022, 19, 5–10. [Google Scholar] [CrossRef]
- Melcher, U. The “30K” Superfamily of Viral Movement Proteins. J. Gen. Virol. 2000, 81, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Karlin, D.G.; Lu, Y.; Wright, K.; Chen, J.; MacFarlane, S. Experimental and Bioinformatic Evidence That Raspberry Leaf Blotch Emaravirus P4 Is a Movement Protein of the 30K Superfamily. J. Gen. Virol. 2013, 94, 2117–2128. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Maejima, K.; Komatsu, K.; Netsu, O.; Keima, T.; Shiraishi, T.; Okano, Y.; Hashimoto, M.; Yamaji, Y.; Namba, S. Fig Mosaic Emaravirus P4 Protein Is Involved in Cell-to-Cell Movement. J. Gen. Virol. 2013, 94, 682–686. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Pan, S.; Peng, Z.; Zhang, Y.; Yang, J. MTM-Align: A Server for Fast Protein Structure Database Search and Multiple Protein Structure Alignment. Nucleic Acids Res. 2018, 46, W380–W386. [Google Scholar] [CrossRef]
- Gupta, A.K.; Hein, G.L.; Graybosch, R.A.; Tatineni, S. Octapartite Negative-Sense RNA Genome of High Plains Wheat Mosaic Virus Encodes Two Suppressors of RNA Silencing. Virology 2018, 518, 152–162. [Google Scholar] [CrossRef]
- Gupta, A.K.; Hein, G.L.; Tatineni, S. P7 and P8 Proteins of High Plains Wheat Mosaic Virus, a Negative-Strand RNA Virus, Employ Distinct Mechanisms of RNA Silencing Suppression. Virology 2019, 535, 20–31. [Google Scholar] [CrossRef]
- Mann, K.; Chisholm, J.; Sanfaçon, H. Strawberry Mottle Virus ( Family Secoviridae, Order Picornavirales ) Encodes a Novel Glutamic Protease To Process the RNA2 Polyprotein at Two Cleavage Sites. J. Virol. 2019, 93, e01679-18. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Gabler, F.; Nam, S.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef]
- Remmert, M.; Biegert, A.; Hauser, A.; Al, E. HHblits: Lightning-Fast Iterative Protein Sequence Searching by HMM-HMM Alignment. Nat. Methods 2012, 9, 173–175. [Google Scholar] [CrossRef]
- Ting, T.-Y.; Baharin, A.; Bazli Ramzi, A.; Ng, C.; Goh, H.-H. Neprosin Belongs to a New Family of Glutamic Peptidase Based on in Silico Evidence. Plant Physiol. Biochem. 2022, 183, 23–35. [Google Scholar] [CrossRef]
- Schräder, C.U.; Lee, L.; Rey, M.; Sarpe, V.; Man, P.; Sharma, S.; Zabrouskov, V.; Larsen, B.; Schriemer, D.C. Neprosin, a Selective Prolyl Endoprotease for Bottom-up Proteomics and Histone Mapping. Mol. Cell. Proteom. 2017, 16, 1162–1171. [Google Scholar] [CrossRef]
- Baharin, A.; Ting, T.-Y.; Goh, H.-H. Post-Proline Cleaving Enzymes (PPCEs): Classification, Structure, Molecular Properties, and Applications. Plants 2022, 11, 1330. [Google Scholar] [CrossRef]
- El-shami, M.; Pontier, D.; Lahmy, S.; Braun, L.; Picart, C.; Vega, D.; Hakimi, M.; Jacobsen, S.E.; Cooke, R.; Lagrange, T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007, 21, 2539–2544. [Google Scholar] [CrossRef]
- Lian, S.L.; Li, S.; Abadal, G.X.; Pauley, B.A.; Fritzler, M.J.; Chan, E.K.L. The C-Terminal Half of Human Ago2 Binds to Multiple GW-Rich Regions of GW182 and Requires GW182 to Mediate Silencing. RNA 2009, 2, 804–813. [Google Scholar] [CrossRef]
- Zielezinski, A.; Karlowski, W.M. Integrative Data Analysis Indicates an Intrinsic Disordered Domain Character of Argonaute-Binding Motifs. Bioinformatics 2015, 31, 332–339. [Google Scholar] [CrossRef]
- Trujillo, J.T.; Mosher, R.A. Identification and Evolutionary Characterization of ARGONAUTE-Binding Platforms. In Methods in Molecular Biology; Carbonell, A., Ed.; Humana Press: New York, NY, USA, 2017. [Google Scholar]
- Walia, J.J.; Willemsen, A.; Elci, E.; Caglayan, K.; Falk, B.W.; Rubio, L. Genetic Variation and Possible Mechanisms Driving the Evolution of Worldwide Fig Mosaic Virus Isolates. Phytopathology 2014, 104, 108–114. [Google Scholar] [CrossRef]
- Patil, B.L.; Dangwal, M.; Mishra, R. Variability of Emaravirus Species Associated with Sterility Mosaic Disease of Pigeonpea in India Provides Evidence of Segment Reassortment. Viruses 2017, 9, 183. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Lemmetty, A.; Latvala, S.; Samuilova, O.; Valkonen, J.P.T. Occurrence and Genetic Diversity of Raspberry Leaf Blotch Virus (RLBV) Infecting Cultivated and Wild Rubus Species in Finland. Ann. Appl. Biol. 2016, 168, 122–132. [Google Scholar] [CrossRef]
- Danesh-Amuz, S.; Rakhshandehroo, F.; Rezaee, S. Prevalence and Genetic Diversity of Fig Mosaic Virus Isolates Infecting Fig Tree in Iran. Acta Virol. 2014, 58, 245–252. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seifers, D.L.; Martin, T.J.; Harvey, T.L.; Fellers, J.P.; Michaud, J.P. Identification of the Wheat Curl Mite as the Vector of Triticum Mosaic Virus. Plant Dis. 2009, 93, 25–29. [Google Scholar] [CrossRef] [PubMed]
- von Bargen, S.; Arndt, N.; Robel, J.; Jalkanen, R.; Büttner, C. Detection and Genetic Variability of European Mountain Ash Ringspot-Associated Virus (EMARaV) in Sweden. For. Pathol. 2013, 43, 429–432. [Google Scholar] [CrossRef]
- Guo, J.; Wang, Y.; Wang, G.; Hong, J.; Yang, Z.; Bai, J.; Hong, N. Molecular Characteristics of Jujube Yellow Mottle-Associated Virus Infecting Jujube (Ziziphus Jujuba Mill.) Grown at Aksu in Xinjiang of China. Viruses 2021, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Valkonen, J.; Rännäli, M. First Report of European Mountain Ash Ringspot-Associated Virus in Sorbus Aucuparia from Eastern Karelia, Russia. Plant Dis. 2010, 94, 921. [Google Scholar] [CrossRef]
- Stewart, L.R. Sequence Diversity of Wheat Mosaic Virus Isolates. Virus Res. 2016, 213, 299–303. [Google Scholar] [CrossRef]
- Garcia-Arenal, F.; Fraile, A.; Malpica, J.M. Variation and Evolution of Plant Virus Populations. Int. Microbiol. 2003, 6, 225–232. [Google Scholar] [CrossRef]
- Current ICTV Taxonomy Release. Available online: https://ictv.global/taxonomy (accessed on 15 August 2022).
- Elbeaino, T.; Whitfield, A.; Sharma, M.; Digiaro, M. Emaravirus-Specific Degenerate PCR Primers Allowed the Identification of Partial RNA-Dependent RNA Polymerase Sequences of Maize Red Stripe Virus and Pigeonpea Sterility Mosaic Virus. J. Virol. Methods 2013, 188, 37–40. [Google Scholar] [CrossRef]
- Emaravirus Rosae(RRV000). Available online: https://gd.eppo.int/taxon/RRV000 (accessed on 15 August 2022).
- Phyllocoptes Fructiphilus(PHYCFR). Available online: https://gd.eppo.int/taxon/PHYCFR (accessed on 16 August 2022).
- Bragard, C.; Baptista, P.; Chatzivassiliou, E.; Di Serio, F.; Gonthier, P.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P.; et al. Pest Categorisation of High Plains Wheat Mosaic Virus. EFSA J. 2022, 20, e07302. [Google Scholar] [CrossRef]
- Patil, B.L. Plant Viral Diseases: Economic Implications. Encycl. Virol. 2021, 3, 81–97. [Google Scholar] [CrossRef]
- Waliczek, T.M.; Byrne, D.; Holeman, D. Opinions of Landscape Roses Available for Purchase and Preferences for the Future Market. Horttechnology 2018, 28, 807–814. [Google Scholar] [CrossRef]
- Vazquez-Iglesias, I.; Ochoa-Corona, F.M.; Tang, J.; Robinson, R.; Clover, G.R.G.; Fox, A.; Boonham, N. Facing Rose Rosette Virus: A Risk to European Rose Cultivation. Plant Pathol. 2020, 69, 1603–1617. [Google Scholar] [CrossRef]
- Büttner, C.; von Bargen, S.; Bandte, M.; Mühlbach, H.P. Forest Diseases Caused by Viruses. In Infectious Forest Diseases; Gonthier, P., Nicolotti, G., Eds.; CABI: Wallingford, UK, 2013; pp. 50–75. ISBN 978-1-78064-040-2. [Google Scholar]
- Büttner, C.; Landgraf, M.; Fernandez, H.; von Bargen, S.; Bandte, M. Virus Diseases of Forest and Urban Trees. In Forest Microbiology—Tree Diseases and Pests; Asiegbu, F.O., Kovalchuk, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; ISBN 9780443186943. [Google Scholar]
- Mielke, N.; Weber, M.; Khan, S.; Muehlbach, H.P. Detection of European Mountain Ash Ringspot-Associated Virus (EMARAV) in Sorbus Aucuparia L. by a Specific Antiserum and Reverse Transcription-PCR. For. Pathol. 2008, 38, 371–380. [Google Scholar] [CrossRef]
- Haji-Hashemi, H.; Safarnejad, M.R.; Norouzi, P.; Ebrahimi, M.; Shahmirzaie, M.; Ganjali, M.R. Simple and Effective Label Free Electrochemical Immunosensor for Fig Mosaic Virus Detection. Anal. Biochem. 2019, 566, 102–106. [Google Scholar] [CrossRef]
- Shahmirzaie, M.; Safarnejad, M.R.; Rakhshandehroo, F.; Safarpour, H.; Rabbani, H.; Zamanizadeh, H.R.; Elbeaino, T. Production of a Polyclonal Antiserum against Recombinant Nucleocapsid Protein and Its Application for the Detection of Fig Mosaic Virus. J. Virol. Methods 2019, 265, 22–25. [Google Scholar] [CrossRef]
- Soliman, H.I.A. Serological and Molecular Detection of Viruses Infecting Fig to Identify the Virus-Free Plants. J. Microbiol. Biotechnol. Food Sci. 2018, 8, 726–731. [Google Scholar] [CrossRef]
- Dobhal, S.; Olson, J.D.; Arif, M.; Garcia Suarez, J.A.; Ochoa-Corona, F.M. A Simplified Strategy for Sensitive Detection of Rose Rosette Virus Compatible with Three RT-PCR Chemistries. J. Virol. Methods 2016, 232, 47–56. [Google Scholar] [CrossRef]
- Jordan, R.; Guaragna, M.A.; Hammond, J. Development of Polyclonal and Monoclonal Antibodies to Rose Rosette Virus Nucleoprotein. Acta Hortic. 2018, 1193, 77–82. [Google Scholar] [CrossRef]
- Di Bello, P.L.; Thekke-Veetil, T.; Druciarek, T.; Tzanetakis, I.E. Transmission Attributes and Resistance to Rose Rosette Virus. Plant Pathol. 2018, 67, 499–504. [Google Scholar] [CrossRef]
- Salazar, A.; Ochoa-Corona, F.M.; Olson, J.D.; Babu, B.; Paret, M. Probing Loop-Mediated Isothermal Amplification (LAMP) Targeting Two Gene-Fragments of Rose Rosette Virus. PLoS ONE 2021, 16, e0256510. [Google Scholar] [CrossRef]
- Babu, B.; Jeyaprakash, A.; Jones, D.; Schubert, T.S.; Baker, C.; Washburn, B.K.; Miller, S.H.; Poduch, K.; Knox, G.W.; Ochoa-Corona, F.M.; et al. Development of a Rapid, Sensitive TaqMan Real-Time RT-PCR Assay for the Detection of Rose Rosette Virus Using Multiple Gene Targets. J. Virol. Methods 2016, 235, 41–50. [Google Scholar] [CrossRef]
- Babu, B.; Washburn, B.K.; Ertek, T.S.; Miller, S.H.; Riddle, C.B.; Knox, G.W.; Ochoa-Corona, F.M.; Olson, J.; Katırcıoğlu, Y.Z.; Paret, M.L. A Field Based Detection Method for Rose Rosette Virus Using Isothermal Probe-Based Reverse Transcription-Recombinase Polymerase Amplification Assay. J. Virol. Methods 2017, 247, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Latha, T.K.S.; Doraiswamy, S. Detection of Pigeonpea Sterility Mosaic Virus, the Causal Agent of Sterility Mosaic Disease of Pigeonpea in Viruliferous Mite Vector by DAS-ELISA and DIBA. Arch. Phytopathol. Plant Prot. 2008, 41, 537–541. [Google Scholar] [CrossRef]
- Lebas, B.; Ochoa-Corona, F.; Elliott, D.; Tang, Z.; Alexander, B. Development of an RT-PCR for High Plains Virus Indexing Scheme in New Zealand Post-Entry Quarantine. Plant Dis. 2005, 89, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Qiu, L.; Yang, T.; Ning, J.; Xu, Q.; Dong, J.; Xi, D. Multiple Reverse Transcription PCR Assay for Simultaneous Detection of Four Main Viruses in Kiwifruit. Eur. J. Plant Pathol. 2020, 156, 1207–1212. [Google Scholar] [CrossRef]
- Hassan, M.; Tzanetakis, I.E. Population Structure, Evolution and Detection of Blackberry Leaf Mottle-Associated Virus, an Emerging Emaravirus. Plant Pathol. 2019, 68, 775–782. [Google Scholar] [CrossRef]
- Mijit, M.; Li, S.F.; Zhang, S.; Zhang, Z.X. First Report of Fig Mosaic Virus Infecting Common Fig (Ficus Carica) in China. Plant Dis. 2015, 99, 422. [Google Scholar] [CrossRef]
- Patil, B.L.; Raghu, R.; Dangwal, M.; Byregowda, M.; Voloudakis, A. Exogenous DsRNA-Mediated Field Protection against Pigeonpea Sterility Mosaic Emaravirus. J. Plant Biochem. Biotechnol. 2021, 30, 400–405. [Google Scholar] [CrossRef]
- Manion, P.D. Tree Disease Concepts; Pearson Education Australia, Ed.; Prentice-Hall: Englewood Cliffs, NJ, USA, 1981. [Google Scholar]




| Species | Common Name | Disease/Symptoms | Host(s) (Botanical Family) | Geographic Occurence | Vector/Transmission | Genomic Segments | Key Reference(s) |
|---|---|---|---|---|---|---|---|
| Established Species (ICTV) | |||||||
| Emaravirus sorbi | European mountain ash ringspot-associated virus (EMARaV) | mosaic and ringspot disease | rowan, serviceberry (Sorbus spp., Amelanchier sp.) and other Rosaceae species | North and Central Europe | Phytoptus pyri, graft transmission and mechanical inoculation to Sorbus aucuparia L. | 6 | [7,14,16,22,23] |
| Emaravirus fici | fig mosaic virus (FMV) | fig mosaic disease (FMD) | fig, cyclamen (Ficus carica, F. pseudocarica, Cyclamen persicum Mill.) (Moraceae, Primulaceae) | North America, Europe, North Africa, Middle East, Asia, New Zealand | Aceria ficus, vegetative propagation and grafting | 6 | [9,24,25] |
| Emaravirus rosae | rose rosette virus (RRV) | rose rosette disease (RRD) | rose (Rosa spp.) (Rosaceae) | USA, Canada, India | Phyllocoptes fructiphilus, grafting | 6 | [26,27] |
| Emaravirus idaeobati | raspberry leaf blotch virus (RLBV) | raspberry leaf blotch disorder (RLBD) | raspberry (Rubus spp.) (Rosaceae) | Scandinavia, UK, Balkans | Phyllocoptes gracilis, mechanical transmission to experimental hosts | 8 | [28,29] |
| Emaravirus cajani | pigeonpea sterility mosaic virus 1 (PPSMV-1) | sterility mosaic disease (SMD) | pigeonpea (Cajanus spp.) (Fabaceae) | South Asia | Aceria cajani, grafting, transmission to experimental hosts by sap inoculation and viruliferous eriophyid mites | 6 | [12,30,31] |
| Emaravirus toordali | pigeonpea sterility mosaic virus 2 (PPSMV-2) | sterility mosaic disease (SMD) | pigeonpea (Cajanus spp.) (Fabaceae) | South Asia | Aceria cajani, grafting, transmission to experimental hosts by sap inoculation and viruliferous eriophyid mites | 6 | [32] |
| Emaravirus tritici | High Plains wheat mosaic virus (HPWMoV) | High Plains disease (HPD) | Sweet grasses (wheat, maize, barley, oat, rye, cheat, green and yellow foxtail) (Poaceae) | Central and Western USA, Canada, Argentina, Australia, Ukraine | Aceria tosichella | 8 | [10,33] |
| Emaravirus cercidis | redbud yellow ringspot- associated virus (RYRaV) | redbud yellow ringspot (RYRS) | redbud (Cercis spp.) (Fabaceae) | USA | graft transmissible to Cercis ‘Canadensis’ and several legume species | 5 | [34] |
| Emaravirus actinidiae | Actinidia chlorotic ringspot- associated virus (AcCRaV) | chlorotic ringspot, mottle and vein yellowing of leaves | kiwifruit (Actinidia spp.) (Actinidiaceae) | China | mechanical transmission to N. benthamiana | 5 | [35] |
| Emaravirus rubi | blackberry leaf mottle- associated virus (BLMaV) | blackberry yellow vein disease (BYVD) | blackberry (Rubus spp.) (Rosaceae) | USA | undescribed gall mite species of Eriophyidae | 5 | [36] |
| Emaravirus pistaciae | Pistacia virus B (PiVB) | unknown | pistachio (Pistacia spp.) (Anacardiaceae) | Turkey | / *1 | 7 | [37] |
| Emaravirus parkinsoniae | palo verde broom virus (PVBV) | witches’ broom disease | blue palo verde (Parkinsonia florida) (Fabaceae) | USA, Mexico | Aculus cercidi? | 4 | [38] |
| Emaravirus kiwi | Actinidia virus 2 (AcV-2) | leaf mottle, chlorotic spots and/or leaf mosaic symptoms | kiwifruit (Actinia spp.) (Actinidiaceae) | China | / | 6 | [39] |
| Emaravirus ziziphi | jujube yellow mottle- associated virus (JYMaV) syn. Chinese date mosaic-associated virus (CDMaV) | jujube yellow mottle disease (JYMD) | jujube (Ziziphus jujuba) (Rhamnaceae) | China | Epitrimerus zizyphagus? | 6 | [40,41] |
| Emaravirus cordylinae | ti ringspot- associated virus (TiRSaV) | ti ringspot | ti (Cordyline fructicosa) (Asparagaceae) | Hawaii | undescribed species of eriophyid mites, mechanical transmission to experimental hosts | 5 | [42] |
| Emaravirus camelliae | Camellia japonica- associated virus 1 (CjaV-1) syn. Camellia chlorotic ringspot virus 1 (CaCRSV-1) | chlorotic ringspots, color- breaking | common camellia (Camellia japonica) (Theaceae) | Italy, China | / | 9 | [43,44,45] |
| Emaravirus verbanni | Camellia japonica- associated virus 2 (CjaV-2) syn. Camellia chlorotic ringspot virus 2 (CaCRSV-2) | chlorotic ringspots | common camellia (Camellia japonica) (Theaceae) | Italy, China | / | 4 | [43] |
| Emaravirus perillae | Perilla mosaic virus (PerMV) | mosaic disease | shiso (Perilla spp.) (Lamiaceae) | Japan | perilla rust mite (Shevtchenkella spp.) mechanical transmission to N. benthamiana | 10 | [46] |
| Emaravirus populi | aspen mosaic- associated virus (AsMaV) | mosaic disease | Euroasien aspen (Populus tremula) (Saliaceae) | Fennoscandinavia | mite vector species unknown, graft-transmissible | 5 | [17] |
| Emaravirus syringae | lilac chlorotic ringspot- associated virus (LiCRaV) | leaf chlorotic ringspots and mottling | common lilac (Syringa vulgaris L.) (Oleaceae) | China | mechanical transmission to N. benthamiana | 5 | [47] |
| Emaravirus pyri | pear chlorotic leaf spot-associated virus (PCLSaV) | chlorotic leaf spot disease | sandy pear (Pyrus pyrifolia) (Rosaceae) | Central and Southern China | / | 5 | [48] |
| Emaravirus quercus | common oak ringspot- associated virus (CORaV) | ringspot disease | common oak (Quercus robur L.) (Fabaceae) | North and Central Europe | graft-transmissible | 5 | [18,19,22] |
| Emaravirus aceris | maple mottle- associated virus (MaMaV) | mottle, mosaic, chlorotic ringspots | sycamore maple (Acer pseudoplatanus) (Sapindaceae) | Germany | graft-transmissible | 6 | [20,49] |
| Emaravirus chrysantemi | chrysanthemum mosaic-associated virus (ChMaV) | ‘Mon-mon’ disease (chlorotic ringspots, mosaic) | Chrysanthemum (Asteraceae) | Japan | Paraphytoptus kikus? | 7 | [50] |
| putative emaravirus based on genomic, biological, and phylogenetic properties | |||||||
| alfalfa ringspot- associated virus (ARaV) | ringspot, yellow mosaic | alfalfa (Medicago sativa L.) (Fabaceae) | Australia, China | / | 4 | [51,52] | |
| Vitis emaravirus (VEV) syn. grapevine emaravirus A (GEVA) | chlorotic mottling | grapevine (Vitis coignetiae, Vitis vinifera L.) (Vitaceae) | Japan, China | graft-transmissible | 5 | [53,54] | |
| ash shoestring- associated virus (ASaV) syn. pea associated emaravirus (PaEV) | shoestring, leaf curling, mottle, chlorotic spots and ringspots | ash, pea (Fraxinus spp., Pisum sativum L.) (Oleaceae, Fabaceae) | Germany, Switzerland, Sweden, Italy | Aceria fraxinivora? | 5 | [21,55] | |
| karaka Okahu purepure virus (KOPV) | chlorotic pale spots | karaka (Corynocarpus laevigatus) (Corynocarpaceae) | New Zealand | Aculus corynocarpi? | 5 | [56] | |
| Japanese star anise ringspot- associated virus (JSARaV) | ringspot disease | Japanese star anise (Illicium anisatum L.) (Schisandraceae) | Japan | eriophyid mites of the family Diptilomiopidae | 5 | [57] | |
| Arceuthobium sichuanense- associated virus 1 (ArSaV1) | unknown | spruce dwarf mistletoe (Arceuthobium sichuanense) (Viscaceae) | China | / | 5 | [58] | |
| Artemisia fimovirus 1 (ArtV1) | unknown | Chinese mugwort (Artemisia verlotiorum) (Asteraceae) | Slovenia | / | 5 | [59] | |
| Ailanthus crinkle leaf-associated emaravirus (ACrLaV) | severe crinkle | tree of heaven (Ailanthus altissima) (Simaroubaceae) | China | / | 4 | [60] | |
| Pueraria lobata- associated emaravirus (PloAEV) | yellow spots, mosaic, mottling | kudzu (Pueraria lobata) (Fabaceae) | China | mechanical transmission to N. benthamiana | 5 | [61] | |
| Orphans * | |||||||
| Woolly burdock yellow vein virus (WBYVV) | yellow vein and leaf mosaic symptoms | wooly burdock (Arctium tomentosum) (Asteraceae) | Finland | / | 2 | [62] | |
| Yunnan emara- like virus | / | / | China | / | 7 | [63] | |
| Illicium anisatum ringspot- associated virus | / | Japanese star anise (Illicium anisatum L.) (Schisandraceae) | / | / | 1 | Shimomoto and Kubota, unpublished | |
| / | / | Solanum lichtensteinii, S. mauritianum (Solanaceae) | South Africa | / | 1 | [64] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehanek, M.; Karlin, D.G.; Bandte, M.; Al Kubrusli, R.; Nourinejhad Zarghani, S.; Candresse, T.; Büttner, C.; von Bargen, S. The Complex World of Emaraviruses—Challenges, Insights, and Prospects. Forests 2022, 13, 1868. https://doi.org/10.3390/f13111868
Rehanek M, Karlin DG, Bandte M, Al Kubrusli R, Nourinejhad Zarghani S, Candresse T, Büttner C, von Bargen S. The Complex World of Emaraviruses—Challenges, Insights, and Prospects. Forests. 2022; 13(11):1868. https://doi.org/10.3390/f13111868
Chicago/Turabian StyleRehanek, Marius, David G. Karlin, Martina Bandte, Rim Al Kubrusli, Shaheen Nourinejhad Zarghani, Thierry Candresse, Carmen Büttner, and Susanne von Bargen. 2022. "The Complex World of Emaraviruses—Challenges, Insights, and Prospects" Forests 13, no. 11: 1868. https://doi.org/10.3390/f13111868
APA StyleRehanek, M., Karlin, D. G., Bandte, M., Al Kubrusli, R., Nourinejhad Zarghani, S., Candresse, T., Büttner, C., & von Bargen, S. (2022). The Complex World of Emaraviruses—Challenges, Insights, and Prospects. Forests, 13(11), 1868. https://doi.org/10.3390/f13111868







