Abstract
The taxonomy of an ancient gymnosperm genus Taxus, with high value in horticulture and medicine, is perplexing because of few reliable morphological characters for diagnosing species. Here, we performed a comprehensive investigation of the evolutionary dynamics of Taxus chloroplast genomes and estimated phylogenetic relationships, divergence times, and ancestral distributions of Taxus species by comparing 18 complete chloroplast genomes. The variations across the chloroplast genome of different Taxus species indicated that remarkably varied genome variations across lineages have reshaped the genome architecture. Our well-resolved phylogeny supported that T. brevifolia Nutt. was basal lineages followed by the other North America lineages. Divergence time estimation and ancestral range reconstruction suggested that the Taxus species originated in North America in the Late Cretaceous and revealed that extant Taxus species shared a common ancestor whose ancestral distribution area was probably in North America and afterwards the earliest members expanded to Southeast Asia from where Chinese Taxus species originated. The predominant European species have more closer relationship with the Eastern Asian species and the speciation of Eurasia species arose from several dispersal and vicariance events in the Miocene. Genome-wide scanning revealed 18 positively selected genes that were involved in translation and photosynthesis system in Taxus, which might be related to the adaptive evolution of Taxus species. The availability of these complete chloroplast genomes not only enhances our understanding of the elusive phylogenetic relationships and chloroplast genome evolution such as conservation, diversity, and gene selection within Taxus genus but also provides excellent templates and genetic bases for further exploration of evolution of related lineages as well as for plant breeding and improvement.
1. Introduction
Taxus, honored as the ‘living fossil of plants’, is the most diverse and widespread genus within Taxaceae [1], and is one of the ancient species originating from the Late Cretaceous [2]. However, most Taxus species are either endangered or near threatened due to slow-growing, weak reproductive capacity, scattered distribution, and the illegal trade and excessive exploitation of its bark and leaves for taxol [3]. All native Taxus species in China have been listed as endangered and national first-class protected plants due to their narrow inhabit niche and decreasing population [4], and India has prohibited/restricted the export of native Taxus [5]. The International Union for Conservation of Nature’s Red List of Threatened Species has recruited several species of Taxus that are threatened to different degrees, including T. floridana Nutt. ex Chapm., T. brevifolia Nutt., T. globosa Schltdl., T. contorta Griff., T. fuana Nan Li & R.R.Mill., T. chinensis (Pilg.) Rehder., and T. wallichiana Zucc. [6,7]. The lack of molecular biology research and genetic information between Taxus and related species seriously hinders the effective conservation and research of this rare and endangered species.
The taxonomic history of Taxus species has been complex and uncertain due to their similar morphological characteristics, whereas Taxus plants usually present high levels of phenotypic plasticity within species [8,9,10]. Moreover, the classifications and phylogenetic relationships between Taxus and Cephalotaxaceae and Podocarpaceae species are not fully understood. Extant Taxus plants are believed to have evolved from a common ancestor whose descendent lineages also includes the oldest recognizable Triassic fossil, Paleotaxus redivia, discovered in 200-million-year-old strata [11]. A mid-Jurassic relative is more identifiable as a member of extant Taxus, named T. jurassica Florin., closely resembling T. baccata L., T. cuspidata Siebold & Zucc., and T. brevifolia [12]. A few authors treated all previously described Taxus species as subspecies of T. baccata [13,14,15]. The monophyly of Taxus has been supported by both chemical and morphological characteristics in seeds and leaves and molecular evidence [16,17], but the phylogenetic relationships among Taxus species have been subjected to various controversies. For example, Pilger [13], Cope [18], and Farjon [19] only admitted 7–12 species in Taxus, with only five in China, whereas a phytogeographical analysis of extensive sampling of Taxus based on leaf anatomical characters resolved 24 species and 55 varieties in the genus, and China contains 16 species and seven varieties [20]. In addition, Liu et al. recognized totally 15 species/lineages in the genus based on a world-wide level of genetic and distribution exploration, which suggested that T. sumatrana (Miq.) de Laub. was assigned as T. chinensis or T. wallichiana according to their geographic position [21]. Li & Fu regarded T. yunnanensis W.C.Cheng & L.K.Fu. as a synonym of T. wallichiana, and they believed that T. wallichiana identified in FRPS as T. fuana [22]. Currently, T. fuana is a synonym of T. contorta [23,24]. Repeated ancient hybridization events were postulated as follows: T. florinii Spjut. = T. wallichiana (♀) × T. chinensis (♂), Emei type = T. chinensis (♀) × T. wallichiana (♂), and Qinling type = T. contorta (♀) × Huangshan type (♂) [23]. The Emei and Qinling type were considered as two additional cryptic species [25]. To data, the genus contains more than 10 species in many different habitats of the northern Hemisphere, covering Europe, North America, and Asia [21,26,27,28], and these species are geographically separable rather than morphologically different. There are at least five species and one variety in Taxus that have been found within China, including T. chinensis, T. yunnanensis, T. cuspidata, T. madia Rehder., T. wallichiana, and T. chinensis var. mairei. Among these five species, T. chinensis and T. chinensis var. mairei originated from China which is one of the modern diversity centers of Taxus.
Molecular phylogenetic have partially resolved the evolutionary relationships between species and reconstructed the phylogeny of different Taxus species. Analysis of one nuclear (ITS) and five chloroplast (matK, psbA-trnH spacer trnL, rbcL, and trnL-trnF spacer) molecular markers clustered T. brevifolia and T. globosa in one single clade with T. baccata, while the endemic T. yunnanensis was included in a group with T. wallichiana, which is distantly related with the other four Taxus species in China; T. floridana is the first-branching taxon in Taxus genus, whereas T. fauna is closer to T. baccata than to other Taxus species [26]. Meanwhile, phylogenetic analysis of ten main Taxus species based on ITS region revealed that three North American species clustered a clade with high support, and T. brevifolia from the Pacific coastal showed a well-supported sister relationship to T. floridana from northwestern Florida and T. globosa from northern Mexico [29]. Plenty of studies have attempted to interpret the genetic diversity and genetic structure, species separation, phylogenetic and interspecific relationships based on nuclear microsatellite markers [30,31,32], random amplified polymorphic DNA [33], chloroplast DNA sequences, and internal transcribed spacer region of nuclear ribosomal DNA [21,26,34] markers. The next-generation sequencing, however, is more cost effective and faster to obtain a high number of gene targets or a whole genome than by traditional Sanger sequencing [35]. Although more conserved in the terms of structures and organization of gene content than nuclear genome in plants, the chloroplast DNA sequences have suffered many mutation events throughout the evolution of vascular plants, including InDels, substitutions, and inversions [36]. Therefore, it is imperative to employ the complete chloroplast genome sequences to resolve the genetic diversity and phylogenetic analysis at high taxonomical levels, and even in lower taxa [37,38].
Numerous phylogenetic studies revealed the phylogenetic origin and backbone relationships of gymnosperms based on whole chloroplast genome sequences [39,40]. Presently, many complete chloroplast genomic sequences of Taxus species have been published, and the unique structure of chloroplast genome such as gene content, genome-scale genomic rearrangements, and gene lost and gain events were identified in them [41,42,43]. However, the global pattern of variation and evolution of the whole chloroplast genomes in this genus have been ignored. Therefore, a further investigation of the phylogenetic relationships and divergence between species, chloroplast genome variations, and evolutionary dynamics of repeat sequences through all Taxus chloroplast genome sequences, is necessary to conserve and manage the germplasm resources, improve breeding, and search for favorable genes. In this study, we newly sequenced four complete chloroplast genomes of T. cuspidata, T. chinensis, T. yunnanensis, and T. wallichiana from China using Illumina technology. Furthermore, our assemblies were integrated with 14 previously assembled chloroplast genomes to yield the most comprehensive phylogenomic study of Taxus. This study aimed to (i) clarify phylogenomic relationships and estimate divergence times between Taxus species; (ii) survey the overall structural variation pattern of the whole chloroplast genome in Taxus species; (iii) investigate the evolutionary dynamics of repetitive sequences across Taxus chloroplast genomes during recent speciation; (iv) estimate the ancestral distribution and migration history of Taxus; (v) and search for Taxus chloroplast genes under positive selection. This study presents the most comprehensive phylogenomic analysis for 18 identified members within Taxus and provides a new paradigm for the investigation of plastome evolution of conifers.
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
Healthy fresh needles of T. cuspidata, T. chinensis, T. yunnanensis, and T. wallichiana were collected from four single individuals located in Northeast Forestry University (Harbin, China), Xi’an Botanical Garden (Xi’an, China), Southwest Forestry University (Kunming, China), and Tibet Agricultural and Animal Husbandry University (Linzhi, Tibet, China), respectively. These leaves were divided into two parts and dried in silica gel and stored at −80 °C for genomic DNA extraction. Total genomic DNA was isolated by using a Dneasy Plant Mini Kit (Qiagen, CA, USA), followed by purification. Subsequently, the DNA concentration of each sample was quantified using a NanoDrop2000 spectrophotometer (Thermo Scientific, Carlsbad, CA, USA). The chloroplast genome sequences of 14 other Taxus species and Pseudotaxus chienii W.C.Cheng. were directly downloaded from NCBI, including T. brevifolia, T. globosa, T. floridana, T. phytonii, T. calcicola, T. mairei, T. canadensis, T. baccata, T. contorta, and T. fuana, and three potential cryptic eco-types, the Huangshan, Qinling and Emei type. All accession numbers are presented in Table S1.
2.2. Chloroplast Genome Sequencing, Assembly and Annotation
Shotgun genomic libraries were generated via fragmentation of qualified DNA samples. A DNA sample was randomly sheared to 250 bp fragments by using Covaris ultrasonicators (Covaris, Inc., Woburn, MA, USA), and the library was constructed by terminal repair, A-tail addition, adaptors ligation, purification, and PCR amplification. Libraries were then multiplexed and sequenced using the Illumina Hiseq 2500 high-throughput sequencing platform (Illumina Inc., San Diego, CA, USA) with 2 × 150 bp paired reads. Firstly, all the raw reads were trimmed using a CLC Genomics Workbench v9 (CLC Bio, Aarhus, Denmark) with the default parameters set. After trimming, high quality PE reads were assembled using the program MITObim v1.7 [44], with the published chloroplast genome of T. mairei (GenBank: KJ123824) [41] as a reference to obtain accurate sequences. The complete chloroplast genomes were annotated using DOGMA program [45], with the homology recognition threshold of protein-coding genes, tRNA, and rRNA coding genes of 60%, 85%, and 85%, respectively. Some genes with large variation and low homology with related species were annotated by manual correction with the homology recognition threshold of 25%–50%. The positions of start and stop codons and intron/exon boundaries were adjusted by BLAST searches by comparison with homologous genes in other chloroplast genomes of many related species, including T. mairei (KJ123824). The obtained tRNA genes were further confirmed using tRNAscan-SE [46], and an open reading frame was detected by ORF-Finder [47]. The Organellar Genome DRAW [48] was used to draw the circular plastid genome maps.
2.3. Phylogenomic Analyses
Phylogenomic analysis was implemented based on three datasets, the complete chloroplast genome sequences, a set of 82 common protein-coding genes and non-coding DNA sequences from 18 Taxus species, including four Chinese species sequenced in the current study, 14 other Taxus species, and one Pseudotaxus species used as an outgroup. The MAFFT v7.0 [49] was used to align the chloroplast genomic sequences from the finalized datasets, with the default parameters set, and adjusted manually where necessary. The best-scoring maximum-likelihood phylogenetic trees were inferred with RAxML v7.2 [50], performing 1000 bootstrap replicates. RAxML searches relied on the general time reversible (GTR) model of nucleotide substitution with the gamma distributed rate variation among sites and a proportion of invariable sites (GTR+G+I model). Occurrence of InDels and SNPs were mapped onto the derived phylogenetic trees constructed by ML analysis according to whole chloroplast genome alignment.
2.4. Inference of Divergence Time and Diversification Pattern
We also estimated the divergence time within Taxus species using Bayesian approach as implemented in BEAST program [51,52]. We tested whether a molecular clock hypothesis could be fitted to our data by employing the log-likelihood value with and without the molecular clock constraints by using MEGA v7.0 [53]. Our dataset strongly rejected a strict molecular clock (with clock, lnL = −286,819.299; without clock, lnL = −278,520.849; p < 0.001), suggesting that the evolutionary process is very complex. Therefore, we used a Yule process tree prior to model speciation and an uncorrelated log-normal relaxed-clock model to account for lineage-specific rate heterogeneity. In this model, genetic distances were transformed into absolute time (in million years) by using five fossil calibration points within the major conifer clades [54,55]. We undertook five independent Markov Chain Monte Carlo (MCMC) runs with 1,000,000 generations to confirm the convergence of the analysis, sampling every 1000 stages, and the initial 20% cycles were discarded as burn-in. The five runs were combined in LogCombiner (part of the BEAST-package). The final result was visualized in Tracer version 1.6 [56] to assess convergence and stationarity of each chain to the posterior distribution. The effective sample size (ESS) of each parameter exceeded 200 sufficient for stable estimates of parameters. The maximum clade credibility tree (MCC) was summarized using TreeAnnotator (part of the BEAST-package) with the initial 10% of trees discarded as burn-in. The derived MCC tree was visualized in Figtree version v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 3 August 2020). To obtain a sense of overall pattern of diversification rate change over time, a lineage-through-time (LTT) analysis was employed to detect temporal shifts in diversification rates, as a function of evolutionary radiations within Taxus genus. The BEAST operations followed the parameters as above, and 1000 trees sampled from the converged Bayesion trees were used to generate the confidence intervals of the LTT plots.
2.5. Ancestral Area Reconstructions (AAR)
In order to reveal the origin and the historical biogeography of the hypothetical ancestor, the mechanism (vicariance/dispersal) underlying the geographically disjunct distribution of Taxus species, and the spatial patterns of biogeographical diversification within Taxus were estimated based on the Bayesian binary method (BBM) applied by RASP v4.0 [57] using the BEAST-derived chronogram of Taxus genus, with the outgroup removed from the tree. All parameters were kept at default. The geographical distribution of Taxus species was broadly categorized into four operational areas (labeled A–D) in accordance with their present distributions: (A) America, (B) Europe, (C) China, and (D) Japan. The current distribution information of Taxus species was mainly derived from our field investigation and GBIF (https://www.gbif.org/, accessed on 5 December 2020).
2.6. Genome-Wide Analyses of Genomic Structural Variants
In order to understand the full spectrum of genetic structural variation in Taxus chloroplast genome, we performed a comprehensive scanning of genome-wide structural variation based on complete chloroplast genome sequences of 18 Taxus species. The phylogeny-aware algorithm PRANK+F [58] was used to align all datasets with the parameters “-showanc -showevents +F”. The phylogeny-aware algorithm performs better than the classical progressive alignment methods regarding the accuracy in InDels-rich sequences as PRANK posterior probabilities take into account the InDel events by distinguishing between the two types of events and retain the high-reliability regions with InDels. Particularly, if the “+F” option is switched on, PRANK flags already infer insertions at their sites as permanent insertions and prevent other insertions from being inferred in an overlapping position during the second round of alignment. This variant algorithm with permanent insertions outperforms the basic algorithm of PRANK provided the correct guide phylogeny and dense sampling of sequences. This indicated that InDels introduced along one branch of the phylogeny are probably introduced from an InDel event occurred in another branch, even if they overlap, and thus generates too long and gappy but potentially more accurate alignments.
2.7. Repeat Sequence Analyses
Three types of repetition were identified, including tandem, dispersed, and palindromic repeats. The minimum repeat sizes were set to 15, 20, and 30 bp for tandem, palindromic, and dispersed repeats, respectively. These three repetitive sequences were identified by first using the program DNAMAN v6.0. (Lynnon Biosoft, Vaudreuil, QC, Canada) and then manually filtering the redundant output.
2.8. Genome-Wide Scan for Protein-Coding Genes under Positive Selected and Accelerated Evolution
To explore the selective pressure of PCGs in Taxus, the CODEML program implemented in the PAML v4.9 package [59] was used to estimate the rate of non-synonymous (dN) and synonymous (dS) substitutions for PCG. We explored the selective pressure of all 82 chloroplast genes in Taxus genus by estimating the rate of non-synonymous (dN) and synonymous (dS) substitutions for all coding sequences with CODEML program [60] implemented in the PAML v4.9 package [59]. The ratio (ω) of dN to dS was used to determine the selective pressure, where ω > 1, ω = 1, and ω < 1 indicate higher positive selection pressure, neutral selection, and higher pressure of negative selection, respectively. PAL2NAL [61] was used to generate the codon-wise alignment of nucleotide sequences as the input sequences for CODEML guided by the peptide alignments. The ML phylogenetic tree based on whole chloroplast genome was used as an input tree. Before calculation, stop codons and gaps between the aligned sequences were deleted. In order to determine whether each shared coding sequence has experienced a different evolutionary force in different lineages, we assigned targeted branch(es) as the foreground branch where positive selection may have occurred and the remains served as background branches where only purifying selection or neutral evolution occurred. We adopted two-ratio models which compare one model that assumes sites to be under positive selection on the foreground branch and sites can evolve either neutrally or under purifying selection on the background branches with the null model in which all branches share the same evolutionary rate. The likelihood ratio test (LRT) with χ2 distribution was used to evaluate the significant difference between the two models with a p < 0.05 significance threshold.
3. Results
3.1. Phylogenomic Analysis
In this study, we explored evolutionary relationships among Taxus species by combining 14 published complete chloroplast genomes and the newly sequenced chloroplast genomes of T. cuspidata, T. chinensis, T. yunnanensis, and T. wallichiana. All Taxus chloroplast genomes displayed discrepant genome sizes ranging from 127,352 bp (T. florinii) to 129,752 bp (T. globosa) but fairly conserved gene content that commonly harbored an identical set of 82 annotated unique protein-coding genes, 25 tRNA genes, and four rRNA genes (Figures S1 and S2). Thus, three datasets, including whole chloroplast sequences, protein-coding genes, and non-coding region, were used to reconstruct phylogenetic trees based on ML method, with P. chienii as an outgroup. The fully resolved phylogenetic trees obtained by using the whole plastome sequences and protein-coding sequences have nearly identical topology to each other with high support values (Figure 1 and Figure S3), whereas the tree constructed based on the non-coding sequences presented disparate topology with that based on the entire chloroplast genome and a total of 82 protein-coding genes (Figure S4), indicating a different evolution pattern between coding and non-coding sequences. In the resulting ML phylogenies based on whole-plastome and coding regions, the four Taxus species originated from New World, T. brevifolia, T. canadensis, T. floridana, and T. globosa did not form a monophyletic clade. T. brevifolia was resolved to be the initial diverged species in Taxus genus and subsequently a sister relationship of T. floridana and T. globosa was well-supported. However, one of the unexpected findings is that the Emei type appear to be more distantly related to the basal lineage with high bootstrap values in comparison with other Old World Taxus species.
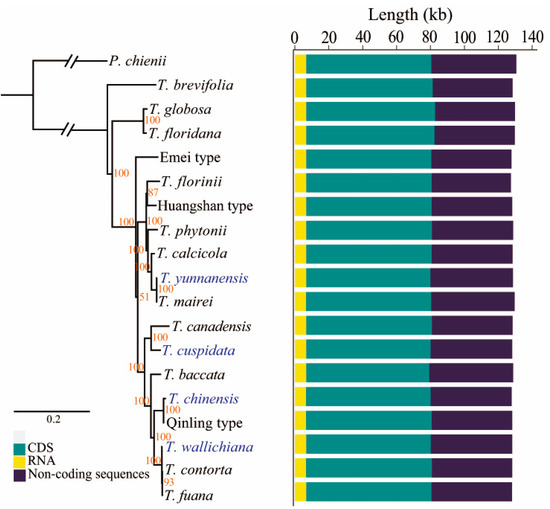
Figure 1.
The phylogeny of Taxus species relied on whole chloroplast sequences and the variation in Taxus chloroplast genome sizes.
The 14 derived Taxus lineages further were divided into two main clades of which one branch was comprised of T. mairei, T. yunnanensis, T. calcicola, T. phytonii, and the Huangshan type and T. florinii, while the other included T. fuana, T. contorta, T. wallichiana, Qingling type, T. chinensis, T. baccata, T. cuspidata, and T. canadensis (Figure 1 and Figure S3). This finding revealed a potential role of Emei type as the ancestral lineage of these clades and was indicative of multiple evolutionary origins of Chinese-specific Taxus species; however, the five Chinese species were strongly supported as a monophyletic clade with T. floridana and T. fuana in the phylogeny based on non-coding regions (Figure S4). The topological differences were also observed when using non-coding sequences, indicating that T. floridana independently formed a monophyletic clade and failed to group with T. globosa within the New World Taxus species. Both phylogenies based on whole-plastome and coding sequences grouped T. contorta into monophyletic clade with T. wallichiana and T. fuana with 100% support, whereas the affinity of T. contorta to T. wallichiana and T. fuana was not confirmed by non-coding sequences: T. contorta formed a close relationship with Qingling type within a clade that is clearly separated from where it was expected to be. These topological differences suggested the need of more plastomes or nuclear sequences to resolve the evolutionary relationships between Taxus species.
3.2. A Genome-Wide Map of Genomic Variation across Taxus Chloroplast Genomes
Four new sequenced Taxus chloroplast genomes, together with the published 14 chloroplast genomes, allowed a comprehensive evaluation of chloroplast genome-wide structural/single nucleotide variation (SNV), indicative of the evolutionary processes in Taxus species. The SNVs, insertions, and deletions (InDels) were characterized in Taxus chloroplast genomes with P. chienii as an outgroup, indicating that genomic variations varied from one species to another. The overview of the distribution of genomic structural/single nucleotide variation across Taxus chloroplast genomes revealed substantial variation but overall conservation of genome architecture across Taxus species (Figure 2 and Figure S5). The uneven distribution of genomic variation highlighted some fast-evolving hotspots across Taxus chloroplast genomes, for example, ycf1, ycf2, accD, and some tRNA gene-rich regions. These highly variable regions can serve as the most promising candidate for DNA barcodes in plastid genome to resolve closely related lineages in phylogenetic analyses. In order to survey the occurrences of genomic variants along divergence branches that account for the evolution of Taxus chloroplast genomes, all genomic structural/single nucleotide variants were mapped onto the phylogenetic tree generated by using 18 Taxus chloroplast genome sequences with the phylogeny-aware algorithm [57], which was proven to outperform traditional approaches in resolving alignment of InDel-rich sequences with regard to the accuracy.
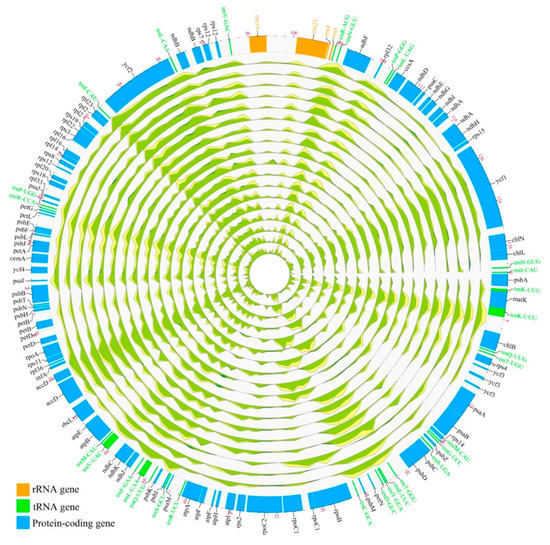
Figure 2.
An overview of variations across the Taxus chloroplast genomes.
On the whole-genome level, we totally identified 1128 and 1904 insertion and deletion events, and the numbers of SNVs and lengths of deletions and insertions varied among evolution branches with average values of 235.37, 569.49, and 1011.49, respectively (Figure 3). The length varied from 1 to 1068 bp in insertions, whereas the length varied from 1 to 1923 bp in deletions, and the length of insertions was summed up to a total of 19,932 bp in length, compared with 35,402 bp of deletions. The majority of discovered Indels in Taxus chloroplast genomes occurred on a small scale, approximately 95% of which were shorter than 100 bp, whereas 70% were smaller than 10 bp (Figure 3 and Figure S6B,C; Table S2). The size-frequency spectrum was consistent with observations from the nuclear genomes of both rice [62] and Arabidopsis lyrata [63] that larger InDels were less abundant than smaller ones. The size distribution of structural variations along lineages indicated that a clear excess of insertions over deletions was detected in most branches except T. florinii and the ancestral lineage leading to the Taxus clade, which suffered significant sequence loss since divergence from Pseudotaxus (Figure 3B and Figure S6B,C). This would account for the slight shrinkage of the Taxus chloroplast genome compared with its close relative Pseudotaxus. Within Taxus species, T. brevifolia had the greatest number of both SNVs and insertions in chloroplast genome due to its deep evolutionary history within Taxus genus, whereas T. florinii chloroplast genome contained the most abundant deletions. SNVs, deletions, and insertions were fewest in T. contorta In Asian species, most chloroplast genomes were affected by deletions, while T. mairei and T. globosa were largely affected by insertions in chloroplast genomes (Figure 3B).
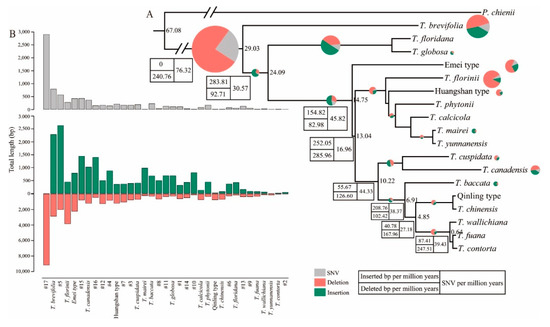
Figure 3.
The occurrence rate of chloroplast genomic variation during diversification in Taxus genus. (A) Total lengths of insertion, deletion and SNV for different lineages of Taxus. (B) Accumulation rates of insertion, deletion and SNV lengths every million years along branches of the Taxus phylogeny. Pie of top 21 branches are scaled proportionally to InDel lengths.
To determine the occurrence rates of these genome variations in Taxus plastomes, we time-calibrated the events of SNVs and InDels by aligning them with our time-scaled phylogeny yielded subsequently from Bayesian relaxed molecular clock method implemented in BEAST2 (Figure 3A). Our results indicated that during the past ~29 Myr in the early Oligocene, the 18 Taxus species and their shared ancestral lineages had an average of ~8.8 insertions and ~7.5 deletions per million years, respectively (Figure S6), whereas the single nucleotide mutation occurred at a speed of 32.8 per million years. The results also suggested that genome variations dramatically varied across lineages. Nonetheless, the estimated occurrence rates of genome variation (both structural and single nucleotide) were virtually constant along these lineages) (Figure 3A and Figure S6A). This result was similar to the observation in the plastome or nuclear genomes of flowering plants such as rice [64,65] that the number of genomic variation events observed along different branches significantly correlated with branch lengths (Figure S6D).
The examination of genomic positions of these predicted structural variations displayed that merely 2% of the detected InDels occurred in exons of protein-coding genes, suggesting their negative role in maintenance of the affected genes that would be constantly swept away by purifying selection during evolution. Further scrutiny of the length distribution of insertions and deletions within the 26 affected chloroplast protein-coding genes revealed that out of the all InDels were with length of multiple of three, suggesting strong negative selection on frameshift InDels that affected the chloroplast protein-coding genes.
3.3. Evolutionary Dynamics of Chloroplast Genome Repetitive DNA
The repetitive sequences contribute to chloroplast genome rearrangement, sequence divergence, as well as evolution [66,67]. Therefore, we explored the evolutionary dynamics of repetitive DNA sequences across 18 chloroplast genomes in order to determine their contribution to Taxus chloroplast genome variation and evolution. Overall, Taxus and P. chienii chloroplast genomes contained 395 repetitive sequences under three categories, namely tandem (Rt), palindromic (Rp), and dispersed (Rd) repeats that located within the gene spacer, coding regions, introns, and other regions (Table S2). Among them, the 282 Rt repeats, ranged from 15 to 406 bp in length, were the most abundant, accounting for 71.4% of total repeats; whereas Rp and Rd were 23.8% and 4.8% of entire repeats, respectively (Figure S7B). These results revealed than Rt repeats were dominant and contributed the most to the Taxus chloroplast genome expansion. Although the repeats were mainly occurred within non-coding regions, a number of protein-coding genes also contained repeat sequences. Comparative genomic analysis demonstrated that the three types of repetitive DNA sequences presented different evolutionary behaviors across Taxus chloroplast genomes. To examine the rate of repeats inserted into or eliminated from the Taxus chloroplast genomes, we approximately mapped the occurrences of these repeat sequences onto the time-calibrated phylogenetic tree and obtained a rough estimation of the speed of their insertion into and/or removal from the Taxus chloroplast genomes (Figure 4A). The identification of 77 insertion and 58 deletion events suggested a certain degree of proliferation of repeat sequences in Taxus chloroplast genome since the lineage divergence from their most common ancestor. Evolutionary dynamics of tandem repeats, represented by the abundance of copy number variation in chloroplast repeats across Taxus lineages, appeared to be more activated than Rd and Rp repeats, that mainly accumulated in the common ancestor and substantially persisted during the evolutionary history (Figure 4A; Table S2). Both Pseudotaxus and the ancestral branch of Taxus underwent extensive loss of repetitive DNA sequences since their split. Our results also revealed that the majority of repeat insertion/deletion events occurred continually across timetabled branches, but the pace of repeat insertions or deletions differed greatly among even closely related Taxus species in their process of speciation. Comparisons of genomic variation (SNVs and InDels) intensity within Rt, Rp, Rd, and their flanking regions with genome background revealed that only the flanking regions of Rp harbored significantly high levels of InDels, whereas the flanking regions of Rt and Rd only displayed high levels of SNVs (Figure 4B). These findings indicated that most chloroplast Rp repeats showed relatively slower substitution rates since their origin in the common ancestor of Taxus species than Rt and Rp repeats and suggested that Rt, Rp, and Rd repeats may display a different impact to accelerate the variation and evolution of Taxus chloroplast genome.
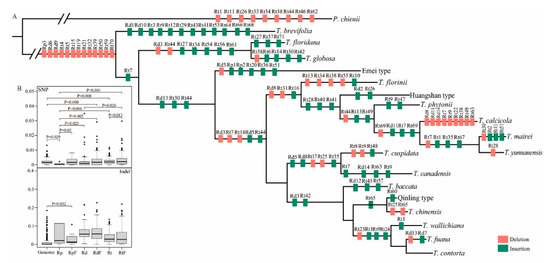
Figure 4.
Evolutionary dynamics of repeat DNA sequences across chloroplast genomes of Taxus species. (A) Occurrences of repeat sequences that are mapped onto different lineages of the Taxus phylogeny with P. chienii as an outgroup. Insertions and deletions of palindromic (Rp), dispersed (Rd) and tandem (Rt) repeats are colored in green and red, respectively. (B) Comparison of InDels and SNVs intensity in repeats with equivalently sized regions randomly sampled from genome. p-value is provided only when the difference is significant. SNV: SNV count per site; Indel: InDel count per site. Genome randomly sampled genome-wide regions, Rd: Rd repeats; Rp: Rp repeats; Rt: Rt repeats; RdF: Rd flanking regions; RpF: Rp flanking regions; RtF: Rt flanking regions. These include regions 50 bp upstream and downstream of repeat sequences, respectively.
3.4. Selective Pressures in the Evolution of the Taxus Genus
To look into the molecular evolution of chloroplast protein-coding genes, all 18 Taxus chloroplast protein-coding genes were used to calculate the non-synonymous/synonymous rate ω (ω = dN/dS). Unsurprisingly, rate heterogeneity existed among these Taxus lineages. Along the terminal branches, T. wallichiana (10.722) exhibited the highest ω value, followed by T. yunnanensis (8.772), while T. mairei (0.577) showed the smallest ω estimates. Meanwhile, T. contorta, the Qinling type, T. floridana, and T. globosa showed the relatively higher ω estimates while the remainder had intermediate values around 1 (Figure S7). The elevation of ω ratios (here ω > 1) provides evidence of adaptation or relaxation of selective constraints of these lineages. Subsequently, positive selection signals on genes along the lineages in the phylogenetic tree were detected by using the p\optimized branch-site model to calculate the ω value changes at each codon on particular branches or clades in the Taxus phylogeny for all 82 chloroplast genes. We convincingly detected 18 genes (ropA, rpl33, rps11, rps19, petD, ycf2, petB, ycf1, chlN, chlL, matK, rps2, rpoC2, rpoC1, rpoB, rbcL, accD, and rps12) under positive selection by the LTR method and a p-value of 5% for statistical significance (FDR < 0.05) (Figure 5), accounting for 22% of all examined chloroplast genes. In the T. floridana and T. globosa clade, only ycf2 gene was under positive pressure, while ycf2 and accD genes were positively selected in all examined clusters (Figure 5), indicating their functional importance. These 18 positively selected genes were functionally assigned as translation genes (rpoA, rpoB, rpoC1, rpoC2, rpl33, rps2, rps11, rps12, and rps19), photosynthesis genes (petB, petD, chlL, chlN, and rbcL), and other genes (ycf1, ycf2, accD, and matK), indicating that the majority of genes under positive selection are relevant to translation and photosynthesis system.
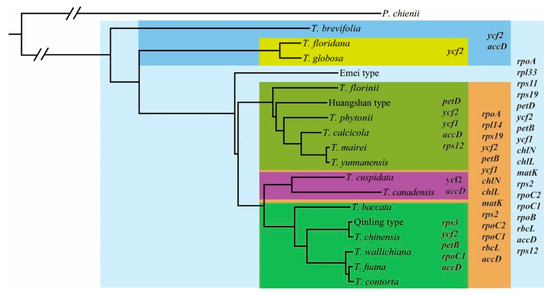
Figure 5.
Genome-wide scanning for lineages-specific positively selected genes along Taxus species phylogeny. Branches in rectangle represent lineages where genes are significantly under positive selection (p < 0.05; FDR < 0.05).
3.5. Diversification History and Divergence Time Estimation in the Genus Taxus
We reconstructed diversification histories of Taxus genus by using the lineage-through-time (LTT) plot to visualize the accumulation of lineages over time. Our visual examination of the diversification rate through time indicated an acceleration in diversification of Taxus lineages occurred since ~29 Mya (ESS > 200) (Figure 6A). Taxus is divergent at the molecular level with several distinct clades resolved in a chloroplast phylogenetic tree. We performed divergence time dating using five calibration points applied to the major conifer’s clades complete chloroplast genome datasets. According to the estimations derived from the time-calibrated tree, the oldest speciation event in conifers was approximately 201.82 Mya with a credible interval ranging from 197.03 to 213.81 Mya (95% HPD) (Figure 6B), suggesting the divergence of conifers in the early Jurassic. Torreya primarily diverged from the shared ancestor of Taxus and Pseudotaxus in the Taxaceae family approximatively 117.70 Mya (95% HPD, 100.02–147.51 Mya; node II). The split between Pseudotaxus and Taxus species was dated back to ~67.08 Mya in the Paleocene (95% HPD, 51.33–85.79 Mya; node III). The diversification of Taxus major lineages began in the Oligocene. The stem age of Taxus was estimated at 29.03 Ma (95% HPD, 18.67–40.51 Ma), after which the first-diverging extant lineage, T. brevifolia, split out from the most recent common ancestor. Subsequently, the divergence event within Taxus, which appears to have been the divergence of T. globosa/T. floridana from the other sections, occurred approximately ~24.09 Mya (95% HPD, 15.12–33.92 Mya; node IV) during the Oligocene. The age estimate of the crown node for T. chinensis/T. cuspidata/T. wallichiana group and T. yunnanensis was dated to be 13.04 Mya (95% HPD, 8.13–18.58 Mya; node V). Subsequently, T. cuspidata diverged from T. chinensis/T. wallichiana ~10.22 Mya (95% HPD, 6.31–15.00 Mya; node VI), and the split between T. chinensis and T. wallichiana likely occurred ~4.85 Mya (95% HPD, 2.26–7.81 Mya; node VII) during the Miocene (Figure 6). Our estimates of most extant species divergence ages within Taxus were often very young, typically concentrated in the last several million years, consistent with the fast accumulation of lineages in the LTT analysis.
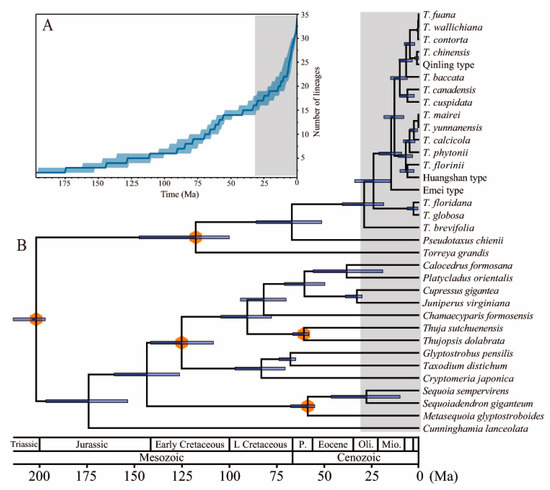
Figure 6.
(A) Lineages through time plot for Taxus genus, showing a diversification of the lineages occurred during the past ~29 million years. (B) Estimated divergence time of Taxus genus derived from BEAST analysis. The orange hexagons on the nods represent five calibration points.
3.6. Ancestral Areas Reconstruction
The results from BBM of biogeographic analysis showed that extant Taxus species shared a common ancestor whose ancestral distribution area was probably North America (A) and the earliest members expanded to Southeast Asia from where Chinese Taxus species originated. Afterwards China became the diversification center that led to a wave of speciation events and harbored at least 15 extant species of Taxus. We identified within Taxus genus three dispersal and two vicariance events that contribution to the spread and speciation of Taxus genus (Figure 7).
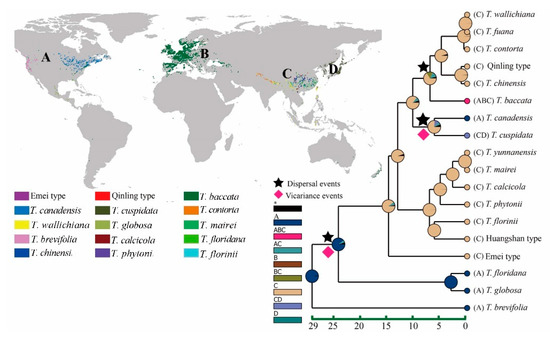
Figure 7.
Reconstructions of ancestral distribution and evolutionary trajectory based on the Bayesian binary Markov chain Monte Carlo (BBM) method implemented in RASP using the BEAST-derived chronogram of Taxus genus (see Figure 5B). The insert map shows the four areas (“A”, “B”, “C”, “D”) used in the analyses and the distribution range of Taxus genus in world. Pie charts indicate the proportion of the ancestral ranges. The color key identifies possible ancestral ranges at different nodes. The black pentagram and red rhombus indicate the dispersal and vicariance events, respectively.
4. Discussion
Unraveling the genomic relationships and evolutionary history of Taxus can help to clarify the phylogeny among Taxus species and infer the origin, evolutionary history, and spread pattern of a given plant clade. In the present study, four complete chloroplast genomes of Taxus species explicitly provided new and valuable information to solve formidable challenges and controversies on the evolution of the genus. Subsequently, we combined 14 other published chloroplast genomes to explore evolutionary relationships that allow drawing a full picture of Taxus chloroplast genome evolution. Our findings laid the foundation for future exploration of more details of the evolution of Taxus, as well as the molecular identification of Taxus species.
The sequenced chloroplast genomes promise a highly reliable phylogenetic tree derived from the whole chloroplast genome sequences, protein-coding genes, and non-coding regions, rooted with P. chienii as an outgroup. The phylogenetic trees, constructed based on the whole chloroplast sequences and protein-coding genes produced similar topological structures but were incongruent with that generated from non-coding regions, possibly because non-coding regions are more variable and provide relatively more variable loci than protein-coding genes. There is a general agreement that Taxus is a monophyletic genus in the Taxaceae family supported by both morphological [68] and molecular evidence [26,34]. In the Taxus genus, T. floridana is resolved as the first diverging species based on five chloroplast (matK, rbcL, trnL, trnL-trnF spacer, and psbA-trnH spacer) and one nuclear (ITS) molecular marker. However, Fu et al. [43] suggested that T. brevifolia is the first-branching species inferred from concatenation of three locally co-linear blocks, which is congruent with our results (Figure 1, Figures S3 and S4). Our analysis resolved T. floridana as the sister of T. globosa with 100% support, which is also supported by Gao et al. [69]. Similar to the results of Fu et al. [43], our results indicated that all New World Taxus (T. brevifolia, T. globosa and T. floridana) except T. canadensis, clustered into a clade separated from New World species. Unlike the phylogeny of Taxus inferred from non-coding sequences, the endemic T. yunnanensis clusters with Huangshan and Emei type were in a subclade and was distantly related with other four Taxus species native to China (Figure 1 and Figure S3). However, the inferred relationship of T. yunnanensis in the study of Hao et al. [34] was quite different from our analysis based on one chloroplast and three nuclear taxadiene synthase, which supported the sister relationship between T. yunnanensis and T. wallichiana. The Huangshan type was shown to be closer to the Emei type than to the Qinling type, by contrast, the concatenated alignment of five barcodes (rbcL, matK, psbA-trnH, trnL-trnF, and ITS) dataset supports a closer relationship between the Qinling and Emei type [21]. Nonetheless, the whole chloroplast genome sequences used by Fu et al. [43] presented a nested clade among the Huangshan type, T. chinensis and T. florinii. Additionally, similar to the result of Fu et al. [43], our results supported that the Qinling type was closer to T. baccata of Europe and T. contorta of West Himalaya (Figure 1, Figures S3 and S4). For a long time, T. floridana was treated as a variety of T. canadensis, suggesting a close relationship between these two species [70]. The examination of stomatal structure and ITS phylogenetic tree [29] in Taxus [71], however, did not group these two species into a monophyletic clade; instead, T. canadensis strongly allied with the Old World species, which was congruent with our results (Figure 1, Figures S3 and S4). T. chinensis has been treated as a variety of either T. wallichiana [72], T. baccata [73], or T. cuspidata [13], while in our phylogenomic tree, T. chinensis and T. wallichiana formed a strongly supported clade, with T. cuspidata more distantly related.
Our ancestral distribution reconstruction analysis 100% support that the ancestor of the extant Taxus lineages originated in North America. The oldest Taxus fossils found in North America can be tracked back to the Late Cretaceous [2] and there seems to be compelling evidence for the existence of Taxus in that area since then. Based on reliable calibration point and our robust phylogenetic estimation, our molecular dating also showed that the ancestor of modern Taxus genus diverged at ~67.08 Mya from its closest relative Pseudotaxus in North America (Figure 6 and Figure 7). However, it should be noted that Pseudotaxus only occurred in China [74], which proposed an alternative scenario of origin of the ancestor leading to modern lineages of Taxus. Moreover, significant older fossils of Taxus have been discovered in Eurasia tracing back to the Lower Cretaceous [75,76,77], suggesting a more ancient origin of Taxus than what we estimated. Nevertheless, the little morphological difference in phyllotaxis and female reproductive structures increased the level of challenge to distinguish their fossil characters. One feasible explanation placed that fossil on the node of the common ancestor of Pseudotaxus and Taxus [23]. Taken together, more extensive fossil and molecular evidence of related lineages are required to confirm the North America origin hypothesis.
Our divergence analyses dated an origin for the Taxus genus back to the Upper (Late) Cretaceous, with the extant lineages diversifying in North America and Asia much later during the Oligocene/early Miocene. This age is in accordance with previous estimate based on a small number of chloroplast and nuclear gene fragments [23,26] but significantly younger than the Eocene epoch inferred by Ran et al. [78]. The timescale of divergence between Taxus and Pseudotaxus overlapped with the Cretaceous-Paleogene extinction event and a subsequent global cooling that might have significantly affected contemporaneous Taxus or Pseudotaxus populations in North America. The surviving populations in refuges might have rapidly diversified in the early Miocene in North America and migrated to Eurasia in the late Lower Miocene via the Bering land bridge. This period coincided with the intensification of the East-Asian summer monsoon and a climate optimum with warmer temperatures [79]. The China population was further divided into two major clades at 13 Mya, a period in line with uplift-driven diversification around the Hengduan Mountains and the intensification of a cooler drier climate. Our research revealed a more recent divergence between European (T. baccata) and Asian (T. cuspidata, T. contorta/T. fuana, T. chinensis, and T. wallichiana) lineages, which support the Asia-to-Europe migration of the species in the late Miocene (~6.9). The Japanese yew T. cuspidata, which mainly occupied Japan and Northeast China, diverged with a common ancestor shared with American yew T. canadensis at that time (~6.15 Mya), after which T. canadensis re-migrated to North America via the Bering land bridge. Additionally, the Qinghai-Tibetan Plateau (QTP) uplift may have been an important factor that contributed to the formation of geographical disjunct distribution of Taxus species [80], which prevented the dispersal of Taxus species between Europe and Asia [81,82] in the late Miocene, a period overlapped with our estimation of the divergence time between the European and East Asian lineages. Similarly, Möller et al. [23] also revealed that the ancestors of extant lineages of Taxus have undergone repeated inter- and intra-continental migrations and linked the diversification/speciation of Taxus with the orogeny of the Qinling and Hengduan Mountains, especially around the Sichuan basin. Similar to our results, Turgai Strait, QTP, and climate change have been proven to serve as main factors affecting the distribution of Forsythia [81]. Thus, our results provide molecular support for the vicariance hypothesis and identify it as the main source of the differentiation in Taxus, upon which, some taxonomists classify the Taxus species according with their geographical distribution [10]. Overall, climate changes have likely provided an opportunity for accelerated diversification of the lineages [83].
The taxonomy of Taxus has been a contentious issue due to the few credible morphological characters for delineating species. In the present study, we successfully used complete chloroplast genomes to reserve of the phylogenetic history of 18 described extant Taxus species, and to produce the first sequence-based map of Taxus chloroplast genomic structural variation and site substitution in the course of recent plant speciation events. Based on phylogenetic reconstruction, molecular dating, ancestral range reconstruction, and fossil records, our study reveals that Taxus were estimated to have originated during the Cretaceous in North America with evidence for rapid diversification in Eurasia coinciding with several geological events ~29 Mya. Our result also supports that the Taxus genus is monophyletic and three North American species do not form a clade and together are inferred as the earliest diverging lineages; the predominantly European species have a closer relationship with the Eastern Asian species.
Furthermore, we estimated the average occurrence rate on average to be ~8.8 insertions and ~7.5 deletions per Myr in the Taxus genus, and we found that the number of insertions and deletions is nearly equivalent among Taxus chloroplast genomes. Comparative genomic analysis of repeat sequences suggests that the Rt repeat was dominant and contributed the most to the Taxus chloroplast genome expansion. Finally, we detected a total of 18 genes under positive selection by accomplishing genome-wide scanning. Moreover, the variation in repetitive sequences is shared among the species of Taxus, supporting that they are effective molecular markers for species identification and discovery in Taxus genus.
5. Conclusions
Many ecological and geographical variants of Taxus species have been formed in the process of long-term effects of the geographic environment and anthropogenic influence. In order to protect and adopt these important species efficiently, greater knowledge of the genetic background and evolutionary dynamics of Taxus is needed. In our study, we reconstructed the phylogenetic history and investigated the genomic variations of Taxus species based on 18 whole chloroplast genomes. The phylogenetic tree supported that T. brevifolia was basal lineages followed by the other North America lineages. We also found evidence that the Taxus species originated in North America in the Late Cretaceous with evidence for rapid diversification in Eurasia coinciding with several geological events, and the earliest members expanded to Southeast Asia. In addition, we found that the number of insertions and deletions is nearly equivalent among Taxus chloroplast genomes. Genome-wide scanning revealed 18 positively selected genes that were involved in translation and photosynthesis system in Taxus, which might be related to the adaptive evolution of Taxus species. These complete chloroplast genome sequences provide valuable resources for investigating the origin, evolution, and plant adaptation, and facilitate future species distinguishing and conservation biology of this ancient globally distributed genus.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13101590/s1, Figure S1: Gene maps of the 18 Taxus species chloroplast genomes. Genes inside the circle are transcribed in a clockwise direction, and genes outside of the circle are transcribed counterclockwise. Genes belonging to different functional groups are marked in different colors. Dashed area in the inner circle indicates GC content of the chloroplast genome; Figure S2: Visualization of sequence alignment of the 18 chloroplast genomes, with P. chienii as an outgroup; Figure S3: ML phylogeny of Taxus inferred from the concatenated sequences of 82 genes of chloroplasts using P. chienii as an outgroup; Figure S4: ML phylogeny of Taxus inferred from the concatenated sequences of non-coding sequences of the chloroplast genome using P. chienii as an outgroup; Figure S5: A map of chloroplast nucleotide variation across the Taxus choloplast genomes; Figure S6: The rate of chloroplast genomic variation in the genus Taxus. (A) Numbers of all Taxus branches to characterize the occurrence of chloroplast genomic variation. (B) The detection of insertion and deletion events occurred in different Taxus plastomes. (C) Overall length distribution of insertions and deletions across the chloroplast genomes. (D) Correlation between branch lengths and indel numbers for the 18 Taxus species and P. chienii chloroplast genomes; Figure S7: The ω (dN/dS) of chloroplast protein-coding genes in Taxus species using P. chienii as an outgroup. Table S1 Accession numbers of Taxus species used in our study. Table S2: Comparisons of repeat sequences across the Taxus choloplast genomes using P. chienii as outgroup.
Author Contributions
Conceptualization, X.L. and X.J.; software, S.F.; formal analysis, S.F.; investigation, H.Z.; writing—original draft preparation, X.J.; writing—review and editing, S.F. and H.Z.; funding acquisition, X.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Shaanxi Forestry Science and technology Innovation Project (SXLK2021-01-03).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fu, L.G.; Li, N.; Mill, R.R. Taxaceae in Flora of China; Wu, Z.Y., Peter, R.H., Eds.; Science Press: Beijing, China, 1999. [Google Scholar]
- Hollick, C.A. The Upper Cretaceous Floras of Alaska; USGS Professional Paper; USGS: Reston, VA, USA, 1930; p. 159.
- IUCN. The IUCN Red List of Threatened Species; Version 2017-2; IUCN: Gland, Switzerland, 2017; Available online: http://www.iucnredlist.org (accessed on 9 September 2017).
- State Forestry Administration and Ministry of Agriculture, P.R. China. List of National Key Protected Wild Species of China; The State Council of the People’s Republic of China: Beijing, China, 1999. Available online: http://www.gov.cn/gongbao/content/2000/content_60072.htm (accessed on 9 September 1999).
- Kala, C.P.; Sajwan, B.S. Conservation of medicinal plants: Conventional and contemporary strategies, regulations and executions. Indian For. 2007, 133, 484–495. [Google Scholar]
- Spector, T.; Thomas, P.; Determann, R. Taxus Floridana. The IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2011. [Google Scholar] [CrossRef]
- Thomas, P.; Farjon, A. Taxus Wallichiana. The IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2011. [Google Scholar] [CrossRef]
- Shah, A.; Li, D.Z.; Mölller, M.; Gao, L.M.; Gibby, H.M. Delimitation of Taxus fuana Nan Li & R.R. Mill (Taxaceae) based on morphological and molecular data. Taxon 2008, 57, 211–222. [Google Scholar]
- Möller, M.; Gao, L.M.; Mill, R.R.; Liu, J.; Zhang, D.Q.; Poudel, R.C.; Li, D.Z. A multidisciplinary approach reveals hidden taxonomic diversity in the morphologically challenging Taxus wallichiana complex. Taxon 2013, 62, 1161–1177. [Google Scholar] [CrossRef]
- Coughlan, P.; Carolan, J.C.; Hook, I.L.I.; Kilmartin, L.; Hodkinson, T.R. Phylogenetics of Taxus using the internal transcribed spacers of nuclear ribosomal DNA and plastid trnL-F regions. Horticulturae 2020, 6, 19. [Google Scholar] [CrossRef]
- Appendino, G. Taxol (paclitaxel): Historical and ecological aspects. Fitoterapia 1993, 64, 2–25. [Google Scholar]
- Hartzell, H.J. The Yew Tree: A Thousand Whispers; Hulogosi: Eugene, OR, USA, 1991; p. 4. [Google Scholar]
- Pilger, R. Taxaceae. In Engler, Das Pflanzenreich [...] [Heft 18] IV. 5"@deu; Engelmann: Leipzig, Germany, 1903; pp. 110–117. [Google Scholar]
- Voliotis, D. Historical and environmental significance of the yew (Taxus baccata L.). Isr. J. Plant Sci. 1986, 35, 47–52. [Google Scholar]
- Jaramillo, A.E. Taxus brevifolia Nutt. Pacific yew. In Silvics of North America; USDA Forest Survey Handbook; Burns, M., Honkala, B.H., Eds.; Forest Service, United States Department of Agriculture: Washington, DC, USA, 1990; Volume 1. [Google Scholar]
- Elwes, H.J.; Henry, A. The Trees of Great Britain; SR Publishers Ltd.: Edinburgh, Scotland, 1906; Volume 1. [Google Scholar]
- Dempsey, D.; Hook, I. Yew (Taxus) species—Chemical and morphological variations. Pharm. Biol. 2000, 38, 274–280. [Google Scholar] [CrossRef]
- Cope, E.A. Taxaceae: The genera and cultivated species. Bot. Rev. 1998, 64, 291–322. [Google Scholar] [CrossRef]
- Farjon, A. World Checklist and Biobiography of Conifers; Royal Botanic Gardens, Kew: Richmond, UK, 1998. [Google Scholar]
- Spjut, R.W. A phytogeographical analysis of Taxus (Taxaceae) based on leaf anatomical characters. J. Bot. Res. Inst. Tex. 2007, 1, 291–332. [Google Scholar]
- Liu, J.; Milne, R.I.; Möller, M.; Zhu, G.F.; Ye, L.J.; Luo, Y.H.; Yang, J.B.; Wambulwa, M.C.; Wang, C.N.; Li, D.Z. Integrating a comprehensive DNA barcode reference library with a global map of yews (Taxus L.) for forensic identification. Mol. Ecol. Resour. 2018, 18, 1115–1131. [Google Scholar] [CrossRef]
- Li, N.; Fu, L.K. Notes on gymnosperms I. Taxonomic treatments of some Chinese conifers. Novon 1997, 7, 261–264. [Google Scholar]
- Möller, M.; Liu, J.; Li, Y.; Li, J.H.; Ye, L.J.; Mill, R.; Thomas, P.; Li, D.Z.; Gao, L.M. Repeated intercontinental migrations and recurring hybridizations characterise the evolutionary history of yew (Taxus L.). Mol. Phylogenet. Evol. 2020, 153, 106952. [Google Scholar] [CrossRef] [PubMed]
- Poudel, R.C.; Möller, M.; Li, D.Z.; Shah, A.; Gao, L.M. Genetic diversity, demographical history and conservation aspects of the endangered yew tree Taxus contorta (syn. Taxus fuana) in Pakistan. Tree Genet. Genomes 2014, 10, 653–665. [Google Scholar] [CrossRef]
- Liu, J.; Möller, M.; Gao, L.M.; Zhang, D.Q.; Li, D.Z. DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol. Ecol. Resour. 2011, 11, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.C.; Pei, G.X.; Huang, B.L.; Ge, G.B.; Yang, L. Interspecific relationships and origins of Taxaceae and Cephalotaxaceae revealed by partitioned Bayesian analyses of chloroplast and nuclear DNA sequences. Plant Syst. Evol. 2008, 276, 89–104. [Google Scholar] [CrossRef]
- Hao, D.C.; Yang, L.; Xiao, P.G. The first insight into the Taxus genome via fosmid library construction and end sequencing. Mol. Genet. Genom. 2011, 285, 197–205. [Google Scholar] [CrossRef]
- Farjon, A.; Filer, D. An Atlas of the World’s Conifers: An Analysis of their Distribution, Biogeography, Diversity and Conservation Status; Brill: Leiden, The Netherlands, 2013. [Google Scholar]
- Li, J.; Davis, C.C.; Donoghue, T. Phylogeny and biogeography of Taxus (Taxaceae) inferred from sequences of the internal transcribed spacer region of nuclear ribosomal DNA. Harv. Pap. Bot. 2001, 6, 267–274. [Google Scholar]
- Mayol, M.; Riba, M.; González-Martínez, S.; Bagnoli, F.; Beaulieu, J.D.; Berganzo, E.; Burgarella, C.; Dubreuil, M.; Krajmerová, D.; Paule, L. Adapting through glacial cycles: Insights from a long-lived tree (Taxus baccata). New Phytol. 2015, 208, 973–986. [Google Scholar] [CrossRef]
- Miao, Y.C.; Su, J.R.; Zhang, Z.J.; Lang, X.D.; Liu, W.D.; Li, S.F. Microsatellite markers indicate genetic differences between cultivated and natural populations of endangered Taxus yunnanensis. Bot. J. Linn. Soc. 2015, 177, 450–461. [Google Scholar] [CrossRef][Green Version]
- Litkowiec, M.; Lewandowski, A.; Wachowiak, W. Genetic variation in Taxus baccata L.: A case study supporting Poland’s protection and restoration program. For. Ecol. Manag. 2018, 409, 148–160. [Google Scholar] [CrossRef]
- Vu, D.D.; Bui, T.; Nguyen, M.T.; Vu, D.G.; Nguyen, M.; Bui, V.T.; Huang, X.; Zhang, Y. Genetic diversity in two threatened species in Vietnam: Taxus chinensis and Taxus wallichiana. J. For. Res. 2017, 28, 265–272. [Google Scholar] [CrossRef]
- Hao, D.C.; Huang, B.L.; Yang, L. Phylogenetic relationships of the genus Taxus inferred from chloroplast intergenic spacer and nuclear coding DNA. Biol. Pharm. Bull. 2008, 31, 260–265. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alkan, C.; Sajjadian, S.; Eichler, E.E. Limitations of next-generation genome sequence assembly. Nat. Methods 2011, 8, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Ingvarsson, P.K.; Ribstein, S.; Taylor, D.R. Molecular evolution of insertions and deletion in the chloroplast genome of Silene. Mol. Biol. Evol. 2003, 20, 1737–1740. [Google Scholar] [CrossRef]
- Pessoa-Filho, M.; Martins, A.M.; Ferreira, M.E. Molecular dating of phylogenetic divergence between Urochloa species based on complete chloroplast genomes. BMC Genom. 2017, 18, 516. [Google Scholar] [CrossRef]
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res.-Engl. 2019, 6, 89. [Google Scholar] [CrossRef]
- Wu, C.S.; Chaw, S.M. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): Evolution towards shorter intergenic spacers. Plant Biotechnol. J. 2014, 12, 344–353. [Google Scholar] [CrossRef]
- Qu, X.J.; Jin, J.J.; Chaw, S.M.; Li, D.Z.; Yi, T.S. Multiple measures could alleviate long-branch attraction in phylogenomic reconstruction of Cupressoideae (Cupressaceae). Sci. Rep.-UK 2017, 7, 41005. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, J.; Yang, B.; Li, R.; Zhu, W.; Sun, L.; Tian, J.; Zhang, L. The complete chloroplast genome sequence of Taxus chinensis var. mairei (Taxaceae): Loss of an inverted repeat region and comparative analysis with related species. Gene 2014, 540, 201–209. [Google Scholar] [CrossRef]
- Jia, X.-M.; Liu, X.-P. Characterization of the complete chloroplast genome of the Chinese yew Taxus chinensis (Taxaceae), an endangered and medicinally important tree species in China. Conserv. Genet. Resour. 2016, 9, 197–199. [Google Scholar] [CrossRef]
- Fu, C.N.; Wu, C.S.; Ye, L.J.; Mo, Z.Q.; Liu, J.; Chang, Y.W.; Li, D.Z.; Chaw, S.M.; Gao, L.M. Prevalence of isomeric plastomes and effectiveness of plastome super-barcodes in yews (Taxus) worldwide. Sci. Rep.-UK 2019, 9, 2773. [Google Scholar] [CrossRef] [PubMed]
- Christoph, H.; Lutz, B.; Bastien, C. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucl. Acids Res. 2013, 41, e129. [Google Scholar]
- Wyman, S.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA Genes in genomic sequence. Nucl. Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S.A. ORF-FINDER: A vector for high-throughput gene identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl. Acids Res. 2013, 41, 575–581. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Drummond, A.J. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Li, M.; Christina, K.; Koichiro, T. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar]
- Mao, K.; Milne, R.I.; Zhang, L.; Peng, Y.; Liu, J.; Thomas, P.; Mill, R.R.; Renner, S.S. Distribution of living Cupressaceae reflects the breakup of Pangea. Proc. Natl. Acad. Sci. USA 2012, 109, 7793–7798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, H.-J.; Landis, J.B.; Deng, T.; Meng, A.-P.; Sun, H.; Peng, Y.-S.; Wang, H.-C.; Sun, Y.-X. Plastome phylogenomic analysis of Torreya (Taxaceae). J. Syst. Evol. 2019, 57, 607–615. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Loytynoja, A.; Goldman, N. A model of evolution and structure for multiple sequence alignment. Philos. Trans. R. Soc. Lond. 2008, 363, 3913–3919. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998, 15, 568–573. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucl. Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef]
- Zhang, Q.J.; Zhu, T.; Xia, E.H.; Shi, C.; Liu, Y.L.; Zhang, Y.; Liu, Y.; Jiang, W.K.; Zhao, Y.J.; Mao, S.Y. Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, 4954–4962. [Google Scholar] [CrossRef]
- Hu, T.T.; Pattyn, P.; Bakker, E.G.; Cao, J.; Cheng, J.F.; Clark, R.M.; Fahlgren, N.; Fawcett, J.A.; Grimwood, J.; Gundlach, H. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011, 43, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Bennetzen, J.L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 12404–12410. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.Z.; Liu, Y.L.; Zhang, D.; Li, W.; Eichler, E.E. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Comms. Biol. 2019, 2, 278. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar]
- Weng, M.L.; Blazier, J.C.; Madhumita, G.; Jansen, R.K. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 2014, 31, 645–659. [Google Scholar] [CrossRef]
- Hart, J.A. A cladistic analysis of conifers: Preliminary results. J. Arnold Arbor. 1987, 68, 269–307. [Google Scholar] [CrossRef]
- Gao, L.M.; Li, D.Z.; Moeller, M. Molecular systematics of Taxus. In Proceedings of the National Symposium on Systematic and Evolutionary Botany and the 9th Youth Symposium on Systematic and Evolutionary Botany, Xi’an, China, 20 August 2006. [Google Scholar]
- Silba, J. An International Census of the Coniferae; Phytologia Memoirs Series; Moldenke: Plainfeld, NJ, USA, 1984; Volume 7, pp. 1–79. [Google Scholar]
- Spjut, R.W. The morphological relationships of Taxus canadensis in North America and Eurasia. Amer. J. Bot. 2000, 87, 159. [Google Scholar]
- Florin, R. On the morphology and relationships on the Taxaceae. Bot. Gaz. 1948, 110, 31–39. [Google Scholar] [CrossRef]
- Henry, J.E. Taxus. In The Tress of Great Britain and Ireland; Henry, J.E., Henry, A., Eds.; Private Printing: Edinburgh, Scotland, 1906; Volume 1. [Google Scholar]
- Kou, Y.; Zhang, L.; Fan, D.; Cheng, S.; Li, D.; Hodel, R.G.J.; Zhang, Z. Evolutionary history of a relict conifer, Pseudotaxus chienii (Taxaceae), in south-east China during the late Neogene: Old lineage, young populations. Ann. Bot. 2020, 125, 105–117. [Google Scholar] [CrossRef]
- Chen, F.; Meng, X.Y.; Ren, S.Q.; Wu, C.L. The Early Cretaceous Flora and Coalbearing Strata of Fuxin Basin and Tiefa Basin; Geological Publishing: Beijing, China, 1988; (In Chinese and English). [Google Scholar]
- Deng, S.H. Early Cretaceous Flora of Huolinhe Basin, Inner Mongolia; Geological Publishing House: Beijing, China, 1995; (In Chinese and English). [Google Scholar]
- Xu, X.H.; Sun, B.N.; Yan, D.F.; Wang, J.; Dong, C. A Taxus leafy branch with attached ovules from the Lower Cretaceous of Inner Mongolia. North China. Cretac. Res. 2015, 54, 266–282. [Google Scholar] [CrossRef]
- Ran, J.H.; Shen, T.T.; Wang, M.M.; Wang, X.Q. Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc. Roy. Soc. B-Biol. Sci. 2018, 285, 20181012. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Gao, L.; Soltis, D.E.; Soltis, P.S.; Yang, J.; Fang, L.; Yang, S.; Li, D. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 2017, 215, 1235–1248. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.-H.; Kim, C.; Choi, K.; Kim, J.-H. Molecular phylogeny and dating of Forsythieae (Oleaceae) provide Insight into the Miocene history of Eurasian temperate shrubs. Front. Plant Sci. 2018, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Mclewin, W.; Fay, M. Molecular phylogeny of Helleborus (Ranunculaceae), with an emphasis on the East Asian-Mediterranean disjunction. Taxon 2001, 50, 1001–1018. [Google Scholar] [CrossRef]
- Zhang, Z.; Fan, L.; Yang, J.; Hao, X.; Gu, Z. Alkaloid polymorphism and ITS sequence variation in the Spiraea japonica complex (Rosaceae) in China: Traces of the biological effects of the Himalaya-Tibet Plateau uplift. Am. J. Bot. 2006, 93, 762–769. [Google Scholar] [CrossRef]
- Hinsinger, D.D.; Basak, J.; Gaudeul, M.; Cruaud, C.; Bertolino, P.; Frascaria-Lacoste, N.; Bousquet, J. The phylogeny and biogeographic history of ashes (Fraxinus, oleaceae) highlight the roles of migration and vicariance in the diversification of temperate trees. PLoS ONE 2013, 8, e80431. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).