Abstract
Camellia japonica is a native tree species with high economic value that is widely cultivated in southern China. In recent years, canker disease has been observed in camellia plantations in Zhejiang Province, China, with the disease incidence rate in some plantations exceeding 20%. Canker disease severely affects the trunks and branches of C. japonica in China, but the causal agent has not yet been identified. In this study, the pathogen was isolated from infected C. japonica tissues through a conventional tissue isolation approach. Species identification was conducted using morphological methods combined with multilocus phylogenetic analysis. Pathogenicity was tested based on Koch’s postulates. The results showed that the pathogen could be isolated from the diseased bark of C. japonica ‘Hongluzhen’. The pathogen was identified as Nectria pseudotrichia based on morphological, cultural, and molecular traits. The inoculation of the pathogen into C. japonica ‘Hongluzhen’ caused necrotic lesions on healthy seedlings, and the fungus N. pseudotrichia could be re-isolated from such lesions. Therefore, N. pseudotrichia is the causal agent of canker disease affecting C. japonica in China.
1. Introduction
Camellia japonica L. is an evergreen shrub or tree in the family Theaceae that is native to China, South Korea, and Japan [1,2]. Over the last 200 years, C. japonica has been gradually introduced into Europe, the Americas, and Oceania, and has become a popular garden plant in most warm-temperate regions of the world [2,3]. C. japonica is a multipurpose tree species with high ornamental, economic, and medicinal value. C. japonica is popular due to its bright color and beautiful flowers, which are responsible for its high ornamental value. The content of unsaturated fatty acids, such as oleic acid, in the seeds of C. japonica accounts for more than 90% of the total fatty acid content, giving this species high economic value [4]. C. japonica plants have been found to contain triterpenes, saponins, flavonoids, tannins, fatty acids, and phenolic compounds, which exhibit various biological activities, including antioxidant, antiaging, antiviral, anti-inflammatory, antihyperuricemic, antiosteoporotic, and antiallergic activities [1,5,6].
Canker disease was first described in C. japonica in the United States in 1924 [7] and was subsequently observed in the United Kingdom [8], Spain [9] and China. The causal agent of this disease was reported to be Glomerella cingulate (Stonem) Spauld & Schrenk in the United States [10] and in the United Kingdom [8] and to be Neofusicoccum luteum (Pennycook & Samuels) Crous, Slippers & A.J.L. Phillips, and N. parvum (Pennycook & Samuels) Crous, Slippers & A.J.L. Phillips in Spain [9]. However, the pathogen causing this disease in China has not yet been reported. In recent years, canker disease in C. japonica has been found in many areas of Zhejiang Province, China. Among these infections, C. japonica plantations in Ningbo and Jinhua are the most seriously impacted, with more than 20% of camellia plants being infected. In Zhejiang Province, canker disease has only been observed on the trunk and branches of C. japonica. Mildly infected plants exhibit large necrotic lesions on the bark, while severe infection can cause the death of the entire branch or even the entire tree. Canker disease has become the most serious disease affecting C. japonica.
This study aims to estimate the incidence rate of canker disease in two camellia plantations in Zhejiang Province, to isolate the pathogen from infected C. japonica tissues through a conventional tissue isolation approach, to identify the fungal species that are associated with camellia canker disease in China based on morphology combined with multilocus phylogenetic analysis, and to determine the pathogenicity of Nectria pseudotrichia Berk. & M.A. Curtis on the stems of C. japonica using Koch’s postulates.
2. Materials and Methods
2.1. Study Site and Field Surveys of Canker Disease in C. japonica
Two camellia plantations, located in Ningbo (29°39′19″ N, 121°29′32″ E) and Jinhua (29°7′36′′ N, 119°35′21′′ E) in Zhejiang Province, China, were selected as the study sites. There are 15 cultivars of camellia planted at the Ningbo camellia plantation, including 8 cultivars of C. japonica, 4 cultivars of C. hiemalis Nakai, 2 cultivars of C. sasanqua Thunb., and 1 cultivar of C. uraku (Mak.) Kitamura. There are eight cultivars of camellia planted at the Jinhua camellia plantation, including three cultivars of C. japonica and five cultivars of C. hiemalis. The area of each camellia plantation is about 2 hectares in size, and the area of C. japonica on each plantation is more than 0.2 hectares. The C. japonica plants range from 10 to 20 years of age, and the planting density is about 4000 plants per hectare. In June 2020, the study investigated the incidence and symptoms of canker disease in two camellia plantations. The preliminary situation of canker disease occurrence in all of the cultivars at the two plantations was obtained by consulting the local managers and through our own systematic observation, and then the incidence and symptoms of canker disease in C. japonica ‘Hongluzhen’ were investigated using a sampling method. Three sampling plots with an area of 20 m × 20 m were randomly selected from a C. japonica ‘Hongluzhen’ planting plot at each plantation, and 20 trees in each plot were investigated to calculate the rate of disease incidence. The disease incidence rate was calculated according to the method proposed by Linaldeddu et al. [11].
A scalpel was used to scrape the diseased C. japonica ‘Hongluzhen’ bark, which was transported to the laboratory so that the surface characteristics could be analyzed and the pathogenic fungi could be isolated.
2.2. Fungal Isolation
A total of 15 bark samples from diseased and healthy plants were collected from two camellia plantations. To isolate the causal agent, the samples were washed under running water for 2 h, and the margin of each infected sample was cut into small sections. The surfaces of these sections were sterilized through immersion in 70% ethanol for 30 s, followed by 10% sodium hypochlorite solution for 10 min, and the sections were then rinsed five times in sterile water and were blotted dry using sterile filter paper [12]. The dry sections were transferred to potato dextrose agar (PDA) medium supplemented with streptomycin (200 mg/L) and were incubated at 25 °C. Mycelial fragments were collected from the growing colony margin, sub-cultured onto synthetic nutrient-poor agar medium (SNA) and PDA, and incubated at 25 °C.
2.3. Morphological Characterization
The morphology of the colonies of the pathogenic fungi that formed on the PDA and SNA plates was observed, and colony sizes were measured with a millimeter ruler. The morphological structures of hyphae and conidia were observed with a microscope. Some samples were stained with Melzer’s reagent [13] or lactophenol cotton blue staining solution (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) prior to observation. The size and morphology of the conidia and synnemata were determined using the Keyence VHX-5000 digital microscope (Keyence, Osaka, Japan).
2.4. DNA Extraction, Amplification, Sequencing, and Phylogenetic Analysis
For genomic DNA extraction, the mycelium was harvested from the PDA plates and was ground into a powder using a TissueLyser-48 grinder. Total genomic DNA was extracted from the powder with a universal genomic DNA Extraction Kit (Takara MiniBEST Universal Genomic DNA Extraction Kit ver. 5.0; Takara Biomedical Technology Co., Ltd., Beijing, China). Polymerase chain reaction (PCR) amplification of the large subunit ribosomal DNA (LSU rDNA), internal transcribed spacer (ITS) rDNA, and the tef1-α region were conducted with the primer pairs LR0R/LR5 [14], ITS5/ITS4 [15,16], and EF1-728F/EF1-986R [17], respectively. The PCR mixture for all regions contained 1 μL genomic DNA, 10 μL 2× PCR Mix (Tiangen Biotech Co., Ltd., Beijing, China), 0.5 μL of each primer, and 8 μL double-distilled sterilized water. The PCR conditions for all of the regions were 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 40 s, followed by a final extension of 72 °C for 10 min. The PCR products were sent to Shangya Biotechnology Co., Ltd. (Hangzhou, China) for sequencing, and representative sequences were deposited in GenBank (Table 1).

Table 1.
Specimens used in phylogenetic analyses and their GenBank accession numbers.
The sequences for the 3 loci (LSU rDNA, ITS rDNA, and tef1-α) that were obtained in this study and 84 sequences of the genus Nectria from GenBank (Table 1) were used for phylogenetic analysis. Cosmospora coccinea (CBS 114050, AR2741) sequences from GenBank were used as an outgroup. Multiple sequence alignment of each locus was performed using MAFFT v.7.313 [18] and were manually adjusted using PhyloSuite v.1.2.2 [19]. Alignments of the three loci were concatenated into a single dataset with PhyloSuite v.1.2.2. Nucleotide substitution models for each locus, which were determined using PartitionFinder v.2 [20], were included for each gene partition. Phylogenetic analysis was performed using MrBayes v.3.2.6 [21]. Two Markov chain Monte Carlo (MCMC) analyses were run from random trees for 1,000,000 generations, with sampling every 100 generations. The initial 25% of the sampled trees were discarded as burn-in, and posterior probabilities were determined from the remaining trees. The resulting tree was visualized in FigTree v.1.4.3 and was edited with Adobe Illustrator CS6.
2.5. Pathogenicity Tests
In October 2020, stems and detached leaves from seedlings (15.7 ± 3.5 mm ground diameter) of C. japonica ‘Hongluzhen’ were inoculated with the representative isolate LPNP of N. pseudotrichia. Fungal agar plugs (5 mm in diameter) from the edge of actively growing cultures were placed into a wounded inoculation region, which were made using a sterile scalpel or dissecting needle, and the inoculation region was then covered with Parafilm to conserve moisture. Negative controls were inoculated with sterile PDA plugs, which were applied to wounds. The inoculated seedlings and detached leaves were maintained in a climate-controlled chamber at 28 °C with a 12 h photoperiod. At 14 and 28 days after inoculation, the necrotic lesions on the detached leaves and stems surrounding each inoculation region were individually observed with a VHX-5000 microscope.
At 28 days after inoculation, re-isolation was conducted from the margin of the necrotic tissue to recover the inoculated fungi and to meet Koch’s postulates. Additional isolation from the negative controls was conducted to verify that no endophytic N. pseudotrichia fungus was present. The re-isolation procedure is described in Section 2.2. The isolates were identified based on morphological characterization and ITS rDNA sequences.
3. Results
3.1. Field Surveys of Canker Disease in C. japonica
Field surveys indicated that canker disease occurred on both camellia plantations, and that the infections only occurred in C. japonica cultivar ‘Hongluzhen’. The disease incidence rates of C. japonica ‘Hongluzhen’ in Ningbo and Jinhua were 46.2% ± 9.4% and 20.0% ± 0.0%, respectively.
The pathogenic fungus can infect the trunk and branches of C. japonica ‘Hongluzhen’. When the pathogen infects the trunk, it often causes the necrosis of inner bark and xylem surface tissues, revealing the xylem (Figure 1a). In natural conditions, it is difficult for a callus to form around the lesion, meaning that it is possible for the lesion to continuously expand, forming longer and larger lesions. When the branches are infected, the bark of the infected regions becomes dark and slightly sunken (Figure 1b), and light red synnematal structures are present on some of the portions of diseased bark. The synnema were erumpent through the epidermis, solitary, slightly spherical at the top, 284.0–414.0 μm in diameter, and had a long (1323.0–1584.0 μm) stipe at the base (Figure 1c).

Figure 1.
Disease symptoms and synnematal morphology of the pathogenic fungus. (a,b) Trunk canker and branch canker observed on C. japonica ‘Hongluzhen’; (c) synnemata on diseased bark.
3.2. Isolation, Morphological Structures and Cultural Characteristics of the Pathogenic Fungus of Camellia Canker
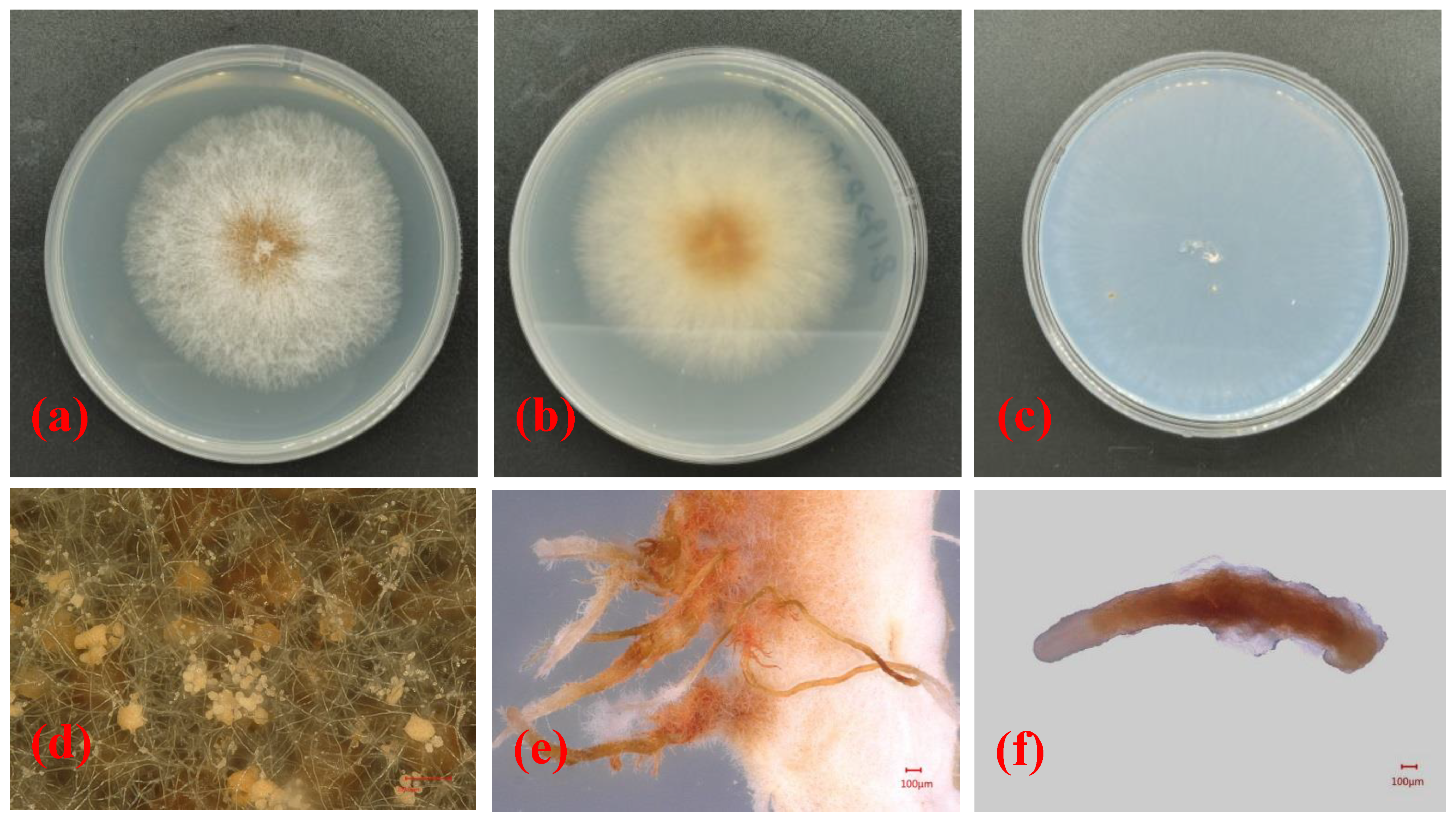
In this study, a total of 15 samples were collected, and Nectriaceae-like fungi were grown on PDA. The isolates grew quickly on PDA, with colony diameters of 8.1 ± 1.6 cm after 6 days of incubation at 25 °C. The upper side of the colonies on PDA was red at the center, and the color gradually lightened from the center to the margin of each colony (Figure 2a). The reverse side of each colony was red at the center and was creamy at the margin (Figure 2b). The isolates formed colorless colonies on SNA (Figure 2c). Vegetative hyphae were observed on SNA, but no aerial hyphae were present (Figure 2c). When the PDA plate was observed using the VHX-5000 microscope, the conidiophores, conidial mass, and conidia were observed in the center of each colony (Figure 2d). The conidia varied in size and morphology on PDA, with young conidia that were ellipsoidal to cylindrical in shape and that were generally 4.5–5.6 μm × 2.6–3.3 μm in size, and mature conidia that were ellipsoidal, cylindrical to allantoid in shape, and approximately 9.1–12.2 μm × 2.9–3.9 μm in size. When the isolate was cultured on PDA for 20 days, a large number of synnemata were produced near the side wall of the Petri dish (Figure 2e). However, only a small number of synnemata were produced on SNA after 60 days (Figure 2f).
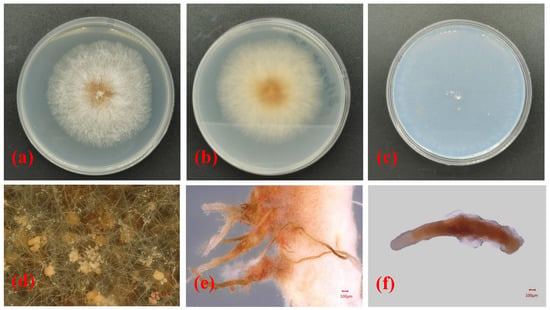
Figure 2.
Morphological structures and cultural characteristics of Nectria pseudotrichia. (a,b) Upper and reverse sides of colonies on PDA, respectively; (c) upper side of colony on SNA; (d) conidial masses produced on PDA; (e,f) synnemata produced on PDA and SNA, respectively.
Based on the morphological characteristics of the mycelia, synnema, conidiophores, conidial mass, and conidia of the isolated pathogenic fungi as well as their cultural characteristics on PDA and SNA, the pathogen affecting camellia C. japonica in Zhejiang Province was preliminarily identified as Nectria pseudotrichia.
3.3. Phylogenetic Analysis
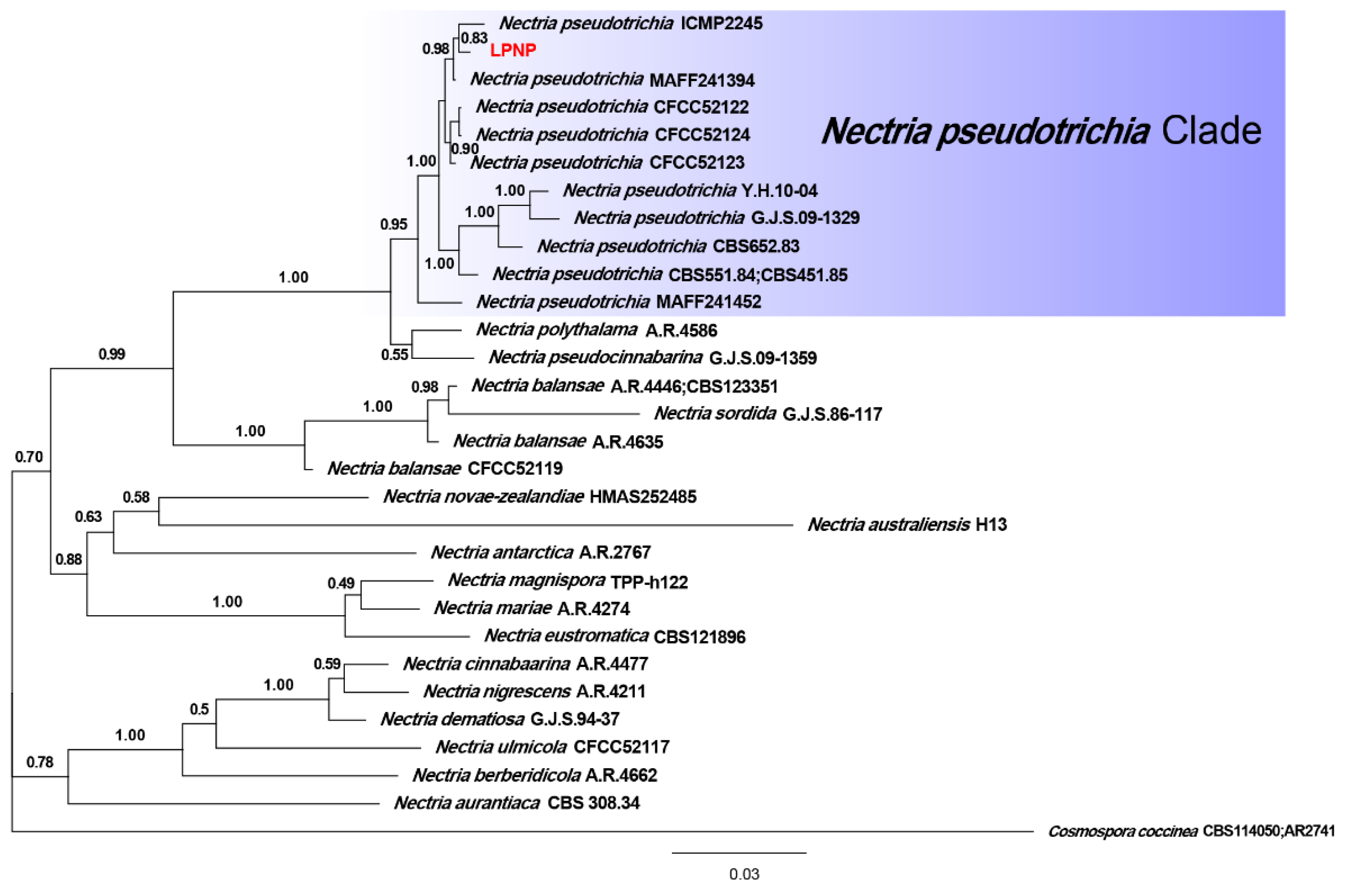
Phylogenetic analyses of 29 strains of Nectria were performed using the concatenated datasets for three loci. A total of 1561 characters (LSU: 1–799, ITS: 800–1308, tef1-α: 1309–1561) were included in the datasets. For Bayesian inference (BI) analysis, GTR + I + G, GTR + I + G, and SYM + G were selected as the best nucleotide substitution models for LSU rDNA, ITS rDNA, and tef1-α, respectively. The phylogenetic tree that was generated from BI analysis showed that the isolate LPNP clustered with 10 strains of N. pseudotrichia from Genbank and had a high posterior probability value (Figure 3). Multilocus phylogenetic analysis confirmed the assignment of the isolate LPNP as a strain of N. pseudotrichia. Multilocus phylogenetic analysis also revealed that within the N. pseudotrichia Clade, the LPNP strain and the two strains ICMP2245 and MAFF241394 from Japan clustered into a subclade, indicating that the genetic distance between these three strains is the closest (Figure 3).
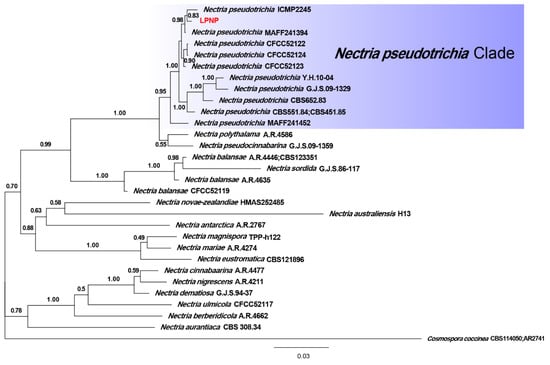
Figure 3.
Phylogenetic tree inferred from BI analysis based on concatenated alignments of LSU rDNA, ITS rDNA, and tef1-α sequences. Cosmospora coccinea was used as an outgroup. Numbers on the nodes represent Bayesian posterior probabilities. The isolate obtained in the current study is highlighted in red.
3.4. Pathogenicity Tests
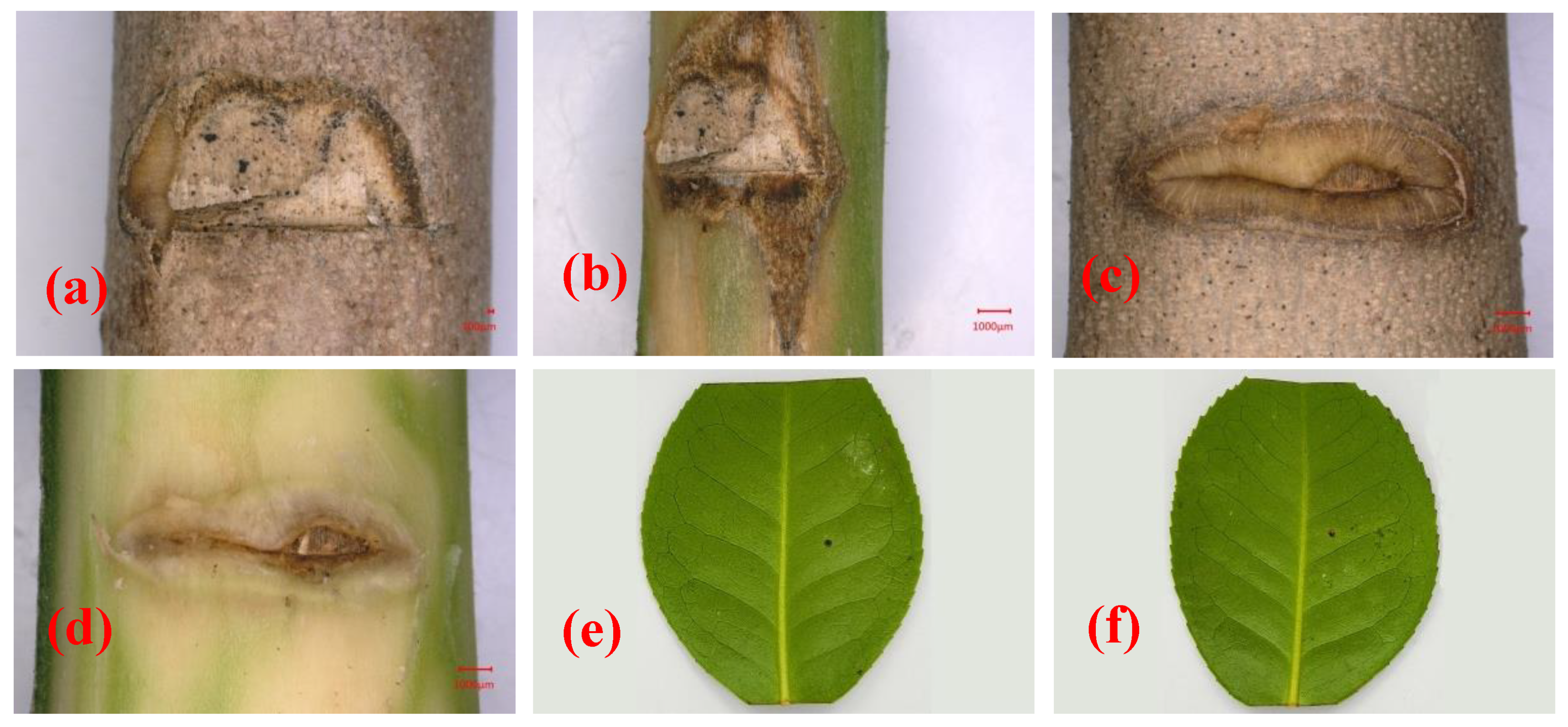
At 28 days after inoculation, symptoms were observed at the inoculation site on the stem of C. japonica ‘Hongluzhen’ (Figure 4a,b). The stems of the seedlings that had been inoculated with sterile PDA plugs produced a large amount of callus at the inoculated site, and the bark surface was flat with no necrotic lesions beneath the bark (Figure 4c,d). However, the stems of seedlings that had been inoculated with N. pseudotrichia produced a negligible callus at the inoculation site; the bark was slightly sunken, and necrotic lesions had formed beneath it (Figure 4a,b). In addition, at 14 days after inoculation, the detached leaves of the seedlings that had been inoculated with N. pseudotrichia showed no lesions (Figure 4e), indicating that N. pseudotrichia had no pathogenicity to the leaves of C. japonica ‘Hongluzhen’. In the re-isolation experiment, a fungus could be isolated from the necrotic tissue of symptomatic seedlings, and the morphological characterization as well as the ITS rDNA sequence of the isolate were consistent with those of LPNP. This pathogen was not isolated from the bark around the inoculation sites of negative control seedlings.
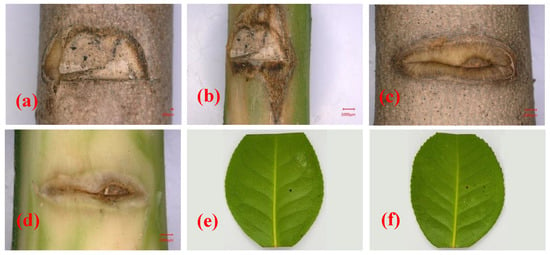
Figure 4.
Pathogenicity analysis of Nectria pseudotrichia on the stems and detached leaves of Camellia japonica ‘Hongluzhen’. (a,b) Lesions on and beneath the bark of a camellia stem inoculated with N. pseudotrichia, respectively; (c,d) lesions on and beneath the bark of a camellia stem inoculated with a sterile PDA plug, respectively; (e,f) lesions on detached leaves inoculated with sterile PDA and N. pseudotrichia, respectively.
N. pseudotrichia can cause necrotic lesions on the stem of C. japonica and can be re-isolated from necrotic tissue, indicating that N. pseudotrichia is the causal agent of camellia canker disease in Zhejiang Province, China.
4. Discussion
Canker disease is characterized by a necrotic region, with swelling surrounding a sunken lesion or blister on the bark of trunks and branches that affect the underlying tissues [22]. The basic characteristic of canker disease is the generation of a visible necrotic area on the bark of infected trees [23]. Most of the pathogenic fungi that cause canker disease belong to a few genera of Ascomycetes, including Nectria, Valsa, Cytospora, Dothiorella, Dothichiza, Botryodiplodia, Macrophoma, Diaporthe, Melanconium, Fusarium, and Trichoscyphella [23,24,25,26,27].
Fungi in the genus Nectria are commonly found on broad-leaved trees and shrubs in the temperate regions of the northern hemisphere [28,29]. These fungi are characterized by the production of brightly colored fleshy perithecia on the bark [28,29]. Approximately 35 species have been described worldwide, with 11 species occurring in China [29]. Nectria causes canker disease on more than 60 species of trees and shrubs, including apple, pear, ash, holly, elm, aspen, birch, dogwood, maple, sassafras, sweet gum, and walnut. Infected trees often exhibit large areas of necrotic bark, which may lead to the death of infected branches or entire trees.
N. pseudotrichia belongs to the genus Nectria, and its anamorph is Tubercularia lateritia (Berk.) Seifert [30]. This fungus is distributed worldwide and has been isolated from more than 40 genera of trees in Asia, Africa, North America, South America, and Oceania [28,30,31,32,33,34]. Early researchers mostly found N. pseudotrichia on dead trees or branches, so it is considered to be a saprophytic tree fungus [30]. In 2003, Becker first reported that N. pseudotrichia can cause canker disease on Pyrus pyrifolia (Burm.) Nakai [31], and the pathogenicity of this fungus has gradually been recognized. This fungus has been reported to cause canker or dieback on P. pyrifolia [31], Macadamia integrifolia Maiden & Betche and M. tetraphylla L.A.S. Johnson [32], Fragaria × ananassa Duch. [33], and Hevea brasiliensis (Willd. ex A. Juss.) Muell. Arg. [35]. In this study, synnemata of N. pseudotrichia were observed on the diseased bark of C. japonica in the field, and this fungus could be isolated from the diseased bark. To our knowledge, this is the first report of N. pseudotrichia infection in Camellia. When we inoculated the isolates onto seedlings of C. japonica, they caused disease in the camellia seedlings, and the fungus could be re-isolated from the diseased bark. Thus, N. pseudotrichia is the causal agent of camellia canker disease in Zhejiang Province, China. Further investigation is required for disease prevention and control measures according to the biological characteristics of N. pseudotrichia to reduce the losses caused by canker disease in C. japonica.
Author Contributions
Conceptualization, X.G. and J.S.; methodology, X.G. and Y.L.; software, X.G.; formal analysis, X.G., Y.L. and Z.L.; investigation, X.G., Y.L. and G.W.; resources, G.W. and J.L.; data curation, X.G. and J.L.; writing—original draft preparation, X.G. and J.L.; writing—review and editing, J.S.; supervision, J.L.; project administration, J.L. and J.S.; funding acquisition, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (Grant number 2019YFD1001005).
Data Availability Statement
The gene sequences have been deposited in GenBank under the accession numbers OK562743, OK562730 and OL440127 for ITS rDNA, LSU rDNA and tef1-α, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lim, T.K. (Ed.) Camellia japonica. In Edible Medicinal and Non Medicinal Plants; Springer: Dordrecht, The Netherlands, 2014; Volume 8, pp. 764–776. [Google Scholar]
- Gao, J.Y.; Parks, C.R.; Du, Y.Q. Collected Species of the Genus Camellia: An Illustrated Outline; Zhejiang Science and Technology Press: Hangzhou, China, 2005; pp. 43–44. [Google Scholar]
- Vela, P.; Salinero, C.; Sainz, M. Phenological growth stages of Camellia japonica. Ann. Appl. Biol. 2012, 162, 182–190. [Google Scholar] [CrossRef]
- Garcia-Jares, C.; Sanchez-Nande, M.; Lamas, J.P.; Lores, M. Profiling the Fatty Acids Content of Ornamental Camellia Seeds Cultivated in Galicia by an Optimized Matrix Solid-Phase Dispersion Extraction. Bioengineering 2017, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-L.; Yin, C.-P.; Kong, L.-C.; Jiang, D.-H. Extraction optimisation, purification and major antioxidant component of red pigments extracted from Camellia japonica. Food Chem. 2011, 129, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Yoon, I.-S.; Park, D.-H.; Kim, J.-E.; Yoo, J.-C.; Bae, M.-S.; Oh, D.-S.; Shim, J.-H.; Choi, C.-Y.; An, K.-W.; Kim, E.-I.; et al. Identification of the biologically active constituents of Camellia japonica leaf and anti-hyperuricemic effect in vitro and in vivo. Int. J. Mol. Med. 2017, 39, 1613–1620. [Google Scholar] [CrossRef]
- Baxter, L.W. Studies on Control of Root Rot and Dieback of Camellias; Louisiana State University: Baton Rouge, FL, USA, 1955. [Google Scholar]
- Dickens, J.S.W.; Cook, R.T.A. Glomerella cingulata on camellia. Plant Pathol. 1989, 38, 75–85. [Google Scholar] [CrossRef]
- Pintos, C.; Redondo, V.; Chaves, M.; Rial, C.; Mansilla, P. Camellia japonica dieback caused by Neofusicoccum luteum and N. parvum in Spain. In Proceedings of the International Camellia Congress, Chuxiong, China, 5–9 February 2012. [Google Scholar]
- Ngo, H.C.; Baxter, L.W.; Fagan, S.G. The status of our knowledge in 1978 of twig blight, canker and dieback of camellias caused by a strain of Glomerella cingulata. Am. Camellia Yearb. 1978, 33, 75–95. [Google Scholar]
- Linaldeddu, B.; Bottecchia, F.; Bregant, C.; Maddau, L.; Montecchio, L. Diplodia fraxini and Diplodia subglobosa: The Main Species Associated with Cankers and Dieback of Fraxinus excelsior in North-Eastern Italy. Forests 2020, 11, 883. [Google Scholar] [CrossRef]
- Geng, X.S.; Shu, J.P.; Sheng, J.L.; Zhang, W.; Peng, H. Isolation and identification of the pathogens causing witches’ broom disease of five bamboo species of non-Phyllostachys. Sci. Silvae Sin. 2020, 56, 82–89. [Google Scholar]
- Leonard, L.M. Melzer’s, lugol’s or iodine for identification of white-spored Agaricales? McIlvainea 2006, 16, 43–51. [Google Scholar]
- Eberhardt, U. Methods for DNA Barcoding of Fungi. In Springer Protocols Handbooks; Springer: Singapore, 2012; Volume 858, pp. 183–205. [Google Scholar]
- White, T.; Bruns, T.; Lee, S.; Taylor, F.J.R.M.; White, T.; Lee, S.H.; Taylor, L.; Shawetaylor, J. Amplification and direct se-quencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, A.M., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Elvira-Recuenco, M.; Cacciola, S.O.; Sanz-Ros, A.V.; Garbelotto, M.; Aguayo, J.; Solla, A.; Mullett, M.; Drenkhan, T.; Oskay, F.; Kaya, A.G.A.; et al. Potential Interactions between Invasive Fusarium circinatum and Other Pine Pathogens in Europe. Forests 2019, 11, 7. [Google Scholar] [CrossRef]
- Agrios, G.N. Plant Pathology, 5th ed.; Elsevier: Burlington, MA, USA, 2005; pp. 473–483. [Google Scholar]
- Bedker, P.J.; Blanchette, R.A. Identification and control of cankers caused by Nectria cinnabarina of honey locust. J. Arboric. 1984, 10, 33–39. [Google Scholar]
- Pan, M.; Zhu, H.; Bonthond, G.; Tian, C.; Fan, X. High Diversity of Cytospora Associated With Canker and Dieback of Rosaceae in China, with 10 New Species Described. Front. Plant Sci. 2020, 11, 690. [Google Scholar] [CrossRef] [PubMed]
- Bartnik, C.; Boroń, P.; Michalcewicz, J.; Ciach, M. The first record of Botryodiplodia canker in Poland. For. Pathol. 2019, 49, e12528. [Google Scholar] [CrossRef]
- Gomes, R.R.; Glienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Pers. Mol. Phylogeny Evol. Fungi 2013, 31, 1–41. [Google Scholar] [CrossRef]
- Yang, Q.; Du, Z.; Liang, Y.-M.; Tian, C.-M. Molecular phylogeny of Nectria species associated with dieback and canker diseases in China, with a new species described. Phytotaxa 2018, 356, 199–214. [Google Scholar] [CrossRef]
- Zhuang, W.Y. Nectriaceae et Bionectriaceae. In Flora Fungorum Sinicorum; Science Press: Beijing, China, 2013; Volume 47, pp. 110–125. [Google Scholar]
- Hirooka, Y.; Rossman, A.; Samuels, G.; Lechat, C.; Chaverri, P. A monograph of Allantonectria, Nectria, and Pleonectria (Nectriaceae, Hypocreales, Ascomycota) and their pycnidial, sporodochial, and synnematous anamorphs. Stud. Mycol. 2012, 71, 1–210. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.F. Nectria pseudotrichia, como agente causal de cancro de ramos, ocorrendo em pereira japonesa no Brasil. Fitopatol. Bras. 2003, 28, 107. [Google Scholar] [CrossRef][Green Version]
- Akinsanmi, O.A.; Drenth, A. First report ofTubercularia lateritiaas the causal agent of canker on macadamia. Australas. Plant Dis. Notes 2006, 1, 49–51. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Duan, K.; Gao, Q.H.; Song, L.L. First Report of Nectria pseudotrichia Causing Crown Rot of Strawberry in China. Plant Dis. 2018, 102, 1655. [Google Scholar] [CrossRef]
- Wanjiku, E.K.; Waceke, J.W.; Wanjala, B.W.; Mbaka, J.N. Identification and Pathogenicity of Fungal Pathogens Associated with Stem End Rots of Avocado Fruits in Kenya. Int. J. Microbiol. 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Jiang, G.Z.; Li, L.; Liu, Y.X.; Zhou, M. Identification of the pathogen causing stem brown spots canker of rubber tree and screening of fungicides. Trop. Agric. Sci. Technol. 2019, 42, 15–20. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).