Development of Genomic SSR for the Subtropical Hardwood Tree Dalbergia hupeana and Assessment of Their Transferability to Other Related Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. De Novo Genome Sequencing
2.3. Genome Survey and Assembly
2.4. Identification and Verification of gSSR Markers
2.5. Transferability to Other Dalbergia Species
2.6. Assessment of Genetic Diversity in Different Species
3. Results
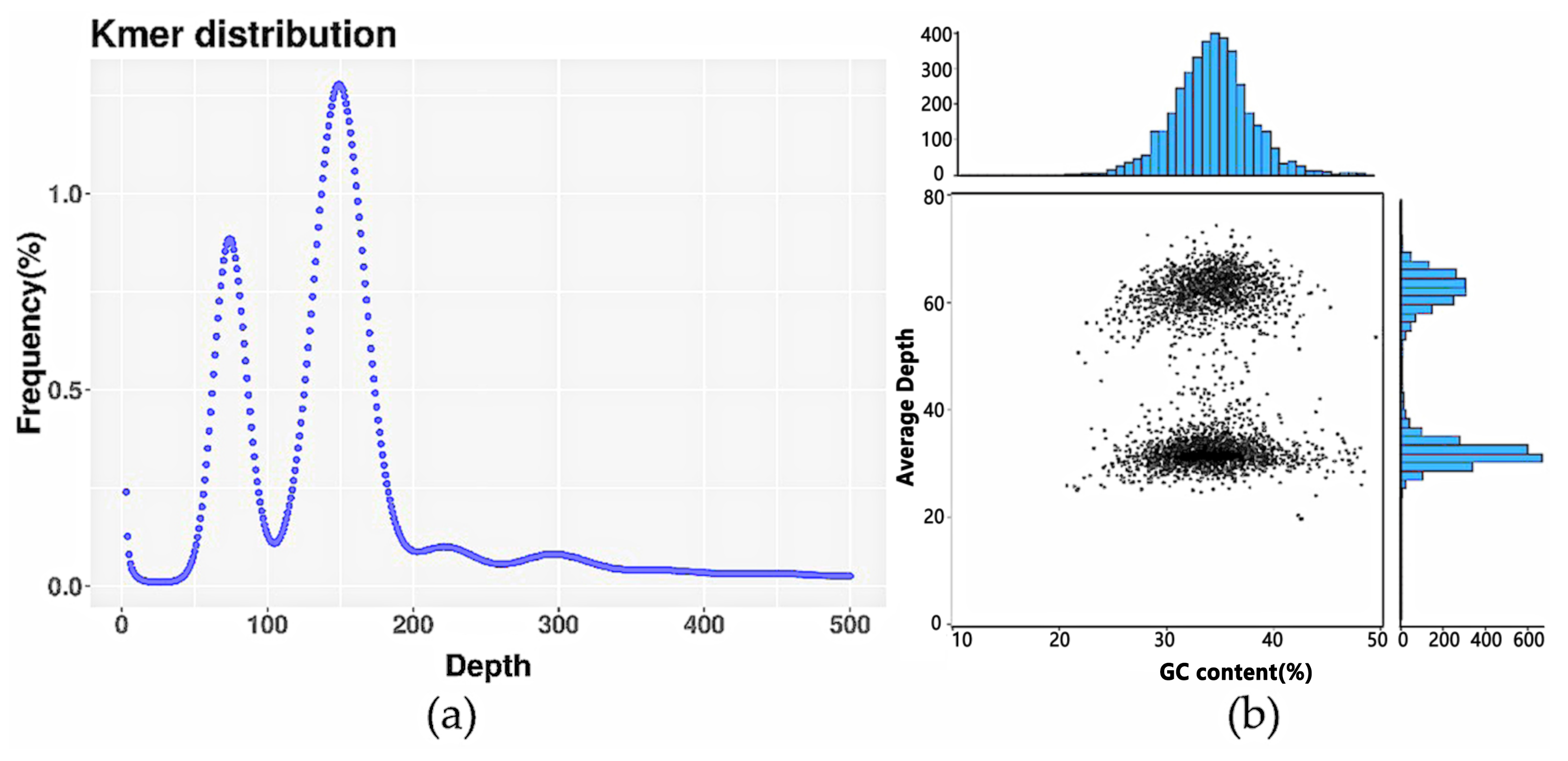
3.1. Genome Survey
3.2. Development and Verification of gSSR Markers
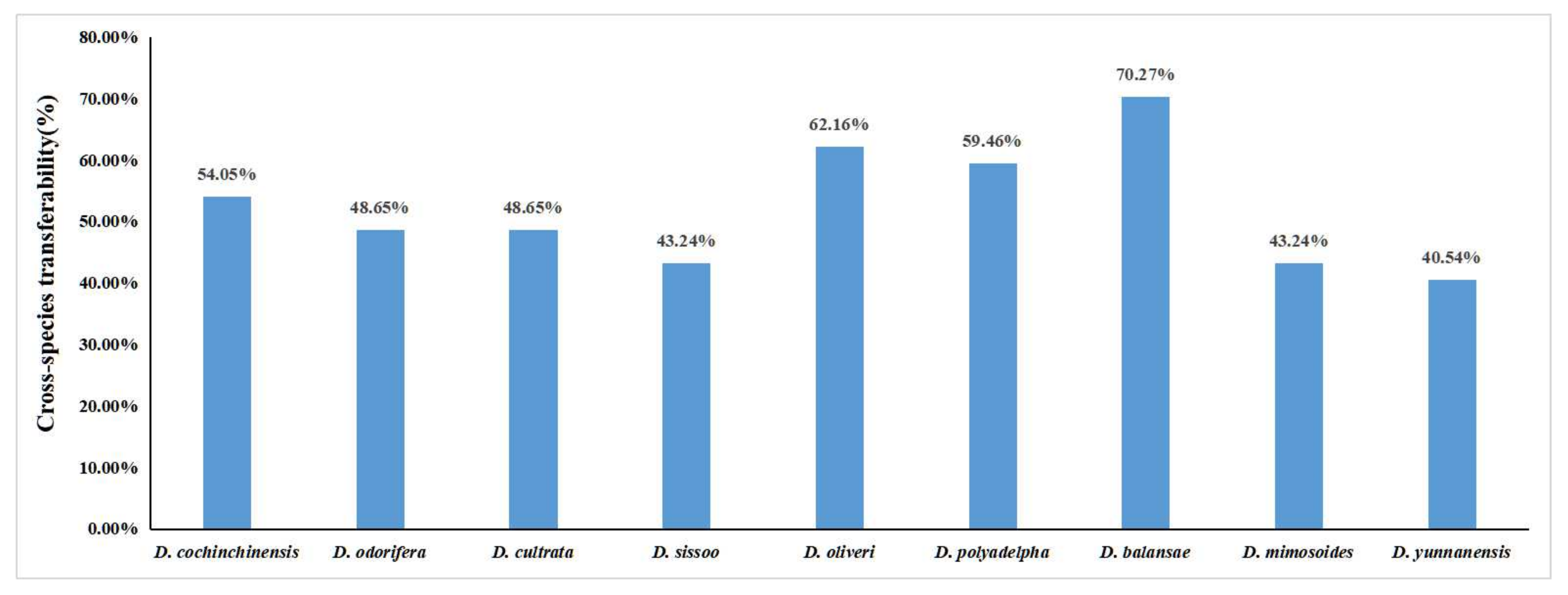
3.3. Transferability of Developed gSSR Markers to Other Dalbergia Species
3.4. Diversity of gSSR Markers in Dalbergia Populations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Klitgaard, B.; Lavin, M. Tribe Dalbergieae sens. Lat. In Legumes of the World; Royal Botanic Gardens: Kew, UK, 2005; pp. 307–335. [Google Scholar]
- Winfield, K.; Grayson, C.; Scott, M. Cites cop17 information document 48—Global status of Dalbergia and Pterocarpus rosewood producing species in trade. In Proceedings of the Convention on International Trade in Endangered Species 17th Conference of Parties–Johannesburg, Johannesburg, South Africa, 24 September–5 October 2016; pp. 1–246. [Google Scholar]
- CITES. Appendices I, II and III. 2017. Available online: https://cites.org/eng/app/appendices.php (accessed on 14 February 2021).
- IUCN. The IUCN Red List of Threatened Species. Version 2021-1. 2021. Available online: Https://www.Iucnredlist.Org (accessed on 1 January 2021).
- Chen, D.; Zhang, D.; Larsen, K. Tribe dalbergieae. Flora China 2010, 10, 121–130. [Google Scholar]
- Ribeiro, R.A.; Rezende, M.F.; Resende, L.C.; LEMOS-FILHO, J.P.; Kalapothakis, E.; Lovato, M.B. Development of polymorphic microsatellite markers for Dalbergia nigra (papilionoideae), an endangered tree from the Brazilian Atlantic forest. Mol. Ecol. Resour. 2009, 9, 203–206. [Google Scholar] [CrossRef]
- Resende, L.C.; Ribeiro, R.A.; Lovato, M.B. Diversity and genetic connectivity among populations of a threatened tree (Dalbergia nigra) in a recently fragmented landscape of the Brazilian Atlantic forest. Genetica 2011, 139, 1159–1168. [Google Scholar] [CrossRef]
- Leite, F.A.B.; Brandão, R.L.; de Oliveira Buzatti, R.S.; de Lemos-Filho, J.P.; Lovato, M.B. Fine-scale genetic structure of the threatened rosewood Dalbergia nigra from the Atlantic forest: Comparing saplings versus adults and small fragment versus continuous forest. Tree Genet. Genomes 2014, 10, 307–316. [Google Scholar] [CrossRef]
- Moritsuka, E.; Chhang, P.; Tagane, S.; Toyama, H.; Sokh, H.; Yahara, T.; Tachida, H. Genetic variation and population structure of a threatened timber tree Dalbergia cochinchinensis in Cambodia. Tree Genet. Genomes 2017, 13, 1–11. [Google Scholar] [CrossRef]
- Phong, D.T.; Hien, V.; Thanh, T.; Tang, D.V. Comparison of rapd and ISSR markers for assessment of genetic diversity among endangered rare Dalbergia oliveri (fabaceae) genotypes in Vietnam. Genet. Mol. Res. GMR 2011, 10, 2382–2393. [Google Scholar] [CrossRef]
- Liu, F.; Hong, Z.; Xu, D.; Jia, H.; Zhang, N.; Liu, X.; Yang, Z.; Lu, M. Genetic diversity of the endangered Dalbergia odorifera revealed by SSR markers. Forests 2019, 10, 225. [Google Scholar] [CrossRef]
- Choi, I.S.; Jin, D.P.; An, S.J.; Choi, B.H. A new distribution of Dalbergia hupeana hance (fabaceae) in Korea and its taxonomic characteristics. Korean J. Plant Taxon. 2015, 45, 22–28. [Google Scholar] [CrossRef]
- Li, S.J. Dalbergia in Asia; Science Press: Beijing, China, 2017; pp. 1–371. [Google Scholar]
- Kong, h. Value utilization and breeding technology of native sandalwood species of Dalbergia hupeana in eastern Anhui. Consum. Guide 2015, 10, 314. [Google Scholar]
- Tang, R.H.; Zhuang, W.J.; Gao, G.Q.; Liang-Qiong, H.E.; Han, Z.Q.; Shan, S.H.; Jiang, J.; Yang-Rui, L.I. Phylogenetic relationships in genus arachis based on SSR and AFLP markers. Agric. Sci. China 2008, 7, 405–414. [Google Scholar] [CrossRef]
- Li, N.; Yao, Y.Y.; Chen, Y.L.; Liang, X.M.; Yang, D.J. Evaluation of genetic diversity and population structure of genus tripterygium based on SSR markers. Acta Pharm. Sin. 2017, 52, 153–161. [Google Scholar]
- Yousaf, Z.; Hu, W.; Zhang, Y.; Zeng, S.; Wang, Y. Exploration of genetic diversity among medicinally important genus epimedium species based on genomic and EST-SSR marker. Nat. Prod. Res. Former. Nat. Prod. Lett. 2015, 29, 1020–1025. [Google Scholar]
- Wu, X.L.; He, C.Y.; Chen, S.Y.; Zhuang, B.C.; Wang, X.C. Phylogenetic analysis of interspecies in genus glycine through SSR markers. Acta Genet. Sin. 2001, 28, 359–366. [Google Scholar]
- Schroeder, H.; Fladung, M. Ssr and snp markers for the identification of clones, hybrids and species within the genus populus. Silvae Genet. 2010, 59, 257–263. [Google Scholar] [CrossRef]
- Sun, R.; Lin, F.; Huang, P.; Zheng, Y. Moderate genetic diversity and genetic differentiation in the relict tree Liquidambar formosana hance revealed by genic simple sequence repeat markers. Front. Plant Sci. 2016, 7, 1411. [Google Scholar] [CrossRef] [PubMed]
- Maguire, T.L.; Saenger, R.P. Comparative analysis of genetic diversity in the mangrove species Avicennia marina (Forsk.) vierh. (Avicenniaceae) detected by AFLPs and SSRs. TAG Theor. Appl. Genet. 2002, 104, 388–398. [Google Scholar] [CrossRef]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef]
- Motalebipour, E.Z.; Kafkas, S.; Khodaeiaminjan, M.; Coban, N.; Gözel, H. Genome survey of pistachio (Pistacia vera L.) by next generation sequencing: Development of novel SSR markers and genetic diversity in pistacia species. BMC Genom. 2016, 17, 3359-X. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Fan, J.; Chang, P.; Zhu, L.; Zhao, M.; Li, L. Genome survey sequencing of acer truncatum bunge to identify genomic information, simple sequence repeat (SSR) markers and complete chloroplast genome. Forests 2019, 10, 87. [Google Scholar] [CrossRef]
- Salgado, L.R.; Koop, D.M.; Pinheiro, D.G.; Rivallan, R.; Le Guen, V.; Nicolás, M.F.; de Almeida, L.G.P.; Rocha, V.R.; Magalhães, M.; Garcia, D.; et al. De novo transcriptome analysis of Hevea brasiliensis tissues by rna-seq and screening for molecular markers. BMC Genom. 2014, 15, 236. [Google Scholar] [CrossRef]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. Genomescope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. Soap: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2010, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6.5: Genetic analysis in excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecology 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Hughes, L. Biological consequences of global warming: Is the signal already apparent? Trends Ecol. Evol. 2000, 15, 56–61. [Google Scholar] [CrossRef]
- Smith, J.B.; Schneider, S.H.; Oppenheimer, M.; Yohe, G.W.; Hare, W.; Mastrandrea, M.D.; Patwardhan, A.; Burton, I.; Corfee-Morlot, J.; Magadza, C.H.D.; et al. Assessing dangerous climate change through an update of the intergovernmental panel on climate change (IPCC) “reasons for concern”. Proc. Natl. Acad. Sci. USA 2009, 106, 4133–4137. [Google Scholar] [CrossRef]
- Vatanparast, M.; Klitgård, B.B.; Adema, F.A.C.B.; Pennington, R.T.; Yahara, T.; Kajita, T. First molecular phylogeny of the pantropical genus Dalbergia: Implications for infrageneric circumscription and biogeography. S. Afr. J. Bot. 2013, 89, 143–149. [Google Scholar] [CrossRef]
- Hassold, S.; Lowry, P.P., II; Bauert, M.R.; Razafintsalama, A.; Ramamonjisoa, L.; Widmer, A. DNA barcoding of malagasy rosewoods: Towards a molecular identification of cites-listed Dalbergia species. PLoS ONE 2016, 11, e0157881. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hunkapiller, M.W. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Nurk, S.; Walenz, B.P.; Rhie, A.; Vollger, M.R.; Koren, S. Hicanu: Accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 2020, 30, 1291–1305. [Google Scholar] [CrossRef]
- Song, X.; Li, N.; Guo, Y.; Bai, Y.; Wu, T.; Yu, T.; Feng, S.; Zhang, Y.; Wang, Z.; Liu, Z.; et al. Comprehensive identification and characterization of simple sequence repeats based on the whole-genome sequences of 14 forest and fruit trees. For. Res. 2021, 1, 7. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar] [PubMed]
- Bazzo, B.R.; Carvalho, L.; Carazzolle, M.F.; Pereira, G.; Colombo, C.A. Development of novel EST-SSR markers in the macaúba palm (Acrocomia aculeata) using transcriptome sequencing and cross-species transferability in arecaceae species. BMC Plant Biol. 2018, 18, 276. [Google Scholar] [CrossRef]
- Wang, C.; Yan, H.; Ji, L.; Zhou, S.; Liu, T.; Zhang, X.; Huang, L. Genome survey sequencing of purple elephant grass (Pennisetum purpureum schum ‘zise’) and identification of its SSR markers. Mol. Breed. 2018, 38, 94. [Google Scholar] [CrossRef]
- Buzatti, R.S.D.O.; Chicata, F.S.L.; Lovato, M.B. Transferability of microsatellite markers across six Dalbergia (fabaceae) species and their characterization for Dalbergia miscolobium. Biochem. Syst. Ecol. 2016, 69, 161–165. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef]
- Cui, F. Molecular Phylogenetics of Dalbergia L. F. (Leguminosae); University of Chinese Academy of Science: Beijing, China, 2014. [Google Scholar]
- Lavin, M.; Pennington, R.T.; Klitgaard, B.B.; Sprent, J.I.; Lima, H.D.; Gasson, P.E. The Dalbergioid legumes (fabaceae): Delimitation of a pantropical monophyletic clade. Am. J. Bot. 2001, 88, 503. [Google Scholar] [CrossRef]
- Ribeiro, R.A.; Lavin, M.; Lemos-Filho, J.P.; Filho, C.V.M.A.; Santos, F.b.C.R.D.; Lovato, M.B. The genus Machaerium (leguminosae) is more closely related to Aeschynomene sect. Ochopodium than to Dalbergia: Inferences from combined sequence data. Syst. Bot. 2007, 32, 762–771. [Google Scholar] [CrossRef]
- Hartvig, I.; So, T.; Changtragoon, S.; Tran, H.T.; Bouamanivong, S.; Theilade, I.; Kjaer, E.D.; Nielsen, L.R. Population genetic structure of the endemic rosewoods Dalbergia cochinchinensis and D. oliveri at a regional scale reflects the indochinese landscape and life-history traits. Ecol. Evol. 2018, 8, 530–545. [Google Scholar] [CrossRef] [PubMed]
| Name | Biotype | Location | No. Individuals |
|---|---|---|---|
| D. balansae | tree | Yingjiang, Yunnan, China | 29 |
| D. cultrata | tree | Puer, Yunnan, China | 2 |
| D. hupeana | tree | Jiangyou, Sichuan, China (JY) | 30 |
| D. hupeana | tree | Xixiang, Shanxi, China (XX) | 30 |
| D. hupeana | tree | Dujiangyan, Sichuan, China (DJY) | 30 |
| D. mimosoides | vine | Tongren, Guizhou, China | 2 |
| D. odorifera | tree | Jiulongling, Fujian, China | 2 |
| D. polyadelpha | tree | Longling, Yunnan, China | 29 |
| D. sissoo | tree | Yunnan, China | 2 |
| D. yunnanensis | shrub | Panzhihua, Sichuan, China | 2 |
| D. cochinchinensis | tree | Jiulongling, Fujian, China | 2 |
| D. oliveri | tree | Jianfengling, Hainan, China | 2 |
| Name | Repeat Motif | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) | Tm(°C) | Size Range (bp) | Fluorescence Dye |
|---|---|---|---|---|---|---|
| Dhup14 | (TCT)5 | TCACAAAGGTATGCATTTCCC | TATACGCTTGGCATTTGCAG | 59 | 207~219 | FAM |
| Dhup61 | (GGA)11 | GGTCGAAGTGGGAATCGAAG | ATTCTCCCTCCCTTGCTCAT | 60 | 102~120 | FAM |
| Dhup64 | (TTA)9 | AGTTAGGTGCACCACAGCCT | TCTATGCCGATGTTTCCCTC | 59 | 158~188 | FAM |
| Dhup70 | (TTA)14 | AAGCAACGGAACAATATGAAAAA | TTATTATCACCCACCGCACA | 59 | 203~248 | FAM |
| Dhup78 | (ATT)10 | AGTATCCATCCCTGGGTTCC | GTGGGTGGCGTACTGTTTTT | 59 | 257~284 | FAM |
| Dhup89 | (TA)20 | TACCGTTGGATGTGATGGTG | ATGCTTCCTGCACCCTAGAA | 59 | 143~177 | FAM |
| Dhup90 | (AAT)12 | TCCGATATGCAAACATGAGC | AATCACGTGGCTTTGATTCC | 59 | 222~255 | FAM |
| Dhup104 | (AG)14 | TGGACCACCCATGATAGTGA | GGAACAATAGAAACAAAACACGG | 59 | 108~154 | FAM |
| Dhup106 | (AT)14 | CATTTGGCATATAACATGTCGG | GCAACAGTGTAACTGATGTTGCT | 59 | 115~165 | FAM |
| Dhup108 | (TC)14 | CCCCCTTAAAACTTGCTTCA | CTGATGAGTGGGAAAACGGT | 59 | 117~153 | FAM |
| Dhup114 | (GGC)8 | TATTTTTGCGATCGTTGCTG | GCTCGCGGTCATGTAGACTT | 59 | 116~143 | FAM |
| Dhup115 | (AAG)10 | TGAAAATGGAGAATGGGGAG | GATACACACGGGAAAATCGG | 59 | 115~142 | FAM |
| Dhup116 | (AAT)8 | ATTTCATTCTCTCGCGCTGT | CAAATTTGTGAGCACATGATGA | 59 | 127~157 | FAM |
| Dhup118 | (AAG)7 | TTGATCCAAGGGCGAGTAAC | GTGGGCTCTTGTTGTTGTGA | 59 | 128~149 | FAM |
| Dhup120 | (AAT)8 | TGAATTTTCTCGGCAAGAGC | TTCCTTCTTTCTCACGGACC | 60 | 124~154 | FAM |
| Dhup122 | (AAT)8 | GAATGAGCTGATGGTGAAGGA | ACATTAATACCTTATTCCTCAATTCTG | 58 | 133~154 | FAM |
| Dhup123 | (AAG)7 | TGAGTGAGAGAAAGATAGATCAACG | CCAACGAAACCTGTTTCACA | 59 | 135~168 | FAM |
| Dhup125 | (AAG)7 | TGTCTCGTTTCTCTCTCTCATCC | TTTGTGCGCTACCAACAGAG | 60 | 133~157 | FAM |
| Dhup129 | (TA)14 | TGTGGTCGATGAAGACTTGG | GCCAGCTATGTGTGAAGTGC | 59 | 174~188 | HEX |
| Dhup130 | (GA)13 | AAGCACGAATTAAATTTCTTGGA | AGAGAGTGTTGGTGGTTGGG | 59 | 228~276 | HEX |
| Dhup132 | (AT)14 | TTGGGTGCATTGTGATTTGT | ACCATCTGGGCAGTGATAGC | 59 | 195~233 | HEX |
| Dhup133 | (AT)14 | CAATACTAATGCAACAAAATTCAAA | CCCGCAATGAGAATAGGAAA | 58 | 199~243 | HEX |
| Dhup139 | (AAT)8 | CCCCGGCACTCTCTATATCA | ACTGCGTATCCTTTGCTGCT | 60 | 181~199 | HEX |
| Dhup141 | (TTG)8 | CCACCGTGAAAGTAGACCAAG | CCTTCCCTTGAGGACCCTAC | 59 | 187~205 | HEX |
| Dhup142 | (CAA)10 | TTTGAGGCCAACCTTCAAAT | ACGAGTGGAGTGAAGTGCAA | 59 | 200~221 | HEX |
| Dhup144 | (AAT)8 | AACAAATTCAAACCAAACCGA | TCCAAAGTAGGGCTTGAAAAA | 59 | 184~217 | HEX |
| Dhup153 | (AG)14 | ACTTGTGTCACGTTCGTCCA | CTGCTTCCTCTGCTTCTGCT | 60 | 251~293 | ROX |
| Dhup159 | (TA)14 | TTTCAGATGCTAGAGTGTGTGTG | CGTGTGCCTAAATTCTTAACCG | 58 | 206~250 | ROX |
| Dhup163 | (TTG)8 | ACGAAACCCTAGCCCTTGTT | CAACGAACCTCTTGAAAGGC | 59 | 228~255 | ROX |
| Dhup164 | (AAG)7 | AATTGCACAACAATCCAGCA | CGGATTATCTCCGAAAACGA | 60 | 233~254 | ROX |
| Dhup167 | (AAT)8 | AAAGGCTGAGAGGAAAAGGC | TCTAAATTAAAGGATTTTTCAGTTGAT | 57 | 232~250 | ROX |
| Dhup171 | (GAG)8 | GTTGCCGGTACCTTCATCAT | GTCACGATCAAAGTCGGGTT | 59 | 228~249 | ROX |
| Dhup181 | (AT)14 | CCACCAGCAAATAGCTCACA | GGATTCCGAACTTGGAAGAA | 59 | 271~293 | TAMRA |
| Dhup183 | (AT)14 | GCAGACATGGTGGTTTCAGA | CGATATTATCACATTTTAAAGACTTGG | 58 | 272~324 | TAMRA |
| Dhup185 | (AT)14 | GTGCCAAGTACAACTGCGAA | TCACACTTAGCCACCCTTATGA | 59 | 258~308 | TAMRA |
| Dhup191 | (AAT)8 | GTGATGGTCACGTCATTTGC | TTTCTCACCGCCGTTAACTT | 59 | 279~303 | TAMRA |
| Dhup194 | (AAT)8 | AACGATTGATCTCTTGGTCATGT | TGTCGGTGTCTATCAATTCCC | 59 | 247~292 | TAMRA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Zheng, Y.; Liu, Y.; Lin, F.; Huang, P. Development of Genomic SSR for the Subtropical Hardwood Tree Dalbergia hupeana and Assessment of Their Transferability to Other Related Species. Forests 2021, 12, 804. https://doi.org/10.3390/f12060804
Li C, Zheng Y, Liu Y, Lin F, Huang P. Development of Genomic SSR for the Subtropical Hardwood Tree Dalbergia hupeana and Assessment of Their Transferability to Other Related Species. Forests. 2021; 12(6):804. https://doi.org/10.3390/f12060804
Chicago/Turabian StyleLi, Changhong, Yongqi Zheng, Yu Liu, Furong Lin, and Ping Huang. 2021. "Development of Genomic SSR for the Subtropical Hardwood Tree Dalbergia hupeana and Assessment of Their Transferability to Other Related Species" Forests 12, no. 6: 804. https://doi.org/10.3390/f12060804
APA StyleLi, C., Zheng, Y., Liu, Y., Lin, F., & Huang, P. (2021). Development of Genomic SSR for the Subtropical Hardwood Tree Dalbergia hupeana and Assessment of Their Transferability to Other Related Species. Forests, 12(6), 804. https://doi.org/10.3390/f12060804






