Genotype–Environment Interactions for Tree Growth and Leaf Phytochemical Content of Cyclocarya paliurus (Batal.) Iljinskaja
Abstract
1. Introduction
2. Materials and Methods
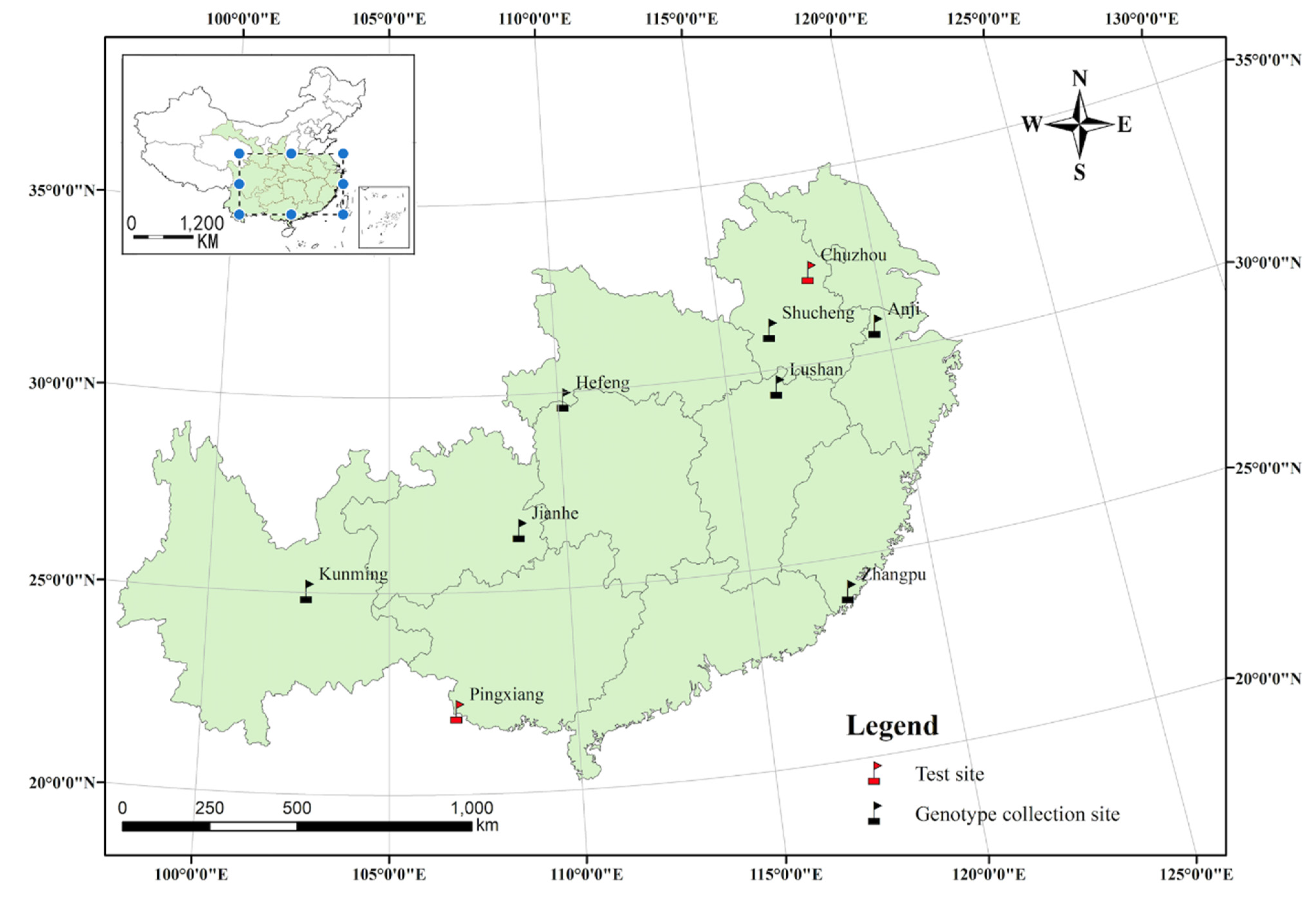
2.1. Plant Materials and Experimental Design
2.2. Determination of Phytochemicals
2.3. Data Analysis
3. Results and Discussion
3.1. Variations in Growth and Leaf Phytochemical Contents
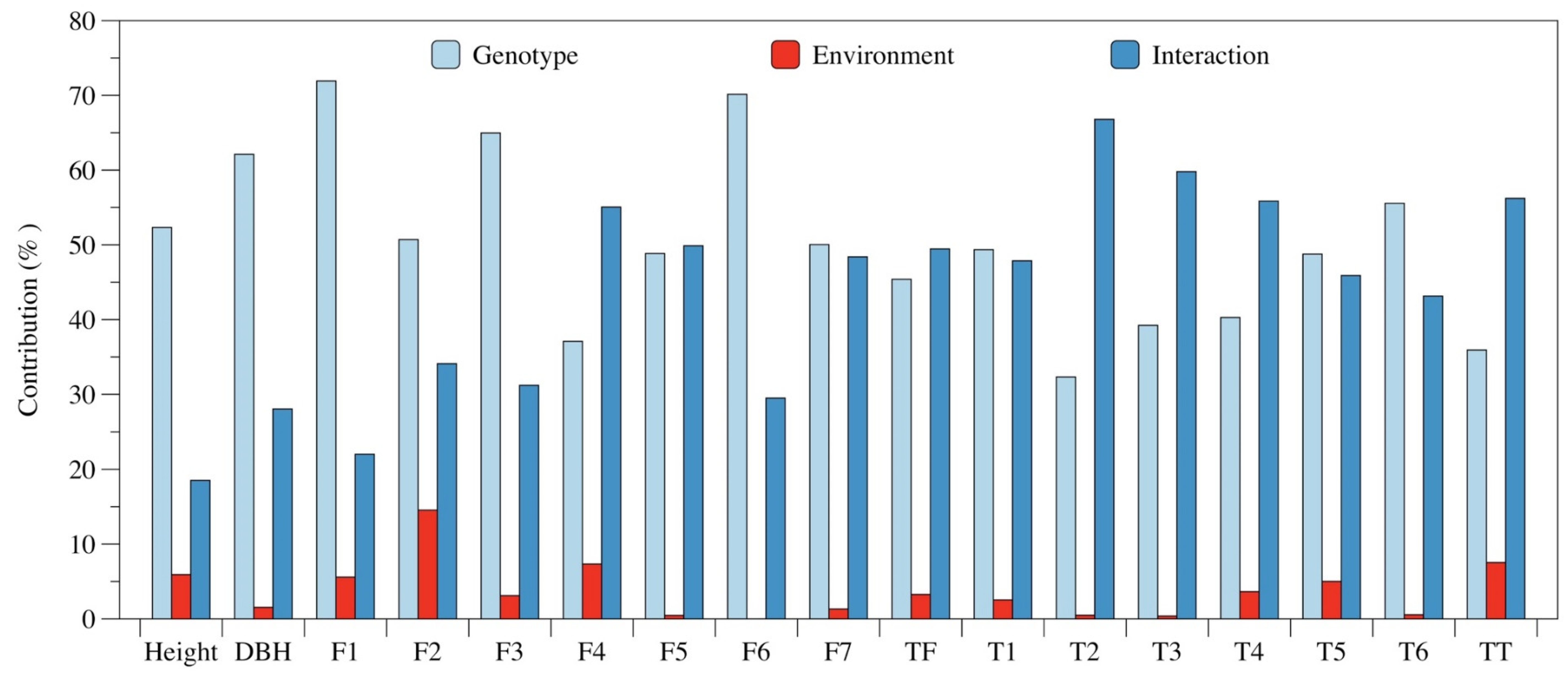
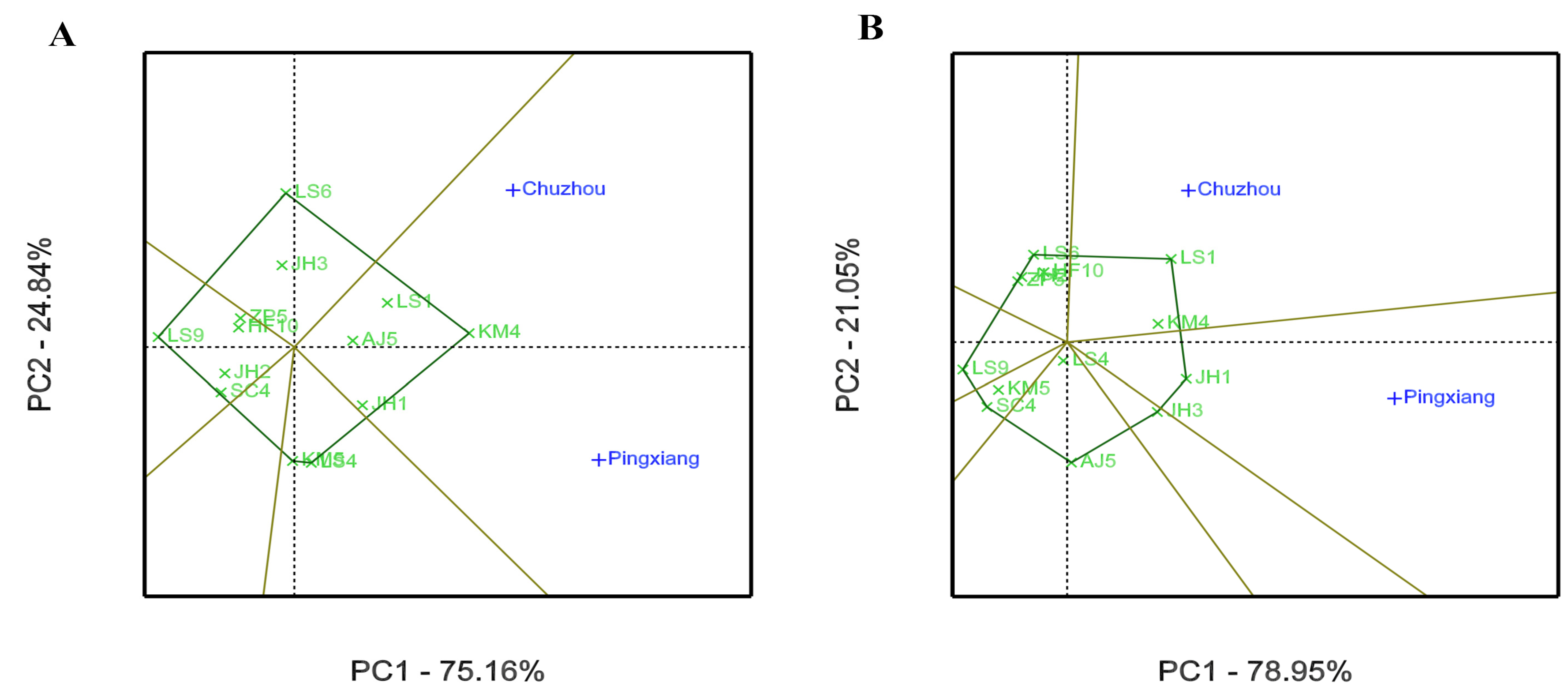
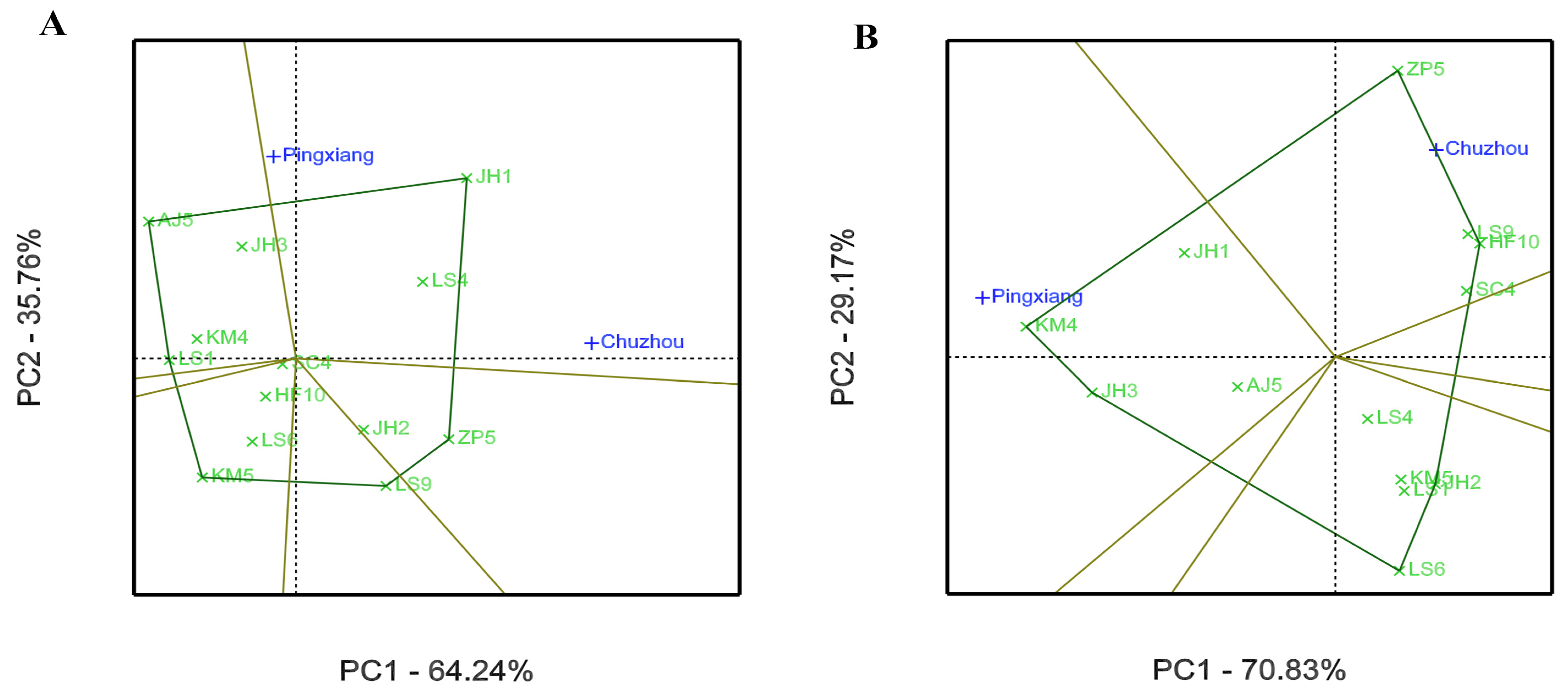
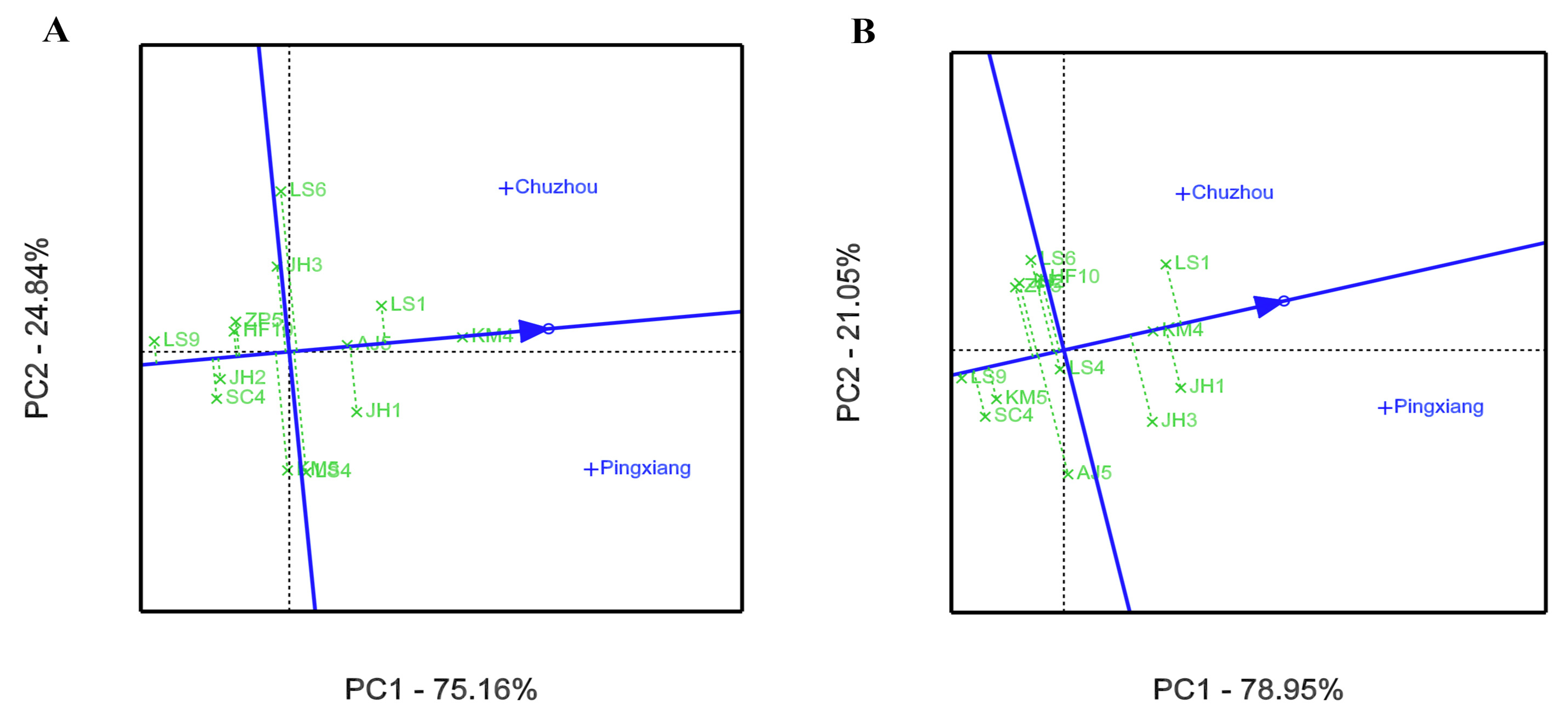
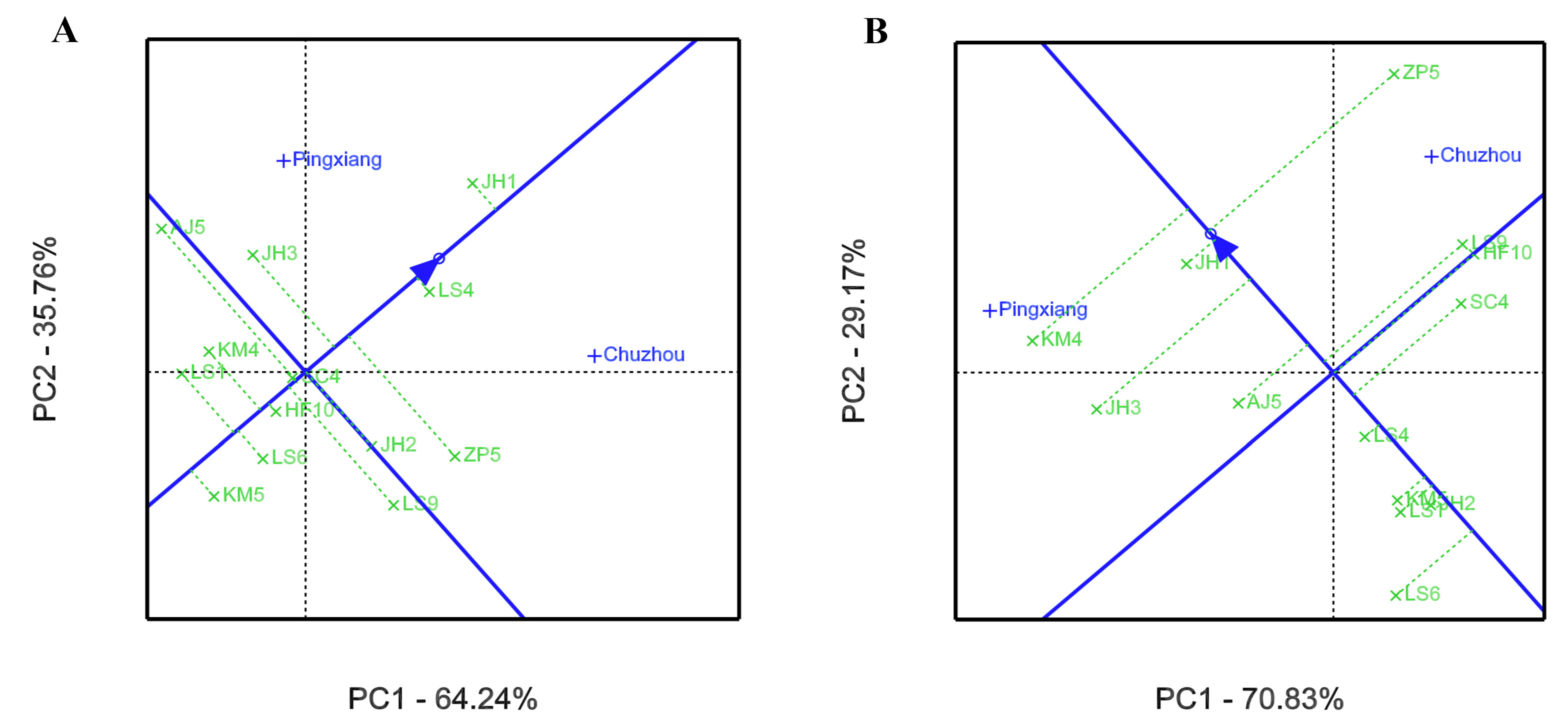
3.2. GEI Analysis for Growth and Leaf Phytochemical Content
3.3. Correlation between Tree Growth and Leaf Phytochemical Content
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Fang, S.Z.; Wang, J.Y.; Wei, Z.Y.; Zhu, Z.X. Methods to break seed dormancy in Cyclocarya paliurus (Batal.) Iljinskaja. Sci. Horti-Amst. 2006, 110, 305–309. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, Y.N.; Fang, S.Z.; Wang, T.L.; Yin, Z.Q.; Shang, X.L.; Yang, W.X.; Fu, X.X. Antidiabetic effects of Cyclocarya paliurus leaves depends on the contents of antihyperglycemic flavonoids and antihyperlipidemic triterpenoids. Molecules 2018, 23, 1042. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.F.; Gao, T.H.; Zhong, R.L.; Lin, Z.; Jiang, C.H.; Ouyang, S.; Zhao, M.; Che, C.T.; Zhang, J.; Yin, Z.Q. Antihyperlipidaemic effect of triterpenic acid-enriched fraction from Cyclocarya paliurus leaves in hyperlipidaemic rats. Pharm. Biol. 2017, 55, 712–721. [Google Scholar] [CrossRef]
- Xiong, L.; Hu, W.B.; Yang, Z.W.; Chen, H.; Ning, W.; Xin, L.; Wang, W.J. Enzymolysis-ultrasonic assisted extraction of flavonoid from Cyclocarya paliurus (Batal) Iljinskaja: HPLC profile, antimicrobial and antioxidant activity. Ind. Crop. Prod. 2019, 130, 615–626. [Google Scholar] [CrossRef]
- Xie, J.H.; Shen, M.Y.; Xie, M.Y.; Nie, S.P.; Chen, Y.; Li, C.; Huang, D.F.; Wang, Y.X. Ultrasonic-assisted extraction, antimicrobial and antioxidant activities of Cyclocarya paliurus (Batal.) Iljinskaja polysaccharides. Carbohyd. Polym. 2012, 89, 177–184. [Google Scholar] [CrossRef]
- Thakur, M.; Bhattacharya, S.; Khosla, P.K.; Puri, S. Improving production of plant secondary metabolites through biotic and abiotic elicitation. J. Appl. Res. Med. Aroma. 2019, 12, 1–12. [Google Scholar] [CrossRef]
- Li, Y.Q.; Kong, D.X.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Bioch. 2020, 148, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.W.; Shang, X.L.; Ding, H.F.; Cao, Y.N.; Fang, S.Z. Natural variations in flavonoids and triterpenoids of Cyclocarya paliurus leaves. J. Forestry Res. 2020. [Google Scholar] [CrossRef]
- Deng, B.; Shang, X.L.; Fang, S.Z.; Li, Q.Q.; Fu, X.X.; Su, J. Integrated effects of light intensity and fertilization on growth and flavonoid accumulation in Cyclocarya paliurus. J. Agric. Food Chem. 2012, 60, 6286–6292. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, S.Z.; Yang, W.X.; Shang, X.L.; Fu, X.X. Light quality affects flavonoid production and related gene expression in Cyclocarya paliurus. J. Photoch. Photobio. B 2018, 179, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Yue, X.L.; Shang, X.L.; Fang, S.Z. Nitrogen forms alter triterpenoid accumulation and related gene expression in Cyclocarya paliurus (Batalin) Iljinsk. Seedl. For. 2020, 11, 631. [Google Scholar] [CrossRef]
- Balakrishnan, D.; Subrahmanyam, D.; Badri, J.; Raju, A.K.; Rao, Y.V.; Beerelli, K.; Mesapogu, S.; Surapaneni, M.; Ponnuswamy, R.; Padmavathi, G.; et al. Genotype × Environment interactions of yield traits in backcross introgression lines derived from Oryza sativa cv. Swarna/Oryza nivara. Front. Plant. Sci. 2016, 7, 1530. [Google Scholar] [CrossRef]
- Chipeta, M.M.; Melis, R.; Shanahan, P.; Sibiya, J.; Benesi, I.R.M. Genotype × environment interaction and stability analysis of Cassava genotypes at different harvest times. J. Anim. Plant Sci. 2017, 27, 901–919. [Google Scholar]
- Wang, R.H.; Hu, D.H.; Zheng, H.Q.; Yan, S.; Wei, R.P. Genotype × environmental interaction by AMMI and GGE biplot analysis for the provenances of Michelia chapensis in South China. J. For. Res. 2015, 27, 659–664. [Google Scholar] [CrossRef]
- Rao, G.R.; Sarkar, B.; Raju, B.M.K.; Sathi-Reddy, P.; Subba-Rao, A.V.M.; Rebecca, J. Genotype × year interaction of pod and seed mass and stability of Pongamia pinnata families in a semi-arid region. J. For. Res. 2019, 31, 1333–1346. [Google Scholar] [CrossRef]
- Deng, B.; Cao, Y.N.; Fang, S.Z.; Shang, X.L.; Yang, W.X. Variation and stability of growth and leaf flavonoid content in Cyclocarya paliurus across environments. Ind. Crop. Prod. 2015, 76, 386–393. [Google Scholar] [CrossRef]
- Deng, B.; Fang, S.Z.; Shang, X.L.; Fu, X.X.; Yang, W.X. Influence of genotypes and environmental factors on leaf triterpenoid content and growth of Cyclocarya paliurus. J. For. Res. 2019, 30, 789–798. [Google Scholar] [CrossRef]
- Eberhart, S.A.; Russell, W.A. Stability parameters for comparing varieties. Crop. Sci. 1966, 6, 36–40. [Google Scholar] [CrossRef]
- Aarthi, S.; Suresh, J.; Leela, N.K.; Prasath, D. Multi environment testing reveals genotype-environment interaction for curcuminoids in turmeric (Curcuma longa L.). Ind. Crop. Prod. 2020, 145, 112090. [Google Scholar] [CrossRef]
- Li, F.M.; Tan, J.; Nie, S.P.; Dong, C.J.; Li, C. The study on determination methods of total flavonoids in Cyclocarya paliurus. Food Sci. Tec-Brazil. 2006, 4, 34–37. [Google Scholar] [CrossRef]
- Fan, J.P.; He, C.H. Simultaneous quantification of three major bioactive triterpene acids in the leaves of Diospyros kaki by high-performance liquid chromatography method. J. Pharmaceut. Biomed. 2006, 41, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.N.; Fang, S.Z.; Yin, Z.Q.; Fu, X.X.; Shang, X.L.; Yang, W.X.; Yang, H.M. Chemical fingerprint and multicomponent quantitative analysis for the quality evaluation of Cyclocarya paliurus leaves by HPLC-Q-TOF-MS. Molecules 2017, 22, 1927. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar] [CrossRef]
- Klápště, J.; Meason, D.; Dungey, H.S.; Telfer, E.J.; Silcock, P.; Rapley, S. Genotype-by-environment interaction in coast redwood outside natural distribution-search for environmental cues. BMC Genet. 2020, 21, 15. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.J.; Xiao, Y.; Hu, J.W.; Wang, F.; Ouyang, F.Q.; Wang, J.H.; Weng, Y.H.; Zhang, H.G. Genotype by environment interaction analysis of growth of Picea koraiensis families at different sites using BLUP-GGE. New For. 2020. [Google Scholar] [CrossRef]
- Xie, P.J.; Huang, L.X.; Zhang, C.H.; Zhang, Y.L. Phenolic compositions, and antioxidant performance of olive leaf and fruit (Olea europaea L.) extracts and their structure-activity relationships. J. Funct. Foods 2015, 16, 460–471. [Google Scholar] [CrossRef]
- Sati, P.; Pandey, A.; Rawat, S.; Rani, A. Phytochemicals and antioxidants in leaf extracts of Ginkgo biloba with reference to location, seasonal variation and solvent system. J. Pharm. Res. 2013, 7, 804–809. [Google Scholar] [CrossRef]
- Yan, W. GGE Biplot-A Windows application for graphical analysis of multi-environment trial data and other types of two-way data. Agron. J. 2001, 93, 1111–1118. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Magaji, U.; Miah, G.; Hussin, G.; Ramli, A. Genotype × Environment interaction and stability analyses of yield and yield components of established and mutant rice genotypes tested in multiple locations in Malaysia. Acta Agric. Scand. Sect. B Siol Plant Sci. 2017, 67, 590–606. [Google Scholar] [CrossRef]
- Dia, M.; Wehner, T.C.; Hassell, R.; Price, D.S.; Boyhan, G.E.; Olson, S.; King, S.; Davis, A.R.; Tolla, G.E. Genotype environment interaction and stability analysis for watermelon fruit yield in the United States. Crop. Sci. 2016, 56, 1645–1661. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE Biplot vs. AMMI analysis of genotype-by-environment data. Crop. Sci. 2007, 47, 643–655. [Google Scholar] [CrossRef]
- Okoth, S.; Rose, L.J.; Ouko, A.; Netshifhefhe, N.E.I.; Sila, H.; Viljoen, A. Assessing genotype-by-environment interactions in Aspergillus ear rot and pre-harvest aflatoxin accumulation in maize inbred lines. Agronomy 2017, 7, 86. [Google Scholar] [CrossRef]
- Mohammadi, R.; Amri, A. Genotype × environment interaction and genetic improvement for yield and yield stability of rainfed durum wheat in Iran. Euphytica 2013, 192, 227–249. [Google Scholar] [CrossRef]
- Navas-Lopez, J.F.; León, L.; Rapoport, H.F.; Moreno-Alías, I.; Lorite, I.J.; Rosa, R.D.I. Genotype, environment and their interaction effects on olive tree flowering phenology and flower quality. Euphytica 2019, 215, 84. [Google Scholar] [CrossRef]
- Kumar, N.; Ajay, B.C.; Dagla, M.C.; Rathnakumar, A.L.; Radhakrishnan, T.; Lal, C.; Samdur, M.Y.; Mathur, R.K.; Manivel, P. Multi-environment evaluation of Spanish bunch groundnut genotypes for fresh seed dormancy. Indian J. Genet. 2019, 79, 572–582. [Google Scholar] [CrossRef]
- Farshadfar, E.; Mahmodi, N.; Yaghotipoor, A. AMMI stability value and simultaneous estimation of yield and yield stability in bread wheat (Triticum aestivum L.). Aust. J. Crop. Sci. 2011, 5, 1837–1844. [Google Scholar] [CrossRef]
- Cai, Z.Q.; Wang, W.H.; Yang, J.; Cai, C.T. Growth, photosynthesis and root reserpine concentrations of two Rauvolfia species in response to a light gradient. Ind. Crop. Prod. 2009, 30, 220–226. [Google Scholar] [CrossRef]
- Cao, Y.N.; Deng, B.; Fang, S.Z.; Shang, X.L.; Fu, X.X.; Yang, W.X. Genotypic variation in tree growth and selected flavonoids in leaves of Cyclocarya paliurus. South For. 2018, 80, 67–74. [Google Scholar] [CrossRef]
- Stowe, K.A.; Marquis, R.J.; Hochwebder, C.G.; Simms, E.L. The evolutionary ecology of tolerance to consumer damage. Ann. Rev. Ecol. Syst. 2000, 31, 565–595. [Google Scholar] [CrossRef]
- Weis, A.; Simms, E.; Hochberg, M.E. Will plants and tolerance be genetically correlated? Effects of intrinsic growth rate and self-limitation on regrowth. Evol. Ecol. 2000, 14, 331–352. [Google Scholar] [CrossRef]
- Coley, P.D. Herbivory and defensive characteristics of tree species in a lowland tropical forest. Ecol. Monogr. 1983, 53, 209–233. [Google Scholar] [CrossRef]
- Strauss, S.Y.; Agrawal, A.A. The ecology and evolution of plant tolerance to herbivory. Trends Ecol. Evol. 1999, 14, 179–185. [Google Scholar] [CrossRef]
| Family | Tree Height (m) | Diameter at Breast Height (cm) |
|---|---|---|
| AJ5 | 7.28 ± 0.55 adc | 6.83 ± 1.95 bc |
| HF10 | 5.97 ± 1.08 cde | 7.50 ± 2.24 b |
| JH1 | 7.32 ± 0.51 abc | 10.83 ± 2.21 a |
| JH2 | 5.75 ± 0.73 de | 6.82 ± 2.34 bc |
| JH3 | 6.55 ± 1.28 bcd | 9.73± 2.45 a |
| KM4 | 8.65 ± 0.54 a | 10.43 ± 1.32 a |
| KM5 | 6.43 ± 0.88 b–e | 5.27 ± 0.69 cde |
| LS1 | 7.73 ± 0.80 ab | 11.32 ± 1.30 a |
| LS4 | 6.65 ± 0.94 bcd | 7.38 ± 0.66 b |
| LS6 | 6.68 ± 1.87 bcd | 7.33 ± 2.57 b |
| LS9 | 5.02 ± 1.05 e | 4.38 ± 1.58 e |
| SC4 | 5.68 ± 0.50 de | 4.80 ± 0.75 de |
| ZP5 | 6.00 ± 1.02 cde | 6.67 ± 2.58 bcd |
| Family | Flavonoid Content (mg/g) | |||||||
|---|---|---|---|---|---|---|---|---|
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | TF | |
| AJ5 | 0.17 ± 0.09 e | 0.05 ± 0.02 g | 0.09 ± 0.06 cd | 0.35 ± 0.27 e | 0.09 ± 0.02 c | 0.06 ± 0.02 e | 0.35 ± 0.38 f | 6.14 ± 4.77 def |
| HF10 | 0.48 ± 0.42 d | 0.18 ± 0.17 f | 0.06 ± 0.06 f | 0.49 ± 0.37 d | 0.08 ± 0.01 d | 0.05 ± 0.03 f | 0.34 ± 0.32 f | 5.48 ± 0.64 efg |
| JH1 | 2.20 ± 0.09 a | 0.49 ± 0.04 a | 0.23 ± 0.00 a | 0.74 ± 0.03 b | 0.13 ± 0.01 a | 0.23 ± 0.07 a | 1.21 ± 0.41 a | 11.03 ± 0.58 a |
| JH2 | 0.63 ± 0.40 c | 0.25 ± 0.04 e | 0.10 ± 0.02 c | 0.49 ± 0.12 d | 0.09 ± 0.01 bc | 0.07 ± 0.03d | 0.58 ± 0.40 d | 6.39 ± 1.61 de |
| JH3 | 0.93 ± 0.42 b | 0.31± 0.06 d | 0.13 ± 0.06 b | 0.80 ± 0.41 b | 0.10 ± 0.02 b | 0.16 ± 0.08 b | 1.02 ± 0.62 b | 7.09 ± 2.87 cd |
| KM4 | 0.66 ± 0.33 c | 0.19 ± 0.13 f | 0.08 ± 0.08 de | 0.47 ± 0.30 d | 0.06 ± 0.07 e | 0.06 ± 0.03 ef | 0.37 ± 0.20 f | 5.29 ± 2.60 fg |
| KM5 | 0.18 ± 0.12 e | 0.08 ± 0.06 g | 0.01 ± 0.01 g | 0.13 ± 0.05 f | 0.06 ± 0.00 e | 0.02 ± 0.03 g | 0.07 ± 0.07 h | 3.58 ± 0.76 h |
| LS1 | 0.22 ± 0.06 e | 0.06 ± 0.02 g | 0.02 ± 0.01 g | 0.16 ± 0.05 f | 0.06 ± 0.00 e | 0.03 ± 0.00 g | 0.15 ± 0.05 g | 4.64 ± 2.74 g |
| LS4 | 0.85 ± 0.26 b | 0.38 ± 0.17 c | 0.13 ± 0.10 b | 0.64 ± 0.26 c | 0.09 ± 0.02 bc | 0.12 ± 0.01 c | 0.53 ± 0.09 de | 9.09 ± 0.75 b |
| LS6 | 0.20 ± 0.14 e | 0.06 ± 0.02 g | 0.01 ± 0.01 g | 0.16 ± 0.05 f | 0.06 ± 0.00 e | 0.02 ± 0.03 g | 0.13 ± 0.12 g | 4.72 ± 0.49 g |
| LS9 | 0.44 ± 0.40 d | 0.24 ± 0.24 e | 0.06 ± 0.07 ef | 0.61 ± 0.57 c | 0.04 ± 0.05 f | 0.06 ± 0.04 e | 0.51 ± 0.54 e | 5.96 ± 2.55 ef |
| SC4 | 0.65 ± 0.64 c | 0.42 ± 0.19 bc | 0.24 ± 0.06 a | 0.48 ± 0.33 d | 0.12 ± 0.01 a | 0.17 ± 0.04 b | 0.48 ± 0.31 e | 6.12 ± 1.06 ef |
| ZP5 | 0.70 ± 0.54 c | 0.43 ± 0.40 b | 0.09 ± 0.09 cd | 0.88 ± 0.68 a | 0.08 ± 0.01 d | 0.13 ± 0.08 c | 0.80 ± 0.48 c | 7.42 ± 3.08 c |
| Family | Triterpenoid Content (mg/g) | ||||||
|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | T5 | T6 | TT | |
| AJ5 | 3.72 ± 2.82 d | 1.47 ± 1.13 b | 0.77 ± 0.03 d | 2.02 ± 0.82 d | 0.78 ± 0.40 c | 0.40 ± 0.03 d | 16.34 ± 6.92 d |
| HF10 | 1.63 ± 1.38 g | 1.19 ± 0.99 cd | 0.44 ± 0.48 f | 1.00 ± 0.98 g | 0.25 ± 0.28 f | 0.24 ± 0.22 fg | 15.04 ± 5.57 ef |
| JH1 | 4.59 ± 2.31 b | 1.81 ± 0.74 a | 0.54 ± 0.38 ef | 2.58 ± 2.31 c | 0.60 ± 0.60 d | 0.65 ± 0.28 a | 21.04 ± 6.99 b |
| JH2 | 2.41 ± 0.32 f | 1.28 ± 0.26 c | 0.47 ± 0.07 f | 1.25 ± 0.17 f | 0.31 ± 0.03 f | 0.29 ± 0.04 ef | 9.55 ± 0.68 h |
| JH3 | 4.31 ± 3.16 c | 1.60 ± 0.85 b | 2.21 ± 1.93 a | 3.52 ± 3.07 b | 0.74 ± 0.49 c | 0.28 ± 0.04 f | 19.26 ± 13.09 c |
| KM4 | 4.56 ± 2.92 bc | 1.84 ± 1.15 a | 1.56 ± 0.92 b | 5.05 ± 3.95 a | 0.88 ± 0.79 b | 0.55 ± 0.36 b | 22.42 ± 14.76 a |
| KM5 | 0.75 ± 0.42 h | 0.55 ± 0.17 f | 0.18 ± 0.15 g | 0.69 ± 0.31 h | 0.14 ± 0.02 g | 0.10 ± 0.04 h | 10.40 ± 1.62 h |
| LS1 | 0.62 ± 0.40 h | 0.39 ± 0.15 g | 0.19 ± 0.21 g | 0.49 ± 0.39 hi | 0.14 ± 0.13 g | 0.13 ± 0.10 h | 10.03 ± 1.69 h |
| LS4 | 2.16 ± 0.21 f | 1.10 ± 0.32 d | 1.00 ± 0.13 c | 1.88 ± 0.52 d | 0.15 ± 0.06 g | 0.21 ± 0.04 g | 12.73 ± 2.08 g |
| LS6 | 0.54 ± 0.13 h | 0.35 ± 0.06 g | 0.07 ± 0.08 g | 0.35 ± 0.11 i | 0.07 ± 0.02 h | 0.09 ± 0.04 h | 7.99 ± 3.15 i |
| LS9 | 1.78 ± 1.36 g | 0.79 ± 0.51 e | 1.74 ± 1.86 b | 1.95 ± 1.82 d | 0.28 ± 0.25 f | 0.33 ± 0.28 e | 15.54 ± 5.22 de |
| SC4 | 2.74 ± 0.82 e | 1.23 ± 0.64 cd | 1.06 ± 0.96 c | 1.50 ± 1.03 e | 0.52 ± 0.02 e | 0.49 ± 0.06 c | 14.05 ± 4.22 f |
| ZP5 | 5.06 ± 1.37 a | 1.84 ± 1.66 a | 0.71 ± 0.35 de | 2.72 ± 0.66 c | 1.04 ± 0.23 a | 0.58 ± 0.21 b | 21.40 ± 4.93 ab |
| Index | F1 | F2 | F3 | F4 | F5 | F6 | F7 | TF | T1 | T2 | T3 | T4 | T5 | T6 | TT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Height | 0.13 | −0.03 | −0.04 | −0.03 | 0.01 | 0.02 | 0.02 | −0.05 | 0.11 | 0.14 | −0.03 | 0.19 | 0.14 | 0.20 | 0.07 |
| DBH | 0.50 ** | 0.23 * | 0.24 * | 0.35 ** | 0.26 * | 0.39 ** | 0.50 ** | 0.35 ** | 0.45 ** | 0.47 ** | 0.25 * | 0.44 ** | 0.37 ** | 0.31 ** | 0.41 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, M.; Chen, P.; Shang, X.; Yang, W.; Fang, S. Genotype–Environment Interactions for Tree Growth and Leaf Phytochemical Content of Cyclocarya paliurus (Batal.) Iljinskaja. Forests 2021, 12, 735. https://doi.org/10.3390/f12060735
Zhou M, Chen P, Shang X, Yang W, Fang S. Genotype–Environment Interactions for Tree Growth and Leaf Phytochemical Content of Cyclocarya paliurus (Batal.) Iljinskaja. Forests. 2021; 12(6):735. https://doi.org/10.3390/f12060735
Chicago/Turabian StyleZhou, Mingming, Pei Chen, Xulan Shang, Wanxia Yang, and Shengzuo Fang. 2021. "Genotype–Environment Interactions for Tree Growth and Leaf Phytochemical Content of Cyclocarya paliurus (Batal.) Iljinskaja" Forests 12, no. 6: 735. https://doi.org/10.3390/f12060735
APA StyleZhou, M., Chen, P., Shang, X., Yang, W., & Fang, S. (2021). Genotype–Environment Interactions for Tree Growth and Leaf Phytochemical Content of Cyclocarya paliurus (Batal.) Iljinskaja. Forests, 12(6), 735. https://doi.org/10.3390/f12060735







