Dissecting Taxonomic Variants within Ulmus spp. Complex in Natural Forests with the Aid of Microsatellite and Morphometric Markers
Abstract
1. Introduction
2. Materials and Methods
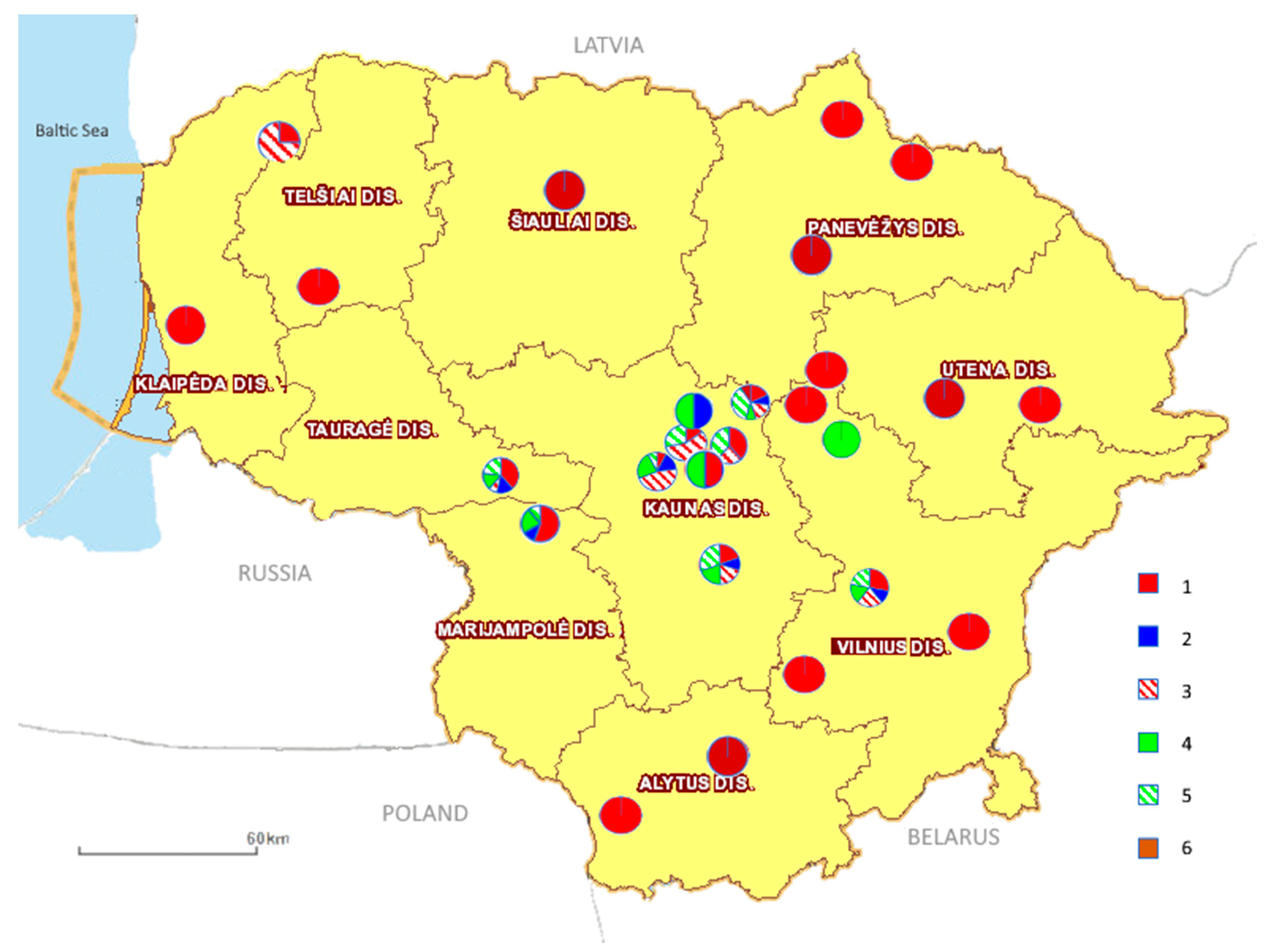
2.1. Objects and Material
2.2. Leaf Characters Studied
2.3. Microsatelite Analysis
2.4. Statistical Analysis of Leaf Characteristics
2.5. Molecular Data Analysis
3. Results
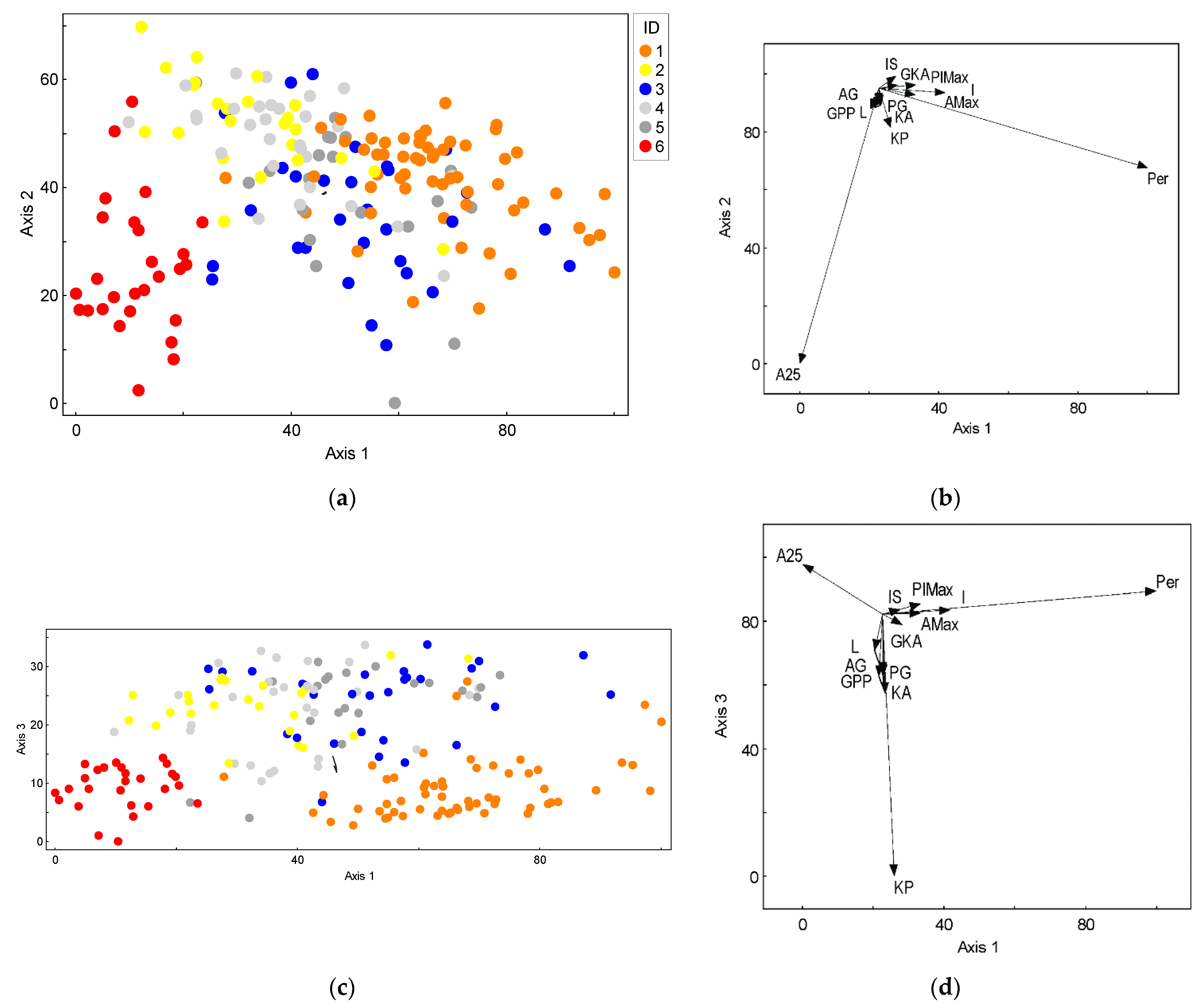
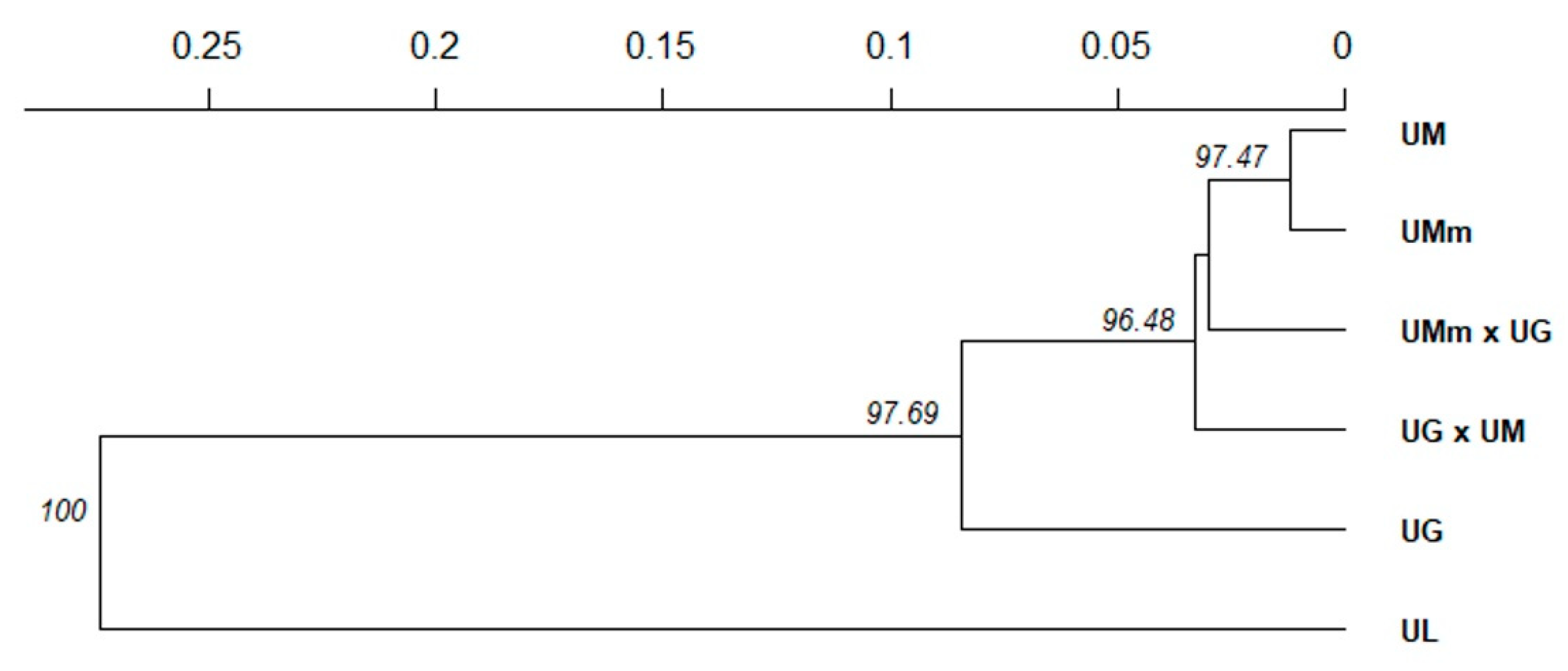
3.1. Leaf Morphology Variation
3.2. Genetic Diversity
3.3. Species Genetic Differentiation
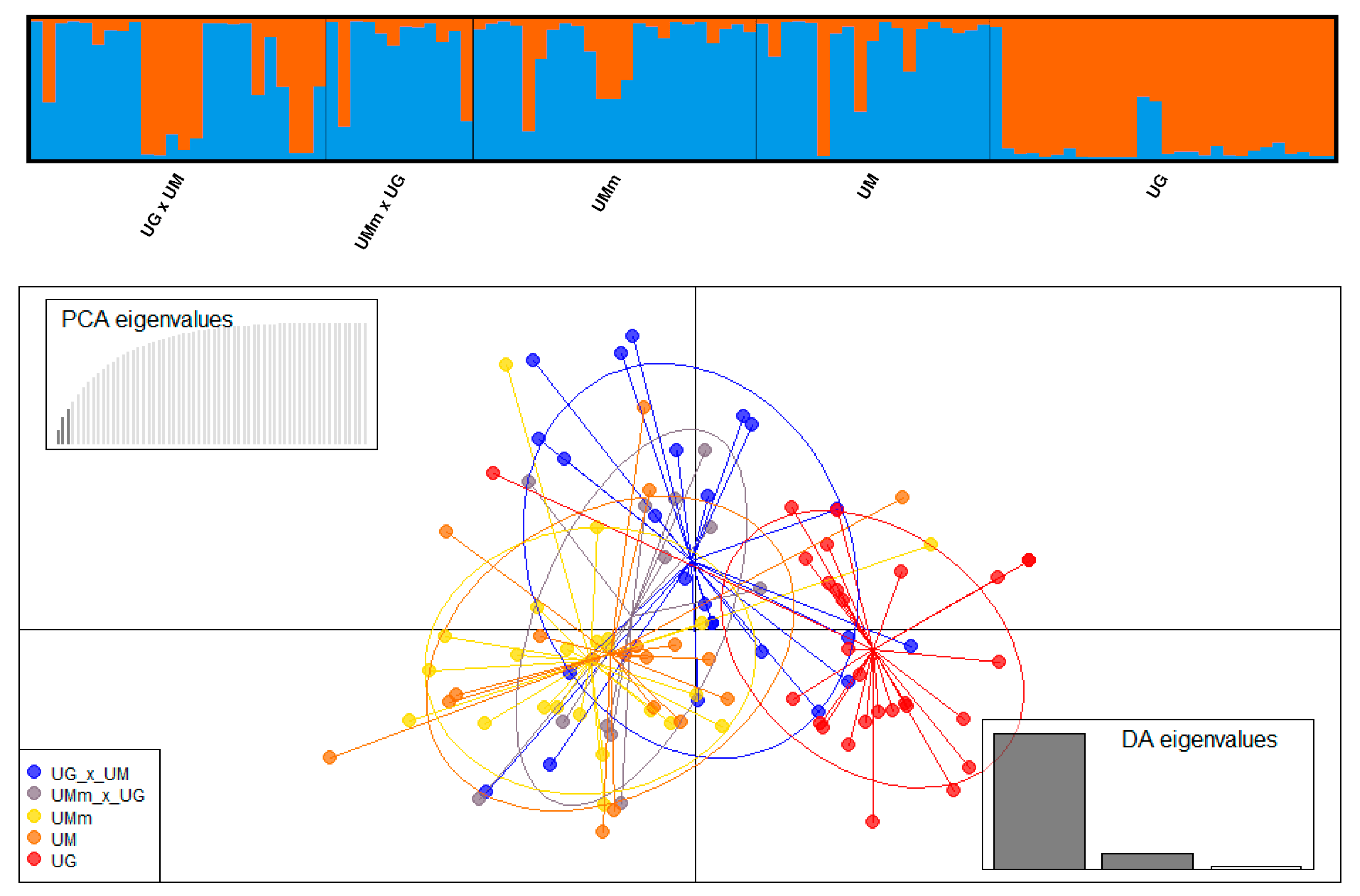
3.4. Hybrid Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Khan, T.N.; Croser, J.S. PEA|Overview. In Encyclopedia of Grain Science; Wrigley, C., Ed.; Elsevier: Oxford, UK, 2004. [Google Scholar]
- Soltis, P.S.; Soltis, D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, H.; Brunet, J.; Zalapa, J.E.; von Wehrden, H.; Hartmann, M.; Kleindienst, C.; Schlautman, B.; Kosman, E.; Wesche, K.; Renison, D.; et al. Intra- and interspecific hybridization in invasive siberian elm. Biol. Invasions 2017, 19, 1889–1904. [Google Scholar] [CrossRef]
- Vít, P.; Wolfová, K.; Urfus, T.; Tájek, P.; Suda, J. Interspecific hybridization between rare and common plant congeners inferred from genome size data: Assessing the threat to the Czech serpentine endemic Cerastium alsinifolium. Preslia 2014, 86, 95–117. [Google Scholar]
- Neale, D.B.; Wheeler, N.C. Hybridization and introgression. In The Conifers: Genomes, Variation and Evolution; Neale, D.B., Wheeler, N.C., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 387–429. [Google Scholar] [CrossRef]
- Abbott, R.J. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol. Evol. 1992, 7, 401–405. [Google Scholar] [CrossRef]
- Futuyma, D.J. Evolutionary biology today and the call for an extended synthesis. Interface Focus 2017, 7, 20160145. [Google Scholar] [CrossRef]
- Whitney, K.D.; Gabler, C.A. Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: Challenges for predicting invasive potential. Divers. Distrib. 2008, 14, 569–580. [Google Scholar] [CrossRef]
- Schierenbeck, K.A.; Ellstrand, N.C. Hybridization and the evolution of invasiveness in plants and other organisms. Biol. Invasions 2008, 11, 1093. [Google Scholar] [CrossRef]
- Blair, A.C.; Hufbauer, R.A. Hybridization and invasion: One of North America’s most devastating invasive plants shows evidence for a history of interspecific hybridization. Evol. Appl. 2010, 3, 40–51. [Google Scholar] [CrossRef]
- Keim, P.; Paige, K.N.; Whitham, T.G.; Lark, K.G. Genetic analysis of an interspecific hybrid swarm of populus: Occurrence of unidirectional introgression. Genetics 1989, 123, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Rieseberg, L.H.; Ellstrand, N.C.; Arnold, D.M. What can molecular and morphological markers tell us about plant hybridization? Crit. Rev. Plant Sci. 1993, 12, 213–241. [Google Scholar] [CrossRef]
- Rieseberg, L.H. The role of hybridization in evolution: Old wine in new skins. Am. J. Bot. 1995, 82, 944–953. [Google Scholar] [CrossRef]
- Kolbe, J.J.; Glor, R.E.; Rodríguez Schettino, L.; Lara, A.C.; Larson, A.; Losos, J.B. Genetic variation increases during biological invasion by a cuban lizard. Nature 2004, 431, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.A.; Overholt, W.A.; Cuda, J.P.; Hughes, C.R. Chloroplast and microsatellite DNA diversities reveal the introduction history of Brazilian Peppertree (Schinus terebinthifolius) in Florida. Mol. Ecol. 2005, 14, 3643–3656. [Google Scholar] [CrossRef]
- Culley, T.M.; Hardiman, N.A. The role of intraspecific hybridization in the evolution of invasiveness: A case study of the ornamental pear tree Pyrus calleryana. Biol. Invasions 2009, 11, 1107–1119. [Google Scholar] [CrossRef]
- Rius, M.; Darling, J.A. How Important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol. Evol. 2014, 29, 233–242. [Google Scholar] [CrossRef]
- Rosenthal, D.M.; Ramakrishnan, A.P.; Cruzan, M.B. Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol. Ecol. 2008, 17, 4657–4669. [Google Scholar] [CrossRef]
- Richardson, D.M.; Rejmánek, M. Trees and shrubs as invasive alien species—A global review. Divers. Distrib. 2011, 17, 788–809. [Google Scholar] [CrossRef]
- Richens, R.H. Studies on Ulmus: III. The Village Elms of Hertfordshire. For. Int. J. For. Res. 1959, 32, 138–154. [Google Scholar] [CrossRef]
- Bell, M. Elm. By R. H. Richens. 25 × 19.5 cm. Pp. 347, 152 Ills. Cambridge: Cambridge University Press, 1983. ISBN 0–521–24916–3. £35.00. Antiqu. J. 1984, 64, 427–428. [Google Scholar] [CrossRef]
- Seehausen, O.; Takimoto, G.; Roy, D.; Jokela, J. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol. Ecol. 2008, 17, 30–44. [Google Scholar] [CrossRef]
- Li, B.; Wu, R. Genetic causes of heterosis in juvenile aspen: A quantitative comparison across intra-and inter-specific hybrids. Theor. Appl. Genet. 1996, 93, 380–391. [Google Scholar] [CrossRef]
- Mejnartowicz, L. Evidence for long-term heterosis phenomenon in the Alnus incana × glutinosa F-1 hybrids. Silvae Genet. Ger. 1999, 48, 100–103. [Google Scholar]
- Richens, R.H.; Jeffers, J.N.R. Multivariate analysis of the elms of northern france. Silvae Genet. 1975, 25, 141–150. [Google Scholar]
- Richens, R.H.; Jeffers, J.N.R. Multivariate analysis of the elms of northern France. II. Pooled analysis of the elm populations of northern France and England. Silvae Genet. 1978, 27, 85–95. [Google Scholar]
- Jeffers, J. Leaf variation in the genus Ulmus. For. Int. J. For. Res. 1999, 72, 183–190. [Google Scholar] [CrossRef][Green Version]
- Jeffers, J.N.R.; Richens, R.H. Multivariate Analysis of the English elm population. Silvae Genet. 1970, 19, 31–38. [Google Scholar]
- Elowsky, C.; Jordon-Thaden, I.; Kaul, R. A Morphological Analysis of a hybrid swarm of native Ulmus rubra and introduced U. Pumila (Ulmaceae) in Southeastern Nebraska. Phytoneuron 2013, 44, 1–23. [Google Scholar]
- Myking, T.; Yakovlev, I. Variation in leaf morphology and chloroplast DNA in Ulmus glabra in the Northern Suture zone: Effects of distinct glacial refugia. Scand. J. For. Res. 2006, 21, 99–107. [Google Scholar] [CrossRef]
- Vander Mijnsbrugge, K.; Le Clercq, R.; Michiels, B. Dissection of leaf morphological traits from isolated and declined relict populations of Ulmus laevis reveals putative random ecotype evolution. Plant Syst. Evol. 2016, 302, 219–229. [Google Scholar] [CrossRef]
- Zebec, M.; Idžojtić, M.; Poljak, I.; Modrić, I. Population variability of wych elm (Ulmus glabra Huds.) in the mountainous region of Croatia according to the leaf morphology. Šumar. List 2015, 139, 429–438. [Google Scholar]
- Zebec, M.; Idžojtić, M.; Poljak, I. Morphological variability of the field elm (Ulmus minor Mill. sensu latissimo) in continental Croatia. Šumar. List 2014, 138, 563–571. [Google Scholar]
- Zebec, M.; Idžojtić, M.; Poljak, I.; Mihaldinec, I. The variability of field elm (Ulmus minor Mill. sensu latissimo) in Croatian Drava river valley according to the leaf morphology. Šumar. List 2010, 134, 569–579. [Google Scholar]
- Petrokas, R.; Baliuckas, V. Natural hybridisation within elms (Ulmus L.) in Lithuania. Balt. For. 2012, 18, 10. [Google Scholar]
- Melville, R. On the Discrimination of species in hybrid swarms with special reference to Ulmus and the nomenclature of U. minor Mill. and U. carpinifolia Gled. Taxon 1978, 27, 345–351. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Pádua, J.G.; Zucchi, M.I.; Vencovsky, R.; Vieira, M.L.C. Origin, evolution and genome distribution of microsatellites. Genet. Mol. Biol. 2006, 29, 294–307. [Google Scholar] [CrossRef]
- Barbará, T.; Palma-Silva, C.; Paggi, G.M.; Bered, F.; Fay, M.F.; Lexer, C. Cross-species transfer of nuclear microsatellite markers: Potential and limitations. Mol. Ecol. 2007, 16, 3759–3767. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.R.; Burke, J.M. EST-SSRs as a Resource for population genetic analyses. Heredity 2007, 99, 125–132. [Google Scholar] [CrossRef]
- Wang, M.L.; Barkley, N.A.; Jenkins, T.M. Microsatellite markers in plants and insects. Part I: Applications of Biotechnology. Genes Genomes Genom. 2009, 3, 54–67. [Google Scholar]
- Zalapa, J.E.; Brunet, J.; Guries, R.P. Isolation and characterization of microsatellite markers for red elm (Ulmus rubra Muhl.) and cross-species amplification with siberian elm (Ulmus pumila L.). Mol. Ecol. Resour. 2008, 8, 109–112. [Google Scholar] [CrossRef]
- Zalapa, J.E.; Brunet, J.; Guries, R.P. Original article: The extent of hybridization and its impact on the genetic diversity and population structure of an invasive tree, Ulmus pumila (Ulmaceae). Evol. Appl. 2010, 3, 157–168. [Google Scholar] [CrossRef]
- Nielsen, L.R.; Kjær, E.D. Fine-scale gene flow and genetic structure in a relic Ulmus laevis population at its northern range. Tree Genet. Genomes 2010, 6, 643–649. [Google Scholar] [CrossRef]
- Venturas, M.; Fuentes-Utrilla, P.; Ennos, R.; Collada, C.; Gil, L. Human-induced changes on fine-scale genetic structure in Ulmus laevis pallas wetland forests at its sw distribution limit. Plant Ecol. 2013, 214, 317–327. [Google Scholar] [CrossRef]
- Bertolasi, B.; Leonarduzzi, C.; Piotti, A.; Leonardi, S.; Zago, L.; Gui, L.; Gorian, F.; Vanetti, I.; Binelli, G. A last stand in the po valley: Genetic structure and gene flow patterns in Ulmus minor and U. pumila. Ann. Bot. 2015, 115, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Buiteveld, J.; Vanden Broeck, A.; Cox, K.; Collin, E. Human impact on the genetic diversity of Dutch field elm (Ulmus minor) populations in the Netherlands: Implications for conservation. Plant Ecol. Evol. 2016, 149, 165–176. [Google Scholar] [CrossRef]
- Martín del Puerto, M.; Martínez García, F.; Mohanty, A.; Martín, J.P. Genetic diversity in relict and fragmented populations of Ulmus glabra Hudson in the central system of the Iberian Peninsula. Forests 2017, 8, 143. [Google Scholar] [CrossRef]
- Zebec, M.; Idžojtić, M.; Šatović, Z.; Poljak, I.; Liber, Z. Alive and kicking, or, living on borrowed time?–microsatellite diversity in natural populations of the endangered Ulmus minor Mill. sensu latissimo from Croatia. Acta Bot. Croat. 2016, 75, 53–59. [Google Scholar] [CrossRef]
- Chudzinska, M.; Litkowiec, M.; Palucka, M.; Paslawska, A.; Lewandowski, A.; Koziol, C. Clonal structure of field elm (Ulmus minor Mill.) in Poland. Sylwan 2019, 163, 839–845. [Google Scholar]
- Chudzińska, M.; Pałucka, M.; Pasławska, A.; Litkowiec, M.; Lewandowski, A.; Kozioł, C. Results of preliminary research on genetic variation and genetic differentiation between wych elm populations (Ulmus glabra Huds.) in Poland. Sylwan 2018, 162, 727–736. [Google Scholar]
- Mittempergher, L.; Porta, N.L. Hybridization studies in the Eurasian species of elm (Ulmus spp.). Silvae Genet. 1991, 40, 237–243. [Google Scholar]
- Brunet, J.; Zalapa, J.E.; Pecori, F.; Santini, A. Hybridization and introgression between the exotic siberian elm, Ulmus pumila, and the native field elm, U. minor, in Italy. Biol. Invasions 2013, 15, 2717–2730. [Google Scholar] [CrossRef]
- Petrokas, R.; Baliuckas, V. Morphological intergradation of native elm species is shown by site-specific parameters. Balt. For. 2014, 20, 238–247. [Google Scholar]
- Richens, R.H. Elm; Cambridge University Press: Cambridge, UK, 1983; 347p. [Google Scholar]
- Mackenthun, G.L. The elms of Co. Cork-a survey of species, varieties and forms. Ir. For. 2007, 64, 44–60. [Google Scholar]
- Elwes, H.J.; Henry, A. The Trees of Great Britain & Ireland; R. & R. Clark, Limited: Edinburgh, UK, 1913; 2022p. [Google Scholar]
- WinFOLIA Image Analysis Software. Leaf Area and Morphology. 2016. Available online: https://regent.qc.ca/assets/winfolia_software.html (accessed on 15 March 2016).
- Dumolin, S.; Demesure, B.; Petit, R.J. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor. Appl. Genet. 1995, 91, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, R.E.; Black-Samuelsson, S.; Clapham, D. Development of microsatellite markers for the European white elm (Ulmus laevis Pall.) and cross-species amplification within the genus Ulmus. Mol. Ecol. Notes 2003, 3, 598–600. [Google Scholar] [CrossRef]
- Collada, C.; Fuentes-Utrilla, P.; Gil, L.; Cervera, M.T. Characterization of microsatellite loci in Ulmus minor Miller and cross-amplification in U. glabra Hudson and U. laevis Pall. Mol. Ecol. Notes 2004, 4, 731–732. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices, Version 2.9.3. 2001. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 5 February 2014).
- Jombart, T.; Collins, C. A Tutorial for discriminant analysis of principal components (DAPC) using adegenet 2.0.0. Lond. Imp. Coll. Lond. MRC Centre Outbreak Anal. Model. 2015, 43p. Available online: http://adegenet.r-forge.r-project.org/files/tutorial-dapc.pdf (accessed on 20 April 2021).
- Jombart, T. An Introduction to Adegent 2.0. 0. R-Tutorial. Lond. Imp. Coll. Lond. MRC Cent. Outbreak Anal. Model. 2015, 79p. Available online: https://adegenet.r-forge.r-project.org/files/tutorial-basics.pdf (accessed on 20 April 2021).
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software Structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. Structure harvester: A website and program for visualizing STRUCTURE output and implementing the evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A Program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Anderson, E.C.; Thompson, E.A. A Model-based method for identifying species hybrids using multilocus genetic data. Genetics 2002, 160, 1217–1229. [Google Scholar] [CrossRef]
- Townsend, A.M. Crossability patterns and morphological variation among elm species and hybrids. Silvae Genet. 1975, 24, 18–23. [Google Scholar]
- Ager, A.A.; Guries, R.P. Barriers to interspecific hybridization in Ulmus americana. Euphytica 1982, 31, 909–920. [Google Scholar] [CrossRef]
- Cox, K.; Broeck, A.V.; Vander Mijnsbrugge, K.; Buiteveld, J.; Collin, E.; Heybroek, H.M.; Mergeay, J. Interspecific hybridisation and interaction with cultivars affect the genetic variation of Ulmus minor and Ulmus glabra in Flanders. Tree Genet. Genomes 2014, 10, 813–826. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Hollingsworth, M.L.; Coleman, M. The European elms: Molecular markers, population genetics, and biosystematics. In The Elms: Breeding, Conservation, and Disease Management; Dunn, C.P., Ed.; Springer: Boston, MA, USA, 2000; pp. 3–20. [Google Scholar] [CrossRef]
- Cogolludo-Agustín, M.Á.; Agúndez, D.; Gil, L. Identification of native and hybrid elms in Spain using isozyme gene markers. Heredity 2000, 85, 157–166. [Google Scholar] [CrossRef][Green Version]
- Fuentes-Utrilla, P.; Valbuena-Carabaña, M.; Ennos, R.; Gil, L. Population clustering and clonal structure evidence the relict state of Ulmus minor Mill. in the Balearic Islands. Heredity 2014, 113, 21–31. [Google Scholar] [CrossRef]
- Vähä, J.-P.; Primmer, C.R. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol. Ecol. 2006, 15, 63–72. [Google Scholar] [CrossRef] [PubMed]
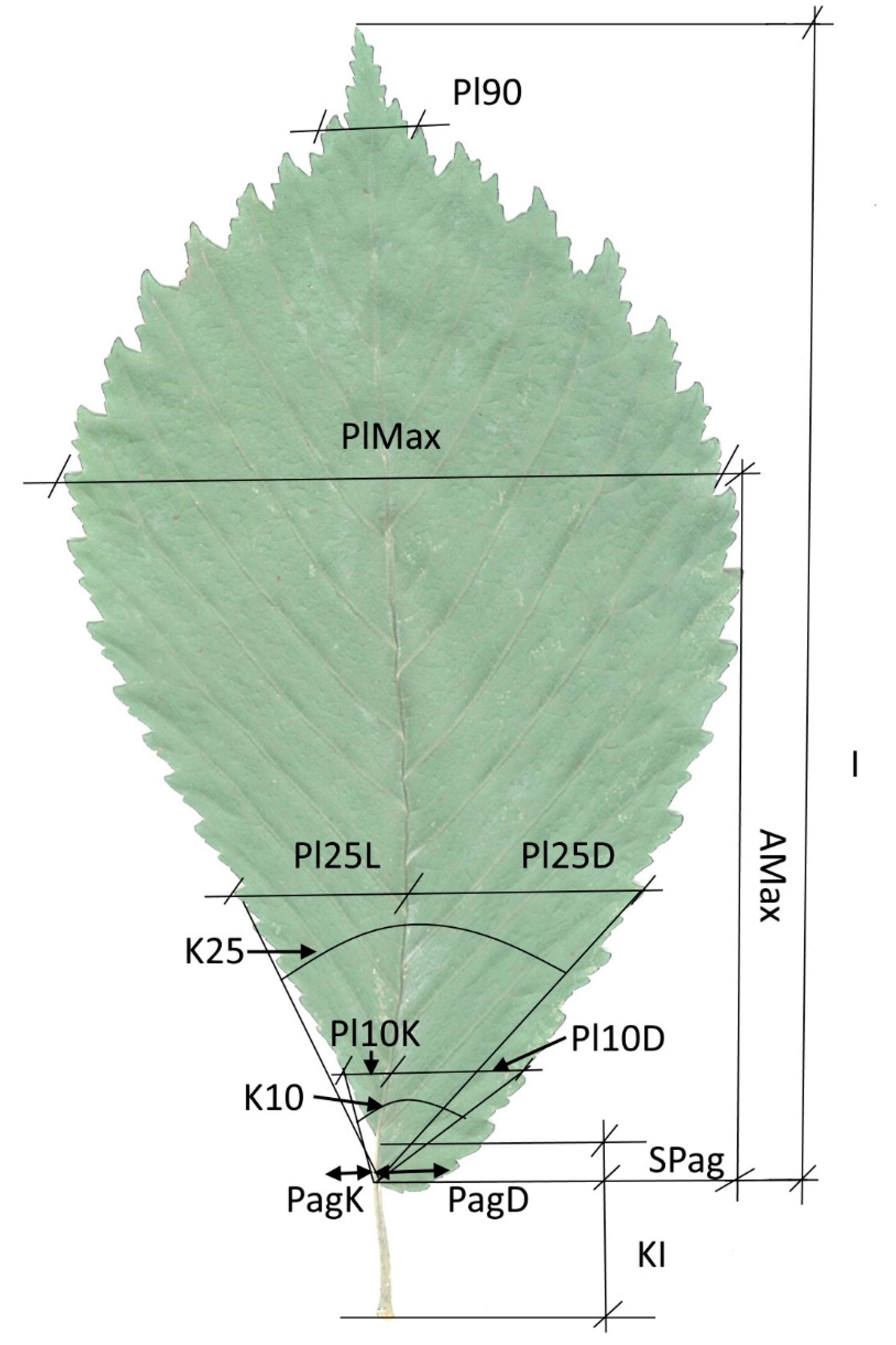

| No. | Leaf Morphology Traits | Abbreviation |
|---|---|---|
| Measured with Win Folia 2016 | ||
| 1 | Perimeter, cm | Per |
| 2 | Form coefficient (=4πA/Per2); A—leaf area; Per—perimeter) | Fk |
| 3 | Blade length, cm | I |
| 4 | Maximum blade width, cm | PlMax |
| 5 | Distance from the base to the point of maximum blade width, cm | AMax |
| 6 | Blade width measured at 90% blade length, cm | Pl90 |
| 7 | The blade lobe angle at 10% blade length | K10 |
| 8 | The blade lobe angle at 25% blade length | K25 |
| 9 | The petiole length, cm | KI |
| 10 | The blade base difference, cm | SPag |
| 11 | The base left width, cm | PagK |
| 12 | The base right width, cm | PagD |
| 13 | The blade width at 10% blade length left side, cm | Pl10K |
| 14 | The blade width at 10% blade length right side, cm | Pl10D |
| 15 | The blade width at 25% blade length left side, cm | Pl25K |
| 16 | The blade width at 25% blade length right side, cm | Pl25D |
| Calculated characteristics | ||
| 17 | Ratio (the blade maximum width and length ratio) (4/3) | Pl/I |
| 18 | The base asymmetry (|11–12|·100)/(11 + 12) | Apag |
| 19 | The asymmetry at 10% blade length (|13–14|·100)/(13 + 14) | A10 |
| 20 | The asymmetry at 25% blade length (|15–16|·100)/(15 + 16) | A25 |
| The visual characteristics set | ||
| 21 | The secondary vein branching (0—not branching; 1—1–4 branching veins; 2—5–10 branching veins; 3—>10 veins) | IS |
| 22 | The main vein angles pubescens (0—no pubescens; 3—a lot of pubescens) | GK |
| 23 | The main vein pubescens (0—no pubescens; 3—a lot of pubescens) | PG |
| 24 | The second veins pubescens (0—no pubescens; 3—a lot of pubescens) | AG |
| 25 | The Blade pubescens (0—no pubescens; 3—a lot of pubescens) | L |
| 26 | The Second veins angles pubescens (0—no pubescens; 3—a lot of pubescens) | GKA |
| 27 | The petiole lower half pubescens (0—no pubescens; 4—a lot of pubescens) | KA |
| 28 | The petiole upper half pubescens (0—no pubescens; 4—a lot of pubescens) | KP |
| 29 | The upper blade half pubescens length (1—short; 2—long) | VP |
| 30 | The main vein pubescens type (0—no pubescens; 1—rare; 2—more; 3—much; 4—overgrown on the sides of vein) | GPP |
| 31 | Pubescens character of corners of main vein (1—closed; 2—of veins sides; 3—from sides and angle) | GKPP |
| No. | Morphology Trait | R2 | F | Pr > F |
|---|---|---|---|---|
| 1 | IS | 0.814 | 159.745 | <0.0001 |
| 2 | L | 0.768 | 121.395 | <0.0001 |
| 3 | KA | 0.761 | 116.689 | <0.0001 |
| 4 | AG | 0.687 | 80.502 | <0.0001 |
| 5 | PG | 0.686 | 80.082 | <0.0001 |
| 6 | GKA | 0.661 | 71.328 | <0.0001 |
| 7 | KP | 0.654 | 69.197 | <0.0001 |
| 8 | AMax | 0.634 | 63.525 | <0.0001 |
| 9 | I | 0.624 | 60.786 | <0.0001 |
| 10 | Per | 0.590 | 52.613 | <0.0001 |
| 11 | PlMax | 0.570 | 48.548 | <0.0001 |
| 12 | GPP | 0.562 | 46.934 | <0.0001 |
| 13 | A25 | 0.533 | 41.765 | <0.0001 |
| 14 | Kilg | 0.462 | 31.453 | <0.0001 |
| 15 | A10 | 0.410 | 25.466 | <0.0001 |
| 16 | GK | 0.299 | 15.635 | <0.0001 |
| 17 | Pl90 | 0.292 | 15.078 | <0.0001 |
| 18 | Apag | 0.278 | 14.123 | <0.0001 |
| 19 | K10 | 0.253 | 12.416 | <0.0001 |
| 20 | Spag | 0.246 | 11.969 | <0.0001 |
| 21 | VP | 0.219 | 10.262 | <0.0001 |
| 22 | Fk | 0.187 | 8.444 | <0.0001 |
| 23 | K25 | 0.088 | 3.535 | 0.004 |
| 24 | Pl/I | 0.042 | 1.599 | 0.163 |
| 25 | GKPP | 0.016 | 0.589 | 0.708 |
| Morphologic Characteristics | PC 1 | PC 2 | PC 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R | R2 | tau | R | R2 | tau | R | R2 | tau | |
| Per | 0.985 * | 0.970 | 0.903 | −0.168 | 0.028 | −0.174 | 0.031 | 0.001 | 0.022 |
| I | 0.945 | 0.893 | 0.791 | 0.033 | 0.001 | −0.104 | 0.023 | 0.001 | 0.017 |
| PlMax | 0.883 | 0.780 | 0.699 | 0.048 | 0.002 | −0.017 | 0.091 | 0.008 | 0.068 |
| AMax | 0.940 | 0.883 | 0.779 | −0.102 | 0.010 | −0.146 | 0.004 | 0.000 | 0.009 |
| A25 | −0.442 | 0.196 | −0.245 | −0.891 | 0.793 | −0.667 | 0.104 | 0.011 | 0.167 |
| PG | 0.031 | 0.001 | 0.049 | −0.323 | 0.104 | −0.233 | −0.850 | 0.723 | −0.637 |
| G | −0.003 | 0.000 | 0.040 | −0.342 | 0.117 | −0.253 | −0.837 | 0.701 | −0.606 |
| L | −0.335 | 0.112 | −0.022 | −0.488 | 0.238 | −0.307 | −0.568 | 0.323 | −0.445 |
| IS | 0.704 | 0.496 | 0.483 | 0.290 | 0.084 | 0.075 | 0.068 | 0.005 | −0.073 |
| GKA | 0.721 | 0.520 | 0.551 | 0.071 | 0.005 | −0.002 | −0.150 | 0.022 | −0.125 |
| KA | 0.076 | 0.006 | 0.085 | −0.285 | 0.081 | −0.199 | −0.881 | 0.776 | −0.674 |
| 2KP | 0.113 | 0.013 | 0.105 | −0.225 | 0.050 | −0.145 | −0.962 | 0.925 | −0.810 |
| GPP | −0.116 | 0.013 | −0.047 | −0.332 | 0.110 | −0.207 | −0.692 | 0.479 | −0.501 |
| Species Based on Leaf Morphology | Abbreviations | N * | Na | Ne | Ar | Ho | uHe | Fis |
|---|---|---|---|---|---|---|---|---|
| U. glabra × U. minor | UG × UM | 24 | 7.67 | 4.06 | 6.51 | 0.554 | 0.725 | 0.209 |
| U. minor subsp. minor × U. glabra | UMm × UG | 12 | 6.50 | 4.35 | 6.50 | 0.583 | 0.768 | 0.183 |
| U. minor subsp. minor | UMm | 23 | 7.33 | 4.14 | 6.28 | 0.648 | 0.726 | 0.063 |
| U. minor | UM | 19 | 7.50 | 4.43 | 6.68 | 0.632 | 0.771 | 0.136 |
| U. glabra | UG | 28 | 8.00 | 4.55 | 6.49 | 0.730 | 0.754 | 0.006 |
| U. laevis | UL | 27 | 4.33 | 2.53 | 3.60 | 0.595 | 0.577 | −0.061 |
| Mean | 6.89 | 4.01 | 6.00 | 0.624 | 0.720 | 0.089 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamošaitis, S.; Jurkšienė, G.; Petrokas, R.; Buchovska, J.; Kavaliauskienė, I.; Danusevičius, D.; Baliuckas, V. Dissecting Taxonomic Variants within Ulmus spp. Complex in Natural Forests with the Aid of Microsatellite and Morphometric Markers. Forests 2021, 12, 653. https://doi.org/10.3390/f12060653
Tamošaitis S, Jurkšienė G, Petrokas R, Buchovska J, Kavaliauskienė I, Danusevičius D, Baliuckas V. Dissecting Taxonomic Variants within Ulmus spp. Complex in Natural Forests with the Aid of Microsatellite and Morphometric Markers. Forests. 2021; 12(6):653. https://doi.org/10.3390/f12060653
Chicago/Turabian StyleTamošaitis, Sigitas, Girmantė Jurkšienė, Raimundas Petrokas, Jurata Buchovska, Ilona Kavaliauskienė, Darius Danusevičius, and Virgilijus Baliuckas. 2021. "Dissecting Taxonomic Variants within Ulmus spp. Complex in Natural Forests with the Aid of Microsatellite and Morphometric Markers" Forests 12, no. 6: 653. https://doi.org/10.3390/f12060653
APA StyleTamošaitis, S., Jurkšienė, G., Petrokas, R., Buchovska, J., Kavaliauskienė, I., Danusevičius, D., & Baliuckas, V. (2021). Dissecting Taxonomic Variants within Ulmus spp. Complex in Natural Forests with the Aid of Microsatellite and Morphometric Markers. Forests, 12(6), 653. https://doi.org/10.3390/f12060653







