Comparative Analysis and Phylogenetic Implications of Plastomes of Five Genera in Subfamily Amyridoideae (Rutaceae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling, DNA Extraction and Sequencing
2.2. Codon Usage and Characterization of Repeat Sequences
2.3. Genome Comparison
2.4. Phylogenetic Analysis
3. Results and Discussion
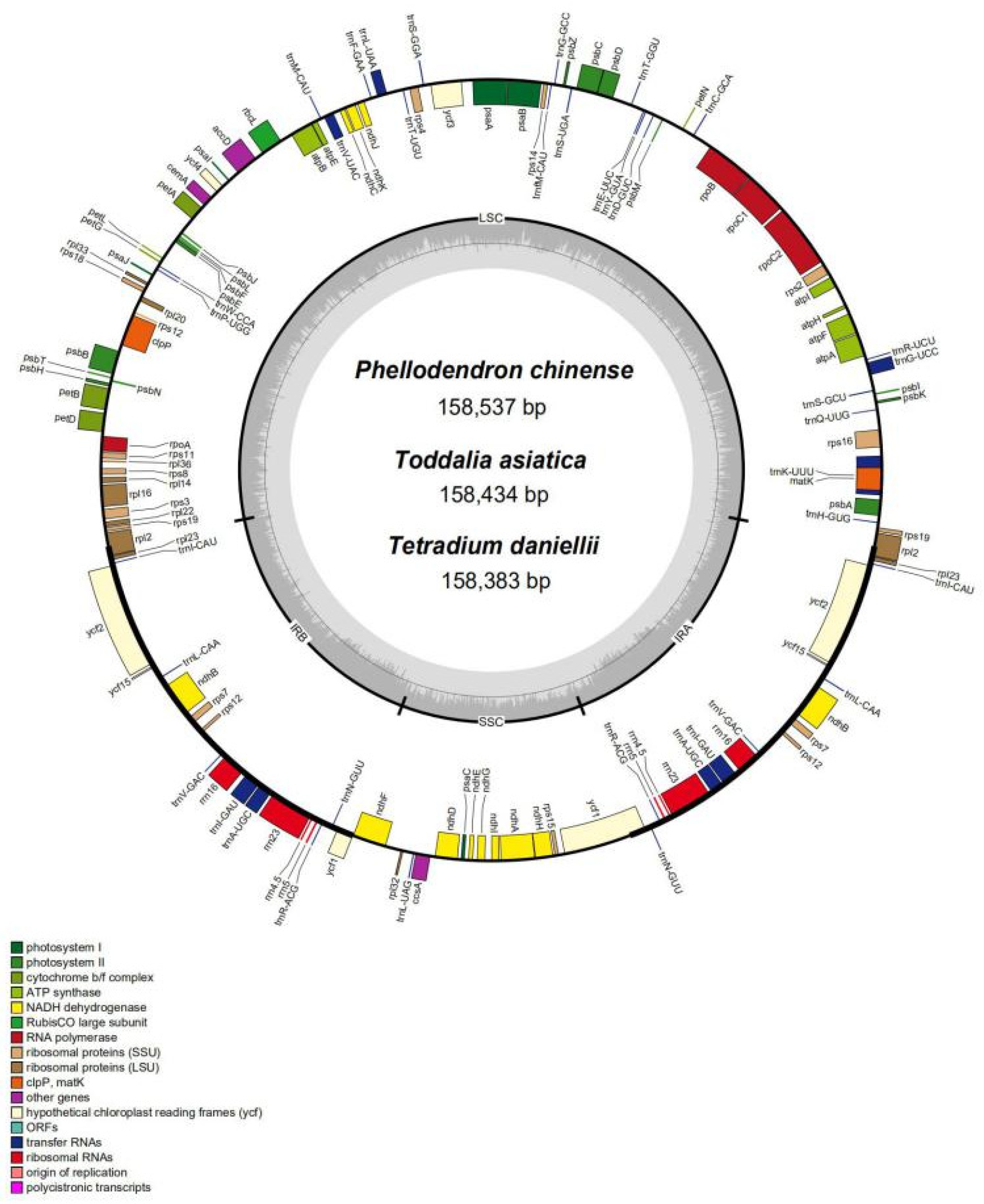
3.1. Structure and Size of Plastomes
3.2. Codon Preference Analysis
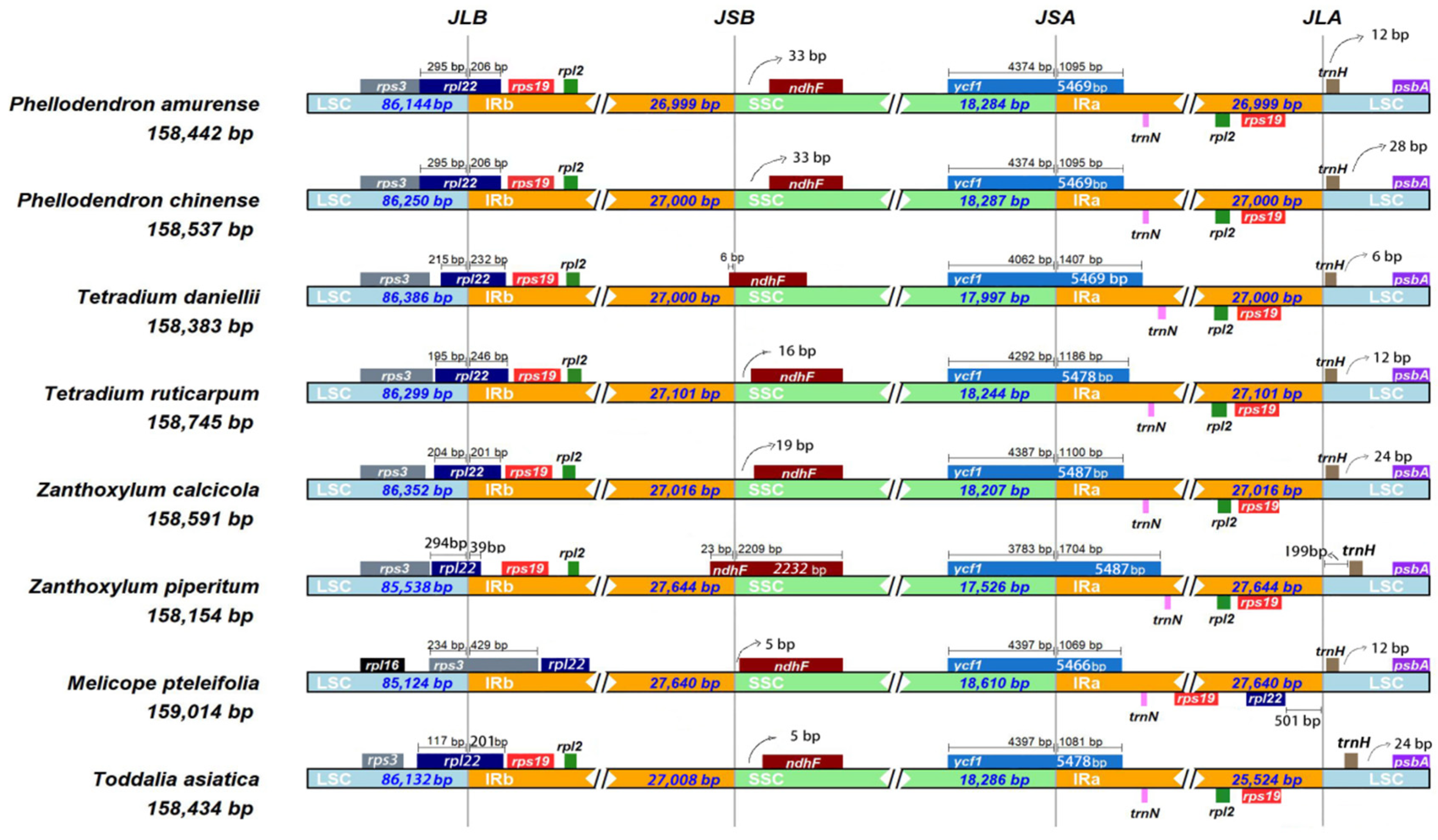
3.3. IR Contraction and Expansion
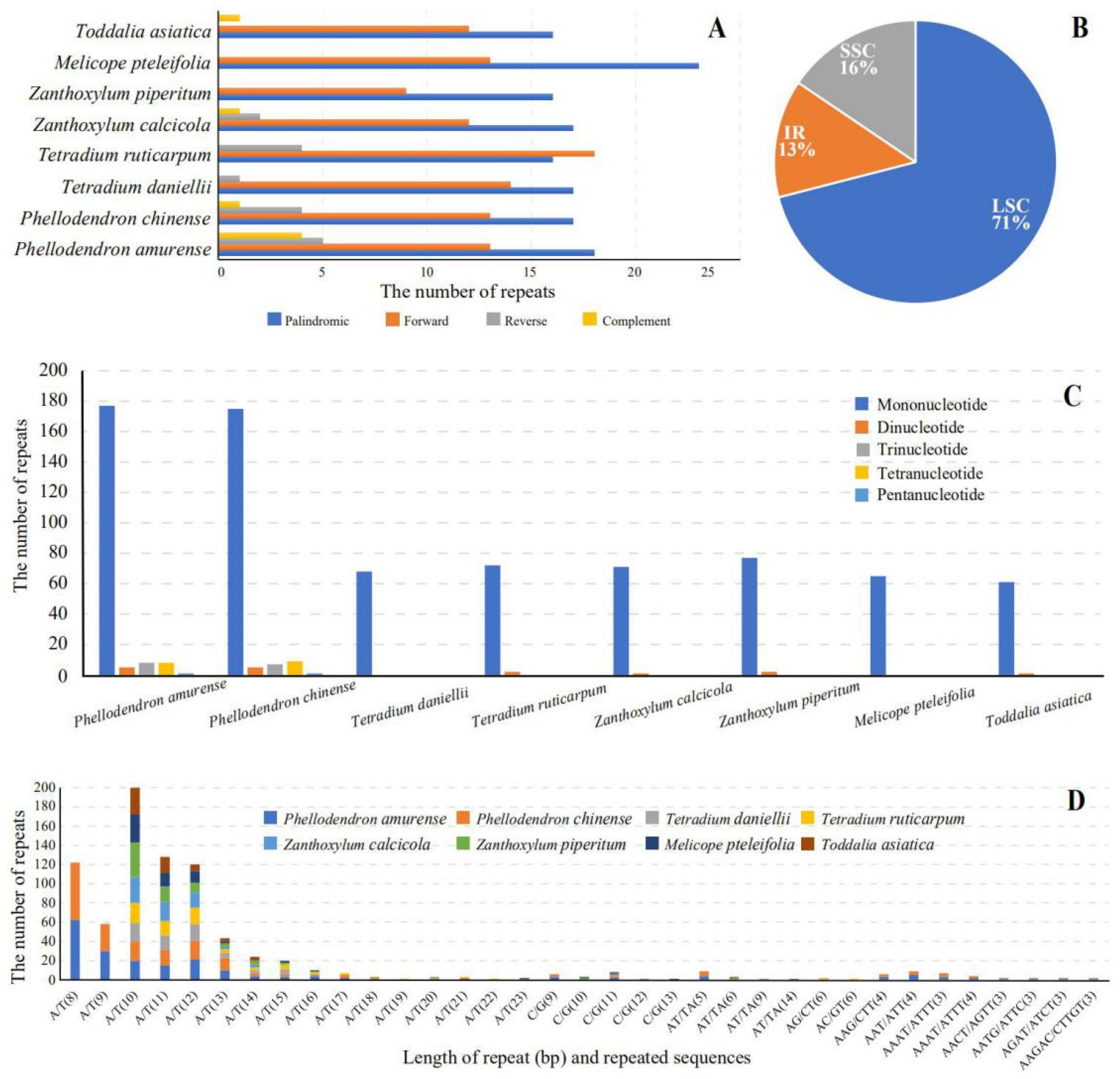
3.4. Repeat and SSR Analysis
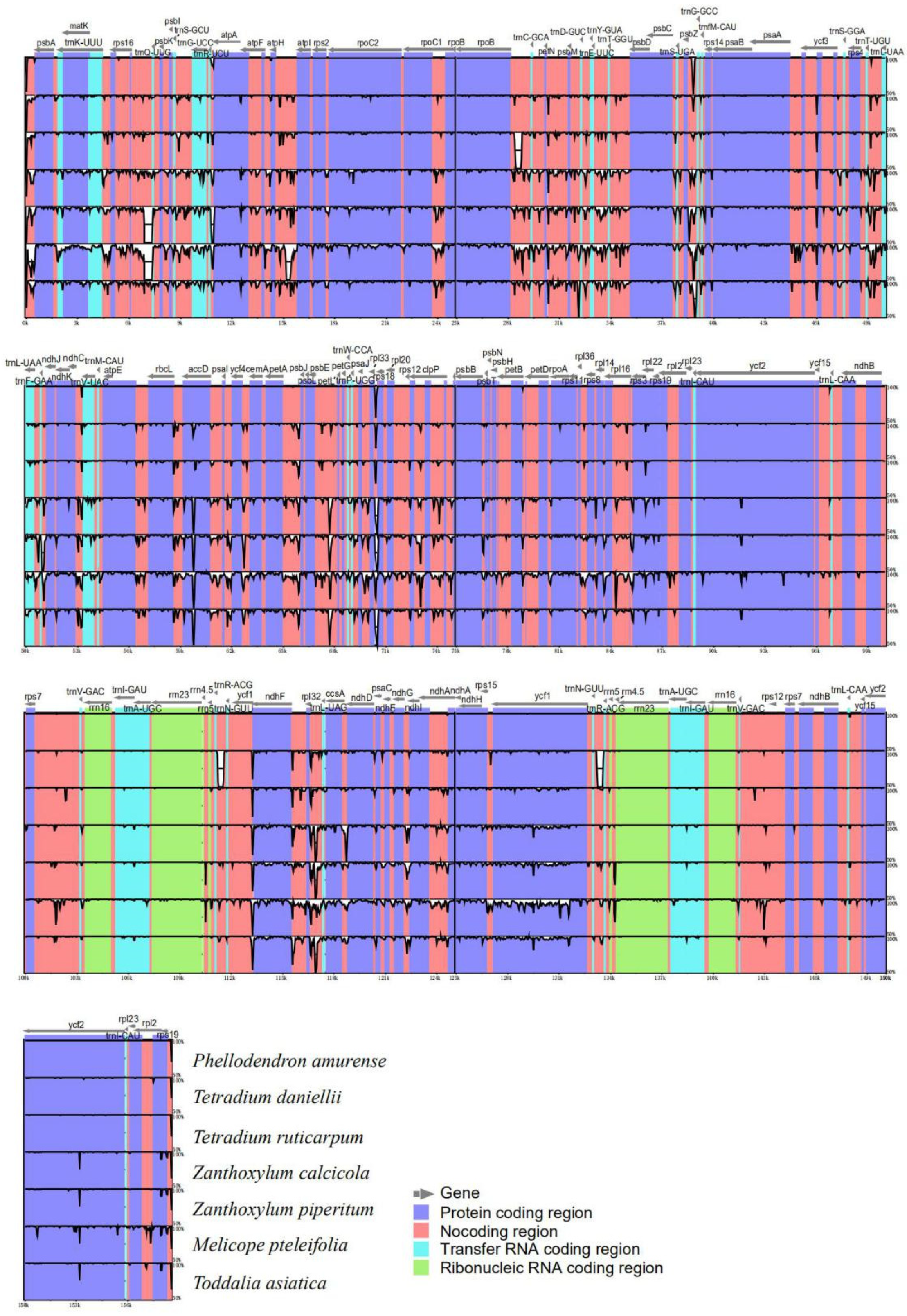
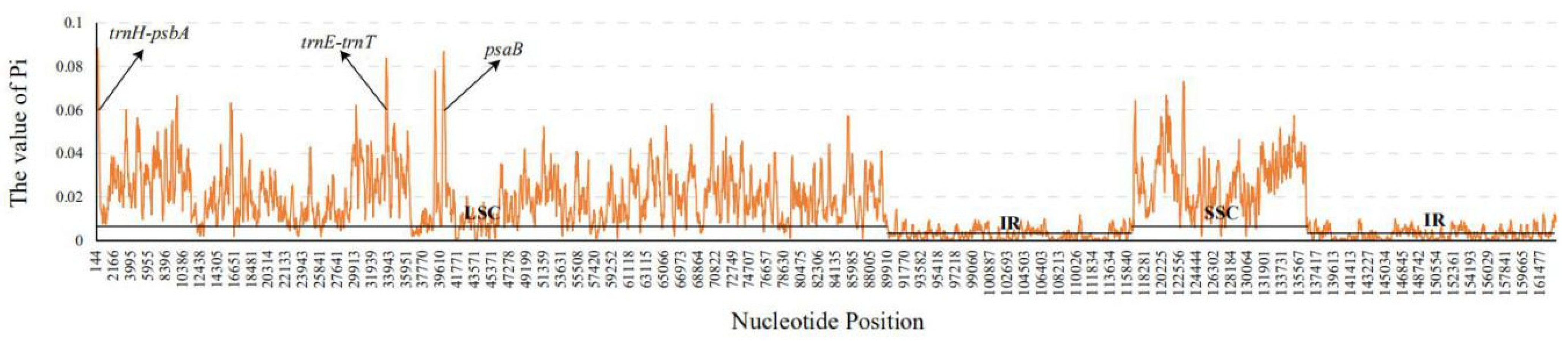
3.5. Sequence Divergence Analysis
3.6. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jansen, R.K. Plastid Genomes of seed plants. In Advances in Photosynthesis and Respiration; Bock, R., Knoop, V., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 103–126. [Google Scholar]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [PubMed] [Green Version]
- Raubeson, L.; Jansen, R. Chloroplast genomes of plants. In Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants, 1st ed.; Robert, J., Ed.; CABI: London, UK, 2005; pp. 53–76. [Google Scholar]
- Ma, P.F.; Zhang, Y.X.; Zeng, C.X.; Guo, Z.H.; Li, D.Z. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable Bamboo tribe Arundinarieae (Poaceae). Syst. Biol. 2014, 63, 933–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, C.S.P.; Henwood, M.J.; Ho, S.Y.W. Plastome sequences and exploration of tree-space help to resolve the phylogeny of riceflowers (Thymelaeaceae:Pimelea). Mol. Phylogenet. Evol. 2018, 127, 156–157. [Google Scholar] [CrossRef]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Jin, J.J.; Wang, B.; Yang, J.B.; Liu, B.; Corlett, R.T. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae. J. Syst. Evol. 2020, 58, 423–439. [Google Scholar] [CrossRef]
- Yang, X.Y.; Wang, Z.F.; Luo, W.C.; Guo, X.Y.; Zhang, C.H.; Liu, J.Q.; Ren, G.P. Plastomes of Betulaceae and phylogenetic implications. J. Syst. Evol. 2019, 57. [Google Scholar] [CrossRef]
- Kubitzki, K.; Kallunki, J.; Duretto, M.; Wilson, P. Rutaceae. In The Families and Genera of Vascular Plants; Kubitzki, K., Ed.; Springer: Berlin, Germany, 2011; Volume 10, pp. 276–356. [Google Scholar]
- Groppo, M.; Kallunki, J.A.; Pirani, J.R.; Antonelli, A. Chilean Pitavia more closely related to Oceania and Old World Rutaceae than to Neotropical groups: Evidence from two cpDNA non-coding regions, with a new subfamilial classifification of the family. PhytoKeys 2012, 19, 9–29. [Google Scholar] [CrossRef]
- Morton, C.M.; Telmer, C. New subfamily classification for the Rutaceae. Ann. Mo. Bot. Gard. 2014, 99, 620–641. [Google Scholar] [CrossRef]
- Engler, A. Rutaceae. In Die natürlichen Pflflanzenfamilien—Band 19a; Engler, A., Harms, H., Eds.; Wilhelm Engelmann: Leipzig, Germany, 1931; pp. 187–359. [Google Scholar]
- Waterman, P.G. Alkaloids of the Rutaceae: Their distribution and systematic significance. Biochem. Syst. Ecol. 1975, 3, 149–180. [Google Scholar]
- Waterman, P.G.; Khalid, S.A. The biochemical systematics of Fagaropsis angolensis and its significance in the Rutales. Biochem. Syst. Ecol. 1981, 9, 45–51. [Google Scholar] [CrossRef]
- Waterman, P. Phylogenetic implications of the distribution of secondary metabolites in the Rutales. In Chemistry and Chemical Taxonomy of the Rutales; Waterman, P., Grundon, M., Eds.; Academic Press: London, UK, 1983; pp. 377–400. [Google Scholar]
- Ng, K.M.; But, P.P.H.; Gray, A.I.; Hartley, T.G.; Kong, Y.C.; Waterman, P.G. The biochemical systematics of Tetradium, Euodia and Melicope and their significance in the Rutaceae. Biochem. Syst. Ecol. 1987, 15, 587–593. [Google Scholar] [CrossRef]
- Ma, J.S.; Cao, W.; Liu, Q.R.; Yu, M.; Han, L.J. A revision of Phellodendron (Rutaceae). Edinb. J. Bot. 2006, 63, 131–151. [Google Scholar] [CrossRef]
- Engler, A. Cneoraceae, Rutaceae. In Die naturlichen Pflflanzenfamilien, 2nd ed.; Engler, A., Prantl, K., Eds.; W. Engelmann: Leipzig, Germany, 1931; Volume Band 19a, pp. 184–359. [Google Scholar]
- Hartley, T.G. A revision of the genus Tetradium (Rutaceae). Gard. Bull. Singap. 1981, 34, 91–131. [Google Scholar]
- Hartley, T.G. On the taxonomy and biogeography of Euodia and Melicope (Rutaceae). Allertonia 2001, 8, 1–341. [Google Scholar]
- Appelhans, M.S.; Reichelt, N.; Groppo, M.; Paetzold, C.; Wen, J. Phylogeny and biogeography of the pantropical genus Zanthoxylum and its closest relatives in the proto-Rutaceae group (Rutaceae). Mol. Phylogenet. Evol. 2018, 126, 31–44. [Google Scholar] [PubMed]
- Poon, W.S.; Shaw, P.S.; Simmons, M.P.; But, P.P.H. Congruence of molecular, morphological, and biochemical profiles in Rutaceae: A cladistic analysis of the subfamilies. Syst. Bot. 2007, 32, 837–846. [Google Scholar] [CrossRef]
- Appelhans, M.S.; Wen, J.; Wagner, W.L. A molecular phylogeny of Acronychia, Euodia, Melicope and relatives (Rutaceae) reveals polyphyletic genera and key innovations for species richness. Mol. Phylogenet. Evol. 2014, 79, 54–68. [Google Scholar] [CrossRef]
- Ling, K.H.; Wang, Y.; Poon, W.S.; Shaw, P.C.; But, P.P.H. The relationship of Fagaropsis and Luvunga in Rutaceae. Taiwania 2009, 54, 338–342. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Huang, D.I.; Cronk, Q.C. Plann: A command-line application for annotating plastome sequences. Appl. Plant Sci. 2015, 3, 1500026. [Google Scholar] [CrossRef] [Green Version]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.N.; Gan, Z.M.; Zhang, F.; Yi, X.Y.; Zhang, J.Z.; Wan, X.H. Analysis of codon usage patterns in citrus based on coding sequence data. BMC Genom. 2020, 21, 234. [Google Scholar] [CrossRef]
- Sau, K.; Gupta, S.K.; Sau, S.; Mandal, S.C.; Ghosh, T.C. Factors influencing synonymous codon and amino acid usage biases in Mimivirus. Biosystems 2006, 85, 107–113. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [Green Version]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Brudno, M.; Do, C.B.; Cooper, G.M.; Kim, M.F.; Davydov, E.; Green, E.D.; Sidow, A.; Batzoglou, S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA outline of algorithms. Genome Res. 2003, 13, 721–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.V.D.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.V.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.R.; Zhang, Y.Y.; Liu, F.; Tian, N.; Pan, D.M.; Bei, X.J.; Cheng, C.Z. Characterization of the complete chloroplast genome of the Hongkong kumquat (Fortunella hindsii Swingle). Mitochondrial DNA Part B 2019, 4, 2612–2613. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.W.; Liu, F.; Tian, N.; Liu, J.P.; Shi, X.B.; Bei, X.J.; Cheng, C.Z. Characterisation of the complete chloroplast genome of Fortunella Crassifolia Swingle and phylogenetic relationships. Mitochondrial DNA Part B 2019, 4, 3538–3539. [Google Scholar] [CrossRef] [Green Version]
- Yoo, Y.H.; Oh, C.J.; Shin, S.C.; Seo, S.; Kim, H.B. Complete chloroplast genome sequence of a medicinal landrace citrus Jinkyool (Citrus sunki Hort. ex Tanaka) in Jeju Island, Korea. Mitochondrial DNA Part B 2020, 5, 3719–3720. [Google Scholar] [CrossRef] [PubMed]
- Angellotti, M.C.; Bhuiyan, S.B.; Chen, G.R.; Wan, X.F. CodonO: Codon usage bias analysis within and across genomes. Nucleic Acids Res. 2007, 35, W132–W136. [Google Scholar] [CrossRef]
- Wright, F. The effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Sueoka, N. Near homogeneity of PR2-Bias fingerprints in the human genome and their implications in phylogenetic analyses. J. Mol. Evol. 2001, 53, 469–476. [Google Scholar] [CrossRef]
- Xu, C.; Dong, J.; Tong, C.F.; Gong, X.D.; Wen, Q.; Qiang, Z.G. Analysis of synonymous codon usage patterns in seven different Citrus species. Evol. Bioinform. 2013, 9, 215–228. [Google Scholar] [CrossRef]
- Yu, X.Y.; Zuo, L.H.; Lu, D.D.; Lu, B.; Yang, M.S.; Wang, J.M. Comparative analysis of chloroplast genomes of five Robinia species: Genome comparative and evolution analysis. Gene 2019, 689, 141–151. [Google Scholar] [CrossRef]
- Cavagnaro, P.F.; Senalik, D.A.; Yang, L.M.; Simon, P.W.; Harkins, T.T.; Kodira, C.D.; Huang, S.W.; Weng, Y.Q. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom. 2010, 11, 569. [Google Scholar] [CrossRef] [Green Version]
- Appelhans, M.S.; Wen, J. Phylogenetic placement of Ivodea and biogeographic affinities of Malagasy Rutaceae. Plant Syst. Evol. 2020, 306, 7. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.C. Rutaceae. In Flora Reipublicae Popularis Sinicae; Huang, C., Ed.; Science Press: Beijing, China, 1997; Volume 43, pp. 1–250. [Google Scholar]
- Zhang, D.; Hartley, T.; Mabberley, D. Rutaceae. In Flora of China; Zhang, D., Hartley, T., Mabberley, D., Eds.; Science Press and St. Louis Missouri Botanical Garden Press: Beijing, China, 2008; Volume 11, pp. 51–97. [Google Scholar]
- Waterman, P.G. The current status of chemical systematics. Phytochemistry 2007, 68, 2896–2903. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Morton, C.M.; Kallunki, J.A. Phylogenetic relationships of Rutaceae: A cladistics analysis of the subfamilies using evidence from rbcL and atpB sequence variation. Am. J. Bot. 1999, 86, 1191–1999. [Google Scholar] [CrossRef]
- Groppo, M.; Pirani, J.R.; Salatino, M.L.F.; Blanco, S.R.; Kallunki, J.A. Phylogeny of Rutaceae based on two noncoding regions from cpDNA. Am. J. Bot. 2008, 95, 985–1005. [Google Scholar] [CrossRef]

| Features | Phellodendron amurense | Phellodendron chinense | Tetradium daniellii | Tetradium ruticarpum | Melicope pteleifolia | Zanthoxylum calcicola | Zanthoxylum piperitum | Toddalia asiatica |
|---|---|---|---|---|---|---|---|---|
| Total cp genome size (bp) | 158,442 | 158,537 | 158,383 | 158,745 | 159,014 | 158,591 | 158,154 | 158,434 |
| Length of LSC (bp) | 86,144 | 86,250 | 86,386 | 86,299 | 85,124 | 86,352 | 85,538 | 86,132 |
| Length of IR (bp) | 26,999 | 27,000 | 27,000 | 27,101 | 27,640 | 27,016 | 27,644 | 27,008 |
| Length of SSR (bp) | 18,284 | 18,287 | 17,997 | 18,244 | 18,610 | 18,207 | 17,526 | 18,288 |
| Coding size (bp) | 91,058 | 91,058 | 90,882 | 92,133 | 91,356 | 91,809 | 92,240 | 92,123 |
| Intron size (bp) | 19,368 | 19,372 | 18,208 | 18,424 | 19,332 | 18,335 | 18,355 | 18,186 |
| Spacer size (bp) | 48,016 | 48,107 | 49,293 | 48,188 | 48,326 | 48,447 | 47,559 | 48,125 |
| Total GC content (%) | 38.4 | 38.4 | 38.4 | 38.3 | 38.6 | 38.4 | 38.5 | 38.5 |
| GC content of LSC (%) | 36.7 | 36.6 | 36.7 | 36.6 | 36.6 | 36.7 | 36.8 | 36.8 |
| GC content of IR (%) | 42.9 | 42.9 | 42.7 | 42.8 | 42.8 | 42.8 | 42.5 | 42.8 |
| GC content of SSC (%) | 33.2 | 33.2 | 33.3 | 33.2 | 32.8 | 33.4 | 33.7 | 33.4 |
| GC content of rRNA (%) | 55.6 | 55.6 | 55.6 | 55.6 | 55.9 | 55.6 | 55.5 | 55.6 |
| GC content of tRNA (%) | 53.2 | 53.2 | 53.1 | 53.1 | 53.3 | 53.0 | 53.0 | 53.1 |
| GC content of Intron (%) | 38.0 | 38.0 | 38.3 | 38.3 | 38.1 | 38.3 | 38.3 | 38.2 |
| Total number of genes | 116 | 116 | 115 | 115 | 115 | 116 | 116 | 115 |
| Protein encoding-genes | 81 | 81 | 81 | 81 | 80 | 81 | 81 | 81 |
| tRNA genes | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 |
| rRNA genes | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Species | T3s | C3s | A3s | G3s | Nc |
|---|---|---|---|---|---|
| Phellodendron amurense | 0.3461 | 0.2424 | 0.3882 | 0.312 | 57.46 |
| Phellodendron chinense | 0.4555 | 0.1816 | 0.4206 | 0.1916 | 51.09 |
| Tetradium daniellii | 0.3982 | 0.2174 | 0.4061 | 0.2481 | 55.36 |
| Tetradium ruticarpum | 0.4539 | 0.1831 | 0.4206 | 0.1924 | 51.23 |
| Zanthoxylum calcicola | 0.4536 | 0.1832 | 0.4207 | 0.1924 | 51.2 |
| Zanthoxylum piperitum | 0.4391 | 0.2002 | 0.4159 | 0.2016 | 52.57 |
| Melicope pteleifolia | 0.4559 | 0.182 | 0.4209 | 0.1912 | 51.18 |
| Toddalia asiatica | 0.4382 | 0.2035 | 0.4122 | 0.2062 | 52.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, K.; Liu, Q.-Y.; Wang, A.; Gao, Y.-W.; Zhao, L.-C.; Guan, W.-B. Comparative Analysis and Phylogenetic Implications of Plastomes of Five Genera in Subfamily Amyridoideae (Rutaceae). Forests 2021, 12, 277. https://doi.org/10.3390/f12030277
Sun K, Liu Q-Y, Wang A, Gao Y-W, Zhao L-C, Guan W-B. Comparative Analysis and Phylogenetic Implications of Plastomes of Five Genera in Subfamily Amyridoideae (Rutaceae). Forests. 2021; 12(3):277. https://doi.org/10.3390/f12030277
Chicago/Turabian StyleSun, Kuo, Qiao-Yun Liu, Ao Wang, Yong-Wei Gao, Liang-Cheng Zhao, and Wen-Bin Guan. 2021. "Comparative Analysis and Phylogenetic Implications of Plastomes of Five Genera in Subfamily Amyridoideae (Rutaceae)" Forests 12, no. 3: 277. https://doi.org/10.3390/f12030277
APA StyleSun, K., Liu, Q.-Y., Wang, A., Gao, Y.-W., Zhao, L.-C., & Guan, W.-B. (2021). Comparative Analysis and Phylogenetic Implications of Plastomes of Five Genera in Subfamily Amyridoideae (Rutaceae). Forests, 12(3), 277. https://doi.org/10.3390/f12030277






