Characterizing Ectomycorrhizal Fungal Community Structure and Function of Two Varieties of Pinus clausa That Differ in Disturbance History
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Sampling Methods
2.2. Field Characterization of the ECM Fungal Community
2.2.1. Fungal Sequencing and OTU Generation
Laboratory Processing of Root Samples and Morphotyping
DNA Extraction and Sequencing
Sequence Assignment
2.2.2. Statistical Analyses
2.3. Greenhouse Experiment
2.3.1. Experiment Description
2.3.2. Statistical Analyses
3. Results
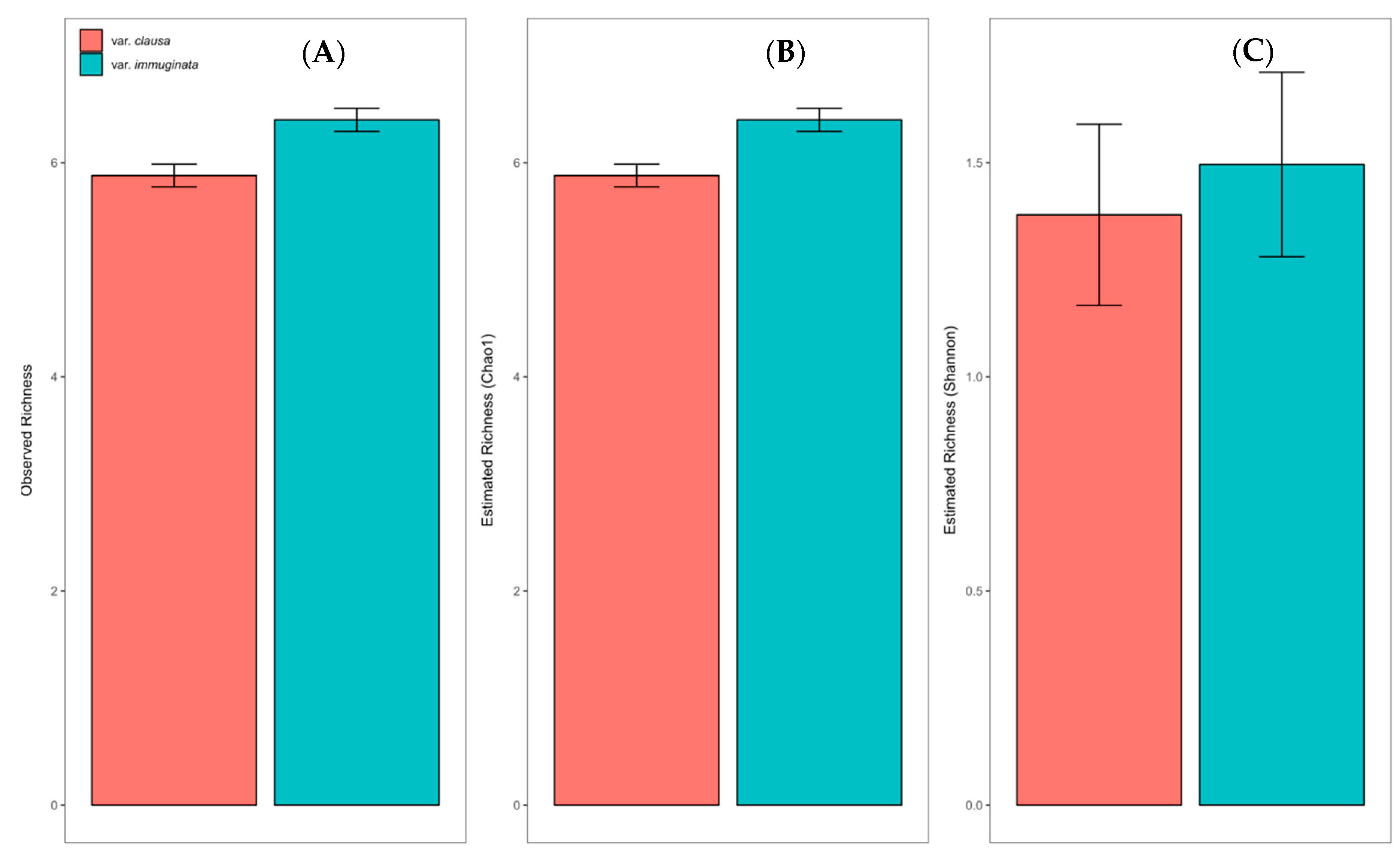
3.1. Field Characterization of the ECM Fungal Community
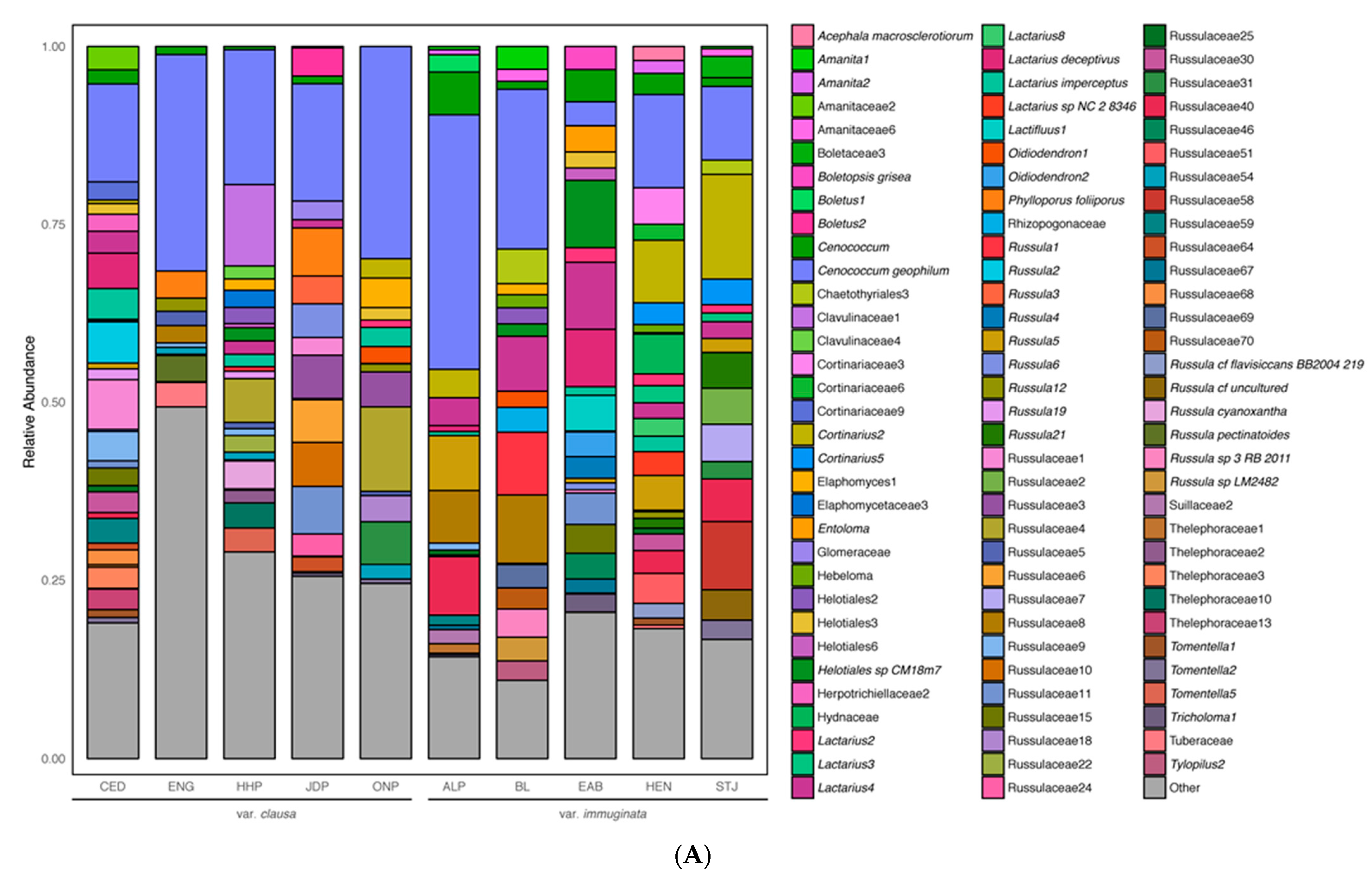
3.1.1. Overall ECM Fungal Community Description
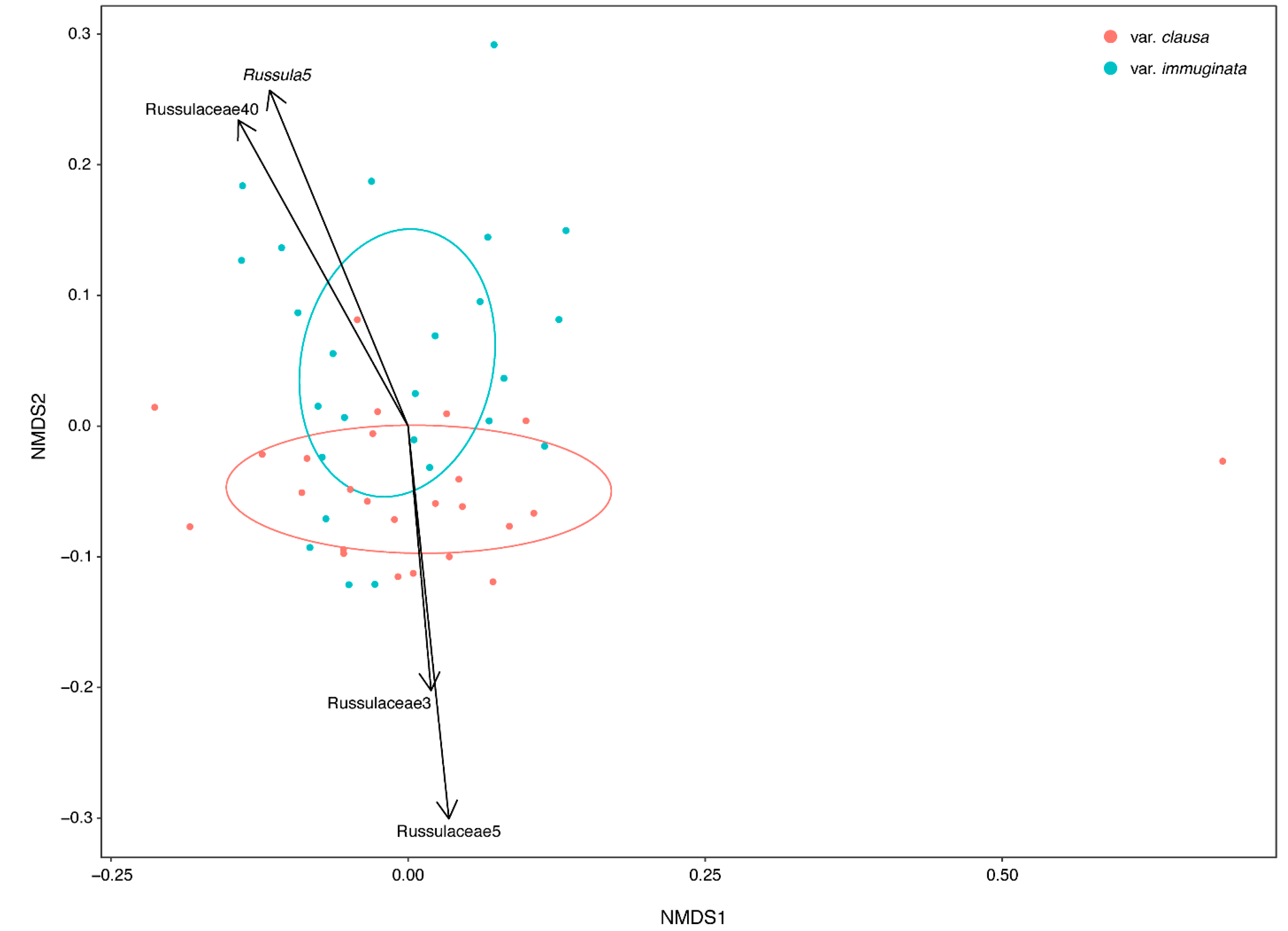
3.1.2. Community Structure as a Function of Variety
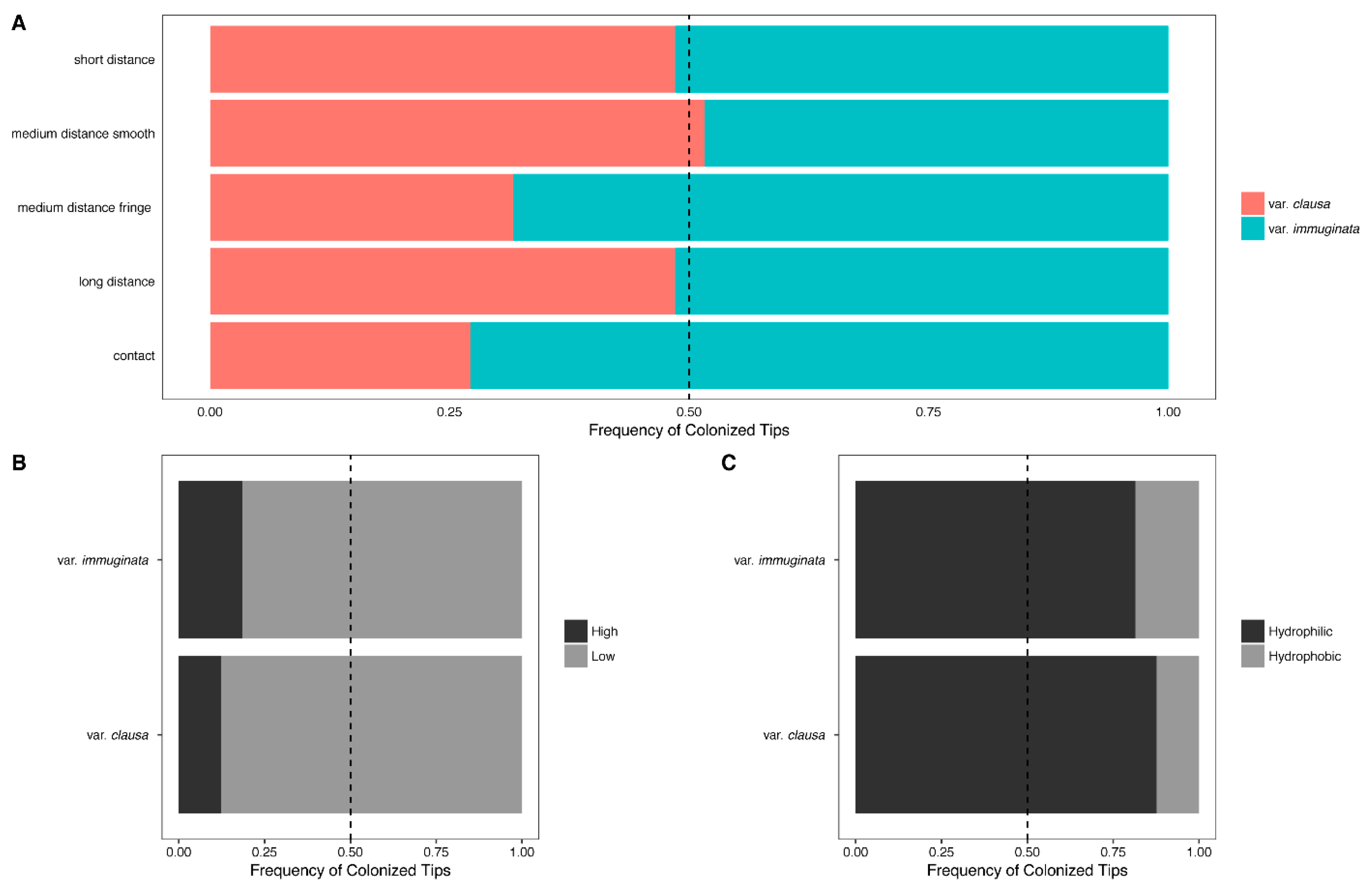
3.1.3. Distribution of Fungal Traits Involved with the Foraging Strategy
3.2. Greenhouse Experiment
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Christian, N.; Whitaker, B.K.; Clay, K. Microbiomes: Unifying animal and plant systems through the lens of community ecology theory. Front. Microbiol. 2015, 6, 869. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef]
- Konopka, A.; Lindemann, S.; Fredrickson, J. Dynamics in microbial communities: Unraveling mechanisms to identify principles. ISME J. 2015, 9, 1488–1495. [Google Scholar] [CrossRef]
- Taylor, A.F.S.; Alexander, I.A.N. The ectomycorrhizal symbiosis: Life in the real world. Mycologist 2005, 19, 102–112. [Google Scholar] [CrossRef]
- Sánchez-Ramírez, S.; Wilson, A.W.; Ryberg, M. Overview of Phylogenetic Approaches to Mycorrhizal Biogeography, Diversity and Evolution. In Biogeography of Mycorrhizal Symbiosis; Tedersoo, L., Ed.; Ecological Studies (Analysis and Synthesis); Springer: Cham, Switzerland, 2017; pp. 1–37. ISBN 978-3-319-56362-6. [Google Scholar]
- Smith, S.E.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Elsevier: London, UK, 2008. [Google Scholar]
- Hasselquist, N.; Germino, M.J.; McGonigle, T.; Smith, W.K. Variability of Cenococcum colonization and it ecophysiological significance to young conifers. New Phytol. 2005, 165, 867–873. [Google Scholar]
- Dale, V.H.; Joyce, L.A.; McNulty, S.; Neilson, R.P.; Ayres, M.P.; Flannigan, M.D.; Hanson, P.J.; Irland, L.C.; Lugo, A.E.; Peterson, C.J.; et al. Climate Change and Forest Disturbances: Climate change can affect forests by altering the frequency, intensity, duration, and timing of fire, drought, introduced species, insect and pathogen outbreaks, hurricanes, windstorms, ice storms, or landslides. BioScience 2001, 51, 723–734. [Google Scholar] [CrossRef]
- Briand, C.H.; Schwilk, D.W.; Gauthier, S.; Bergeron, Y. Does fire regime influence life history traits of jack pine in the southern boreal forest of Québec, Canada? Plant Ecol. 2015, 216, 157–164. [Google Scholar] [CrossRef]
- Leclerc, T.; Vimal, R.; Troispoux, V.; Périgon, S.; Scotti, I. Life after disturbance (I): Changes in the spatial genetic structure of Jacaranda copaia (Aubl.) D. Don (Bignonianceae) after logging in an intensively studied plot in French Guiana. Ann. For. Sci. 2015, 72, 509–516. [Google Scholar] [CrossRef]
- Parker, K.C.; Hamrick, J.L.; Parker, A.J.; Nason, J.D. Fine-scale genetic structure in Pinus clausa (Pinaceae) populations: Effects of disturbance history. Heredity 2001, 87, 99–113. [Google Scholar]
- Parker, K.C.; Hamrick, J.L. Genetic variation in sand pine (Pinus clausa). Can. J. For. Res. 2013, 26, 244–254. [Google Scholar] [CrossRef]
- Gehring, C.A.; Sthultz, C.M.; Flores-Rentería, L.; Whipple, A.V.; Whitham, T.G. Tree genetics defines fungal partner communities that may confer drought tolerance. Proc. Natl. Acad. Sci. USA 2017. [Google Scholar] [CrossRef]
- Lamit, L.J.; Holeski, L.M.; Flores-Rentería, L.; Whitham, T.G.; Gehring, C.A. Tree genotype influences ectomycorrhizal fungal community structure: Ecological and evolutionary implications. Fungal Ecol. 2016, 24, 124–134. [Google Scholar] [CrossRef]
- Mousseau, T.A.; Fox, C.W. The adaptive significance of maternal effects. Trends Ecol. Evol. 1998, 13, 403–407. [Google Scholar] [CrossRef]
- Bingham, M.A.; Simard, S.W. Seedling genetics and life history outweigh mycorrhizal network potential to improve conifer regeneration under drought. For. Ecol. Manag. 2013, 287, 132–139. [Google Scholar] [CrossRef]
- Agerer, R. Exploration types of ectomycorrhizae. Mycorrhiza 2001, 11, 107–114. [Google Scholar] [CrossRef]
- Hobbie, E.A.; Agerer, R. Nitrogen isotopes in ectomycorrhizal sporocarps correspond to belowground exploration types. Plant Soil 2010, 327, 71–83. [Google Scholar] [CrossRef]
- Lilleskov, E.A.; Hobbie, E.A.; Horton, T.R. Conservation of ectomycorrhizal fungi: Exploring the linkages between functional and taxonomic responses to anthropogenic N deposition. Fungal Ecol. 2011, 4, 174–183. [Google Scholar] [CrossRef]
- Knicker, H. How does fire affect the nature and stability of soil organic nitrogen and carbon? A review. Biogeochemistry 2007, 85, 91–118. [Google Scholar] [CrossRef]
- Lilleskov, E.A.; Fahey, T.J.; Horton, T.R.; Lovett, G.M. Belowground ectomycorrhizal fungal community change over a nitrogen deposition gradient in Alaska. Ecology 2002, 83, 104–115. [Google Scholar] [CrossRef]
- Lodge, D.J.; McDowell, W.H. Summary of Ecosystem-Level Effects of Caribbean Hurricanes. Biotropica 1991, 23, 373–378. [Google Scholar] [CrossRef]
- Aguilar-Trigueros, C.A.; Hempel, S.; Powell, J.R.; Anderson, I.C.; Antonovics, J.; Bergmann, J.; Cavagnaro, T.R.; Chen, B.; Hart, M.M.; Klironomos, J.; et al. Branching out: Towards a trait-based understanding of fungal ecology. Fungal Biol. Rev. 2015, 29, 34–41. [Google Scholar] [CrossRef]
- Krause, S.; Le Roux, X.; Niklaus, P.A.; Van Bodegom, P.M.; Lennon, J.T.; Bertilsson, S.; Grossart, H.-P.; Philippot, L.; Bodelier, P.L.E. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 2014, 5, 251. [Google Scholar] [CrossRef]
- Moeller, H.V.; Peay, K.G.; Fukami, T. Ectomycorrhizal fungal traits reflect environmental conditions along a coastal California edaphic gradient. FEMS Microbiol. Ecol. 2014, 87, 797–806. [Google Scholar] [CrossRef]
- Rúa, M.; Moore, B.; Hergott, N.; Van, L.; Jackson, C.; Hoeksema, J. Ectomycorrhizal Fungal Communities and Enzymatic Activities Vary across an Ecotone between a Forest and Field. J. Fungi 2015, 1, 185–210. [Google Scholar] [CrossRef]
- Parker, K.C.; Parker, A.J.; Beaty, R.M.; Fuller, M.M.; Faust, T.D. Population Structure and Spatial Pattern of Two Coastal Populations of Ocala Sand Pine (Pinus Clausa (Chapm. Ex Engelm.) Vasey Ex Sarg. Var. Clausa D. B. Ward). J. Torrey Bot. Soc. 1997, 124, 22–33. [Google Scholar] [CrossRef]
- Myers, R.L. Scrub and High Pine. In Ecosystems of Florida; Myers, R.L., Ewel, J.J., Eds.; University of Central Florida Press: Orlando, FL, USA, 1990; pp. 150–193. [Google Scholar]
- Ruehle, J.L.; Brendemuehl, R.H. Performance of Choctawhatchee Sand Pine Seedlings Inoculated with Ectomycorrhizal Fungi and Outplanted in the Sandhills of North Florida; US Department of Agriculture, Forest Service, Southeastern Forest Experiment Station: Asheville, NC, USA, 1981; Volume 301. [Google Scholar]
- Parker, A.J.; Parker, K.C.; Faust, T.D.; Fuller, M.M. The effects of climatic variability on radial growth of two varieties of sand pine (Pinus clausa) in Florida, USA. Ann. For. Sci. 2001, 58, 333–350. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar]
- Rúa, M.A.; Wilson, E.C.; Steele, S.; Munters, A.R.; Hoeksema, J.D.; Frank, C. Associations between Ectomycorrhizal Fungi and Bacterial Needle Endophytes in Pinus radiata: Implications for Biotic Selection of Microbial Communities. Front. Microbiol. 2016, 7, 399. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. Cap3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar]
- Izzo, A.; Agbowo, J.; Bruns, T.D. Detection of plot-level changes in ectomycorrhizal communities across years in an old-growth mixed-conifer forest. New Phytol. 2005, 166, 619–630. [Google Scholar]
- O’Brien, H.E.; Parrent, J.L.; Jackson, J.A.; Moncalvo, J.M.; Vilgalys, R. Fungal Community Analysis by Large-Scale Sequencing of Environmental Samples. Appl. Environ. Microbiol. 2005, 71, 5544–5550. [Google Scholar]
- Smith, M.D.; Douhan, G.; Rizzo, D. Ectomycorrhizal community structure in a xeric Quercus woodland based on rDNA sequence analyzsis of sporocarps and pooled roots. New Phytol. 2007, 174, 847–863. [Google Scholar]
- Horton, T.R. Molecular approaches to ectomycorrhizal diversity studies: Variation in ITS at a local scale. Plant Soil 2002, 244, 29–39. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Tedersoo, L.; Smith, M.E. Ectomycorrhizal Fungal Lineages: Detection of Four New Groups and Notes on Consistent Recognition of Ectomycorrhizal Taxa in High-Throughput Sequencing Studies. In Biogeography of Mycorrhizal Symbiosis; Tedersoo, L., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 125–142. [Google Scholar]
- Tedersoo, L.; Smith, M.E. Lineages of ectomycorrhizal fungi revisited: Foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol. Rev. 2013, 27, 83–99. [Google Scholar] [CrossRef]
- Unestam, T. Water repellency, mat formation, and leaf-stimulated growth of some ectomycorrhizal fungi. Mycorrhiza 1991, 1, 13–20. [Google Scholar] [CrossRef]
- Agerer, R. Fungal relationships and structural identity of their ectomycorrhizae. Mycol. Prog. 2006, 5, 67–107. [Google Scholar] [CrossRef]
- R. Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- McCune, B.; Grace, J.B. Analysis of Ecological Communities; MjM Software Design: Gleneden Beach, OR, USA, 2002; Volume 28. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; et al. Vegan: Community Ecology Package R Package Version 2.5-6. 2019. Available online: https://cran.r-project.org/ (accessed on 22 October 2020).
- Wickham, H. GGplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- De Caceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar]
- Newman, E.I. A Method of Estimating the Total Length of Root in a Sample. J. Appl. Ecol. 1966, 3, 139–145. [Google Scholar]
- Bates, D.; Maechler, M.; Bolker, B.; Steve, W. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric Models. Biometr. J. 2008, 50, 346–363. [Google Scholar]
- Chemidlin Prévost-Bouré, N.; Dequiedt, S.; Thioulouse, J.; Lelièvre, M.; Saby, N.P.A.; Jolivet, C.; Arrouays, D.; Plassart, P.; Lemanceau, P.; Ranjard, L. Similar Processes but Different Environmental Filters for Soil Bacterial and Fungal Community Composition Turnover on a Broad Spatial Scale. PLoS ONE 2014, 9, e111667. [Google Scholar] [CrossRef]
- Ranjard, L.; Dequiedt, S.; Chemidlin Prévost-Bouré, N.; Thioulouse, J.; Saby, N.P.A.; Lelievre, M.; Maron, P.A.; Morin, F.E.R.; Bispo, A.; Jolivet, C.; et al. Turnover of soil bacterial diversity driven by wide-scale environmental heterogeneity. Nat. Commun. 2013, 4, 1434. [Google Scholar] [CrossRef]
- Karst, J.; Burns, C.; Cale, J.; Antunes, P.M.; Woods, M.J.; Lamit, L.J.; Hoeksema, J.; Zabinski, C.; Gehring, C.; La Fleche, M.; et al. Tree species with limited geograbec ranges show extreme responses to ectomycorrhizas. Glob. Ecol. Biogeogr. 2018, 27, 839–848. [Google Scholar] [CrossRef]
- Last, F.T.; Dighton, J.; Mason, P.A. Successions of sheathing mycorrhizal fungi. Trends Ecol. Evol. 1987, 2, 157–161. [Google Scholar] [CrossRef]
- Visser, S. Ectomycorrhizal fungal succession in jack pine stands following wildfire. New Phytol. 1995, 129, 389–401. [Google Scholar] [CrossRef]
- Kirk, P.M. Species Fungorum (Version Oct 2017). 2019. Available online: www.catalogueoflife.org/annual-checklist/2019 (accessed on 22 October 2020).
- Looney, B.P.; Meidl, P.; Piatek, M.J.; Miettinen, O.; Francis, M.M.; Matheny, P.B.; Labbe, J.L. Russulaceae: A New Genomic Dataset to Study Ecosystem Function and Evolutionary Diversification of Ectomycorrhizal Fungi with Their Tree Associates. New Phytol. 2018, 218, 54–65. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Kõljalg, U.; Abarenkov, K. Identifying the ‘Unidentified’ Fungi: A Global-Scale Long-Read Third-Generation Sequencing Approach. Fungal Divers. 2020, 103, 273–293. [Google Scholar] [CrossRef]
- Freeman, J.E.; Kobziar, L.N. Tracking postfire successional trajectories in a plant community adapted to high-severity fire. Ecol. Appl. 2011, 21, 61–74. [Google Scholar] [CrossRef]
- McNulty, S.G. Hurricane impacts on US forest carbon sequestration. Environ. Pollut. 2002, 116 (Suppl. 1), S17–S24. [Google Scholar] [CrossRef]
- Sanford, R.L.; Parton, W.J.; Ojima, D.S.; Lodge, D.J. Hurricane Effects on Soil Organic Matter Dynamics and Forest Production in the Luquillo Experimental Forest, Puerto Rico: Results of Simulation Modeling. Biotropica 1991, 23, 364–372. [Google Scholar] [CrossRef]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
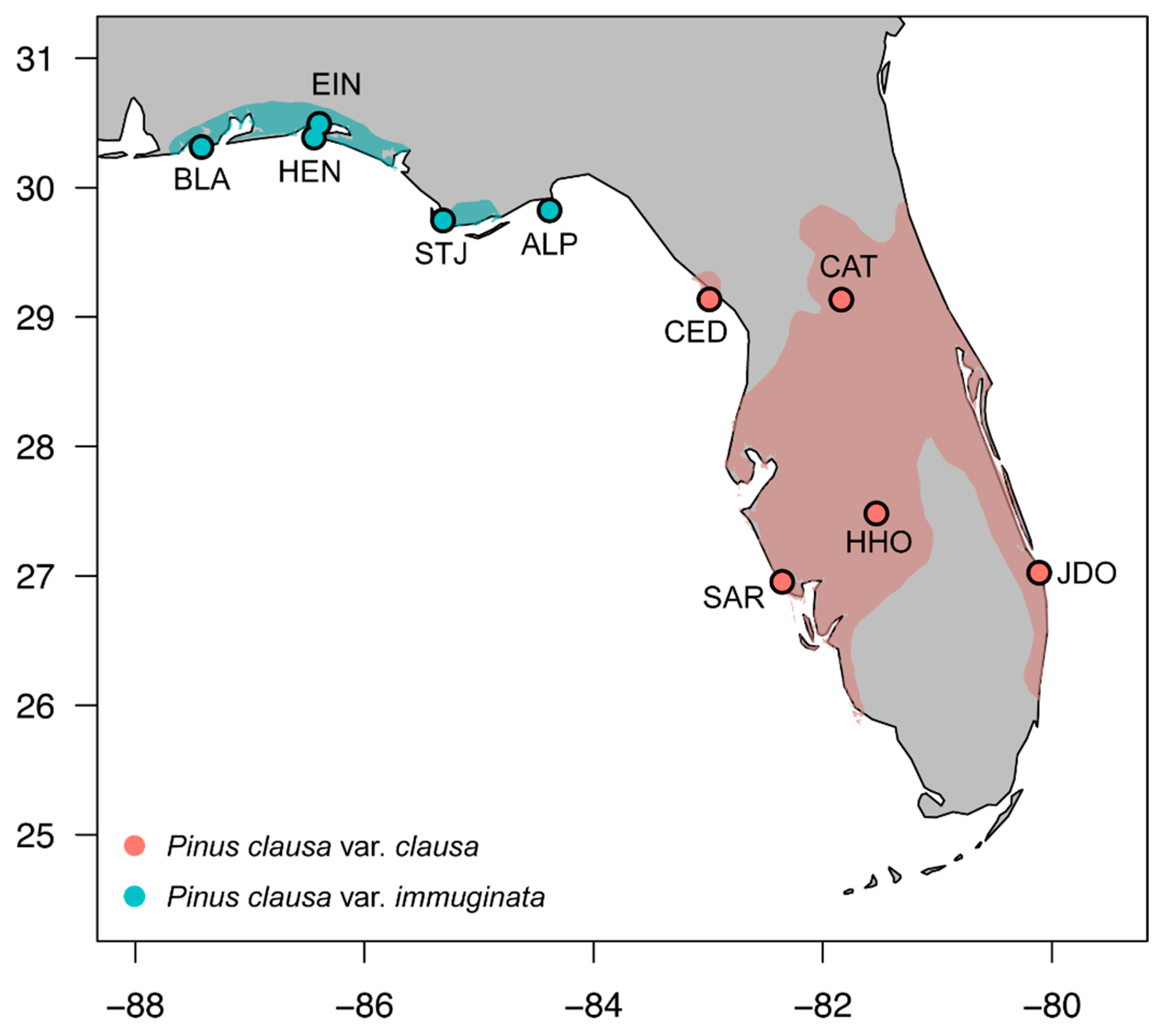



| Site Abbreviation | Location Name | County | Latitude | Longitude | Average DBH |
|---|---|---|---|---|---|
| Pinus clausa var. clausa | |||||
| CAT | Ocala National Forest | Marion | 29°03′ N | 81°51′ W | 92 |
| CED | Cedar Key Scrub State Reserve | Levy | 29°11′ N | 83°01′ W | 89 |
| HHO | Highlands Hammock State Park | Highlands | 27°29′ N | 81°31′ W | 88 |
| JDO | Jonathan Dickinson State Park | Martin | 27°01′ N | 80°07′ W | 94 |
| SAR | Englewood Water District | Sarasota | 26°57′ N | 82°21′ W | 136 |
| Pinus clausa var. immuginata | |||||
| STJ | St. Joseph Peninsula State Park | Gulf | 29°48′ N | 85°25′ W | 76 |
| HEN | Henderson Beach State Recreation Area | Okaloosa | 30°23′ N | 86°28′ W | 66 |
| EIN | Eglin Air Force Base | Okaloosa | 30°29′ N | 86°24′ W | 81 |
| BLA | Big Lagoon State Recreation Area | Santa Rosa | 30°18′ N | 87°24′ W | 72 |
| ALP | Alligator Point | Franklin | 29°56′ N | 84°22′ W | 70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rúa, M.A. Characterizing Ectomycorrhizal Fungal Community Structure and Function of Two Varieties of Pinus clausa That Differ in Disturbance History. Forests 2021, 12, 219. https://doi.org/10.3390/f12020219
Rúa MA. Characterizing Ectomycorrhizal Fungal Community Structure and Function of Two Varieties of Pinus clausa That Differ in Disturbance History. Forests. 2021; 12(2):219. https://doi.org/10.3390/f12020219
Chicago/Turabian StyleRúa, Megan A. 2021. "Characterizing Ectomycorrhizal Fungal Community Structure and Function of Two Varieties of Pinus clausa That Differ in Disturbance History" Forests 12, no. 2: 219. https://doi.org/10.3390/f12020219
APA StyleRúa, M. A. (2021). Characterizing Ectomycorrhizal Fungal Community Structure and Function of Two Varieties of Pinus clausa That Differ in Disturbance History. Forests, 12(2), 219. https://doi.org/10.3390/f12020219






