Abstract
Riparian forests were frequently cleared and converted to agricultural pastures, but in recent times these pastures are often revegetated in an effort to return riparian forest structure and function. We tested if there is a change in the soil bacterial taxonomy and function in areas of riparian forest cleared for agricultural pasture then revegetated, and if soil bacterial taxonomy and function is related to vegetation and soil physicochemical properties. The study was conducted in six riparian areas in south-eastern Australia, each comprising of three land-use types: remnant riparian forest, cleared forest converted to pasture, and revegetated pastures. We surveyed three strata of vegetation and sampled surface soil and subsoil to characterize physicochemical properties. Taxonomic and functional composition of soil bacterial communities were assessed using 16S rRNA gene sequences and community level physiological profiles, respectively. Few soil physiochemical properties differed with land use despite distinct vegetation in pasture relative to remnant and revegetated areas. Overall bacterial taxonomic and functional composition of remnant forest and revegetated soils were distinct from pasture soil. Land-use differences were not consistent for all bacterial phyla, as Acidobacteria were more abundant in remnant soils; conversely, Actinobacteria were more abundant in pasture soils. Overall, bacterial metabolic activity and soil carbon and nitrogen content decreased with soil depth, while bacterial metabolic diversity and evenness increased with soil depth. Soil bacterial taxonomic composition was related to soil texture and soil fertility, but functional composition was only related to soil texture. Our results suggest that the conversion of riparian forests to pasture is associated with significant changes in the soil bacterial community, and that revegetation contributes to reversing such changes. Nevertheless, the observed changes in bacterial community composition (taxonomic and functional) were not directly related to changes in vegetation but were more closely related to soil attributes.
1. Introduction
Land-use change can strongly affect vegetation, soil physicochemical properties, and soil bacterial communities [1,2,3,4]. Soil bacteria are essential to the biogeochemical cycles of terrestrial ecosystems, as they return nutrients and organic matter to the soil, carrying out metabolic activities key to decomposition processes [5,6]. At the same time, bacterial community abundance, composition, and activity may be critical in improving soil properties and increasing plant establishment after land-use change [7,8,9]. Understanding the relationships between soil bacterial communities and plant community structure during the conversion of agricultural land to native vegetation is essential for increasing revegetation success and cost-effectiveness [10].
Soil bacterial communities can be copiotrophic where their abundance increases in environments rich in nutrients [11], or oligotrophic and thrive in areas with low nutrients. Bacterial community structure has been shown to covary with soil texture and salinity [12], soil fertility (nitrogen, phosphorus, potassium, pH), and soil organic matter [13]. For instance, increases in organic matter have been shown to increase relative abundance of Actinobacteria and decrease that of Acidobacteria, while reducing organic matter has the opposite effect [14]. Furthermore, vegetation properties can influence soil bacterial abundance, activity, and taxonomic composition [15,16]. Previous studies have shown differences in the soil bacterial communities were related to exotic and native ground-cover vegetation [17]. These vegetation-driven changes in soil bacteria may be a bacterial response to plant root exudations, which are carbon-based compounds comprised of organic acids, carbohydrates, and amino acids [18,19].
The clearing of forest and conversion to agricultural pasture is associated with a range of changes which can have implications for soil bacterial communities. Disturbance from land-use change due to agricultural intensification is known to be especially detrimental to soil bacterial communities as it causes imbalances in soil nutrient pools [20,21,22]. Agricultural intensification in grasslands can have a greater impact on bacterial taxonomic composition than on function [1]. Forest-to-pasture conversion altered bacterial community composition in the Amazon rainforest; specifically, alpha diversity increased while beta diversity decreased [23]. In addition to changes in soil bacterial taxonomic composition, Rodrigues, et al. [23] found pasture communities were linked to increases in soil carbon, nitrogen, and magnesium concentrations. The shift from woody vegetation to pasture species decreases rooting depth, affecting bacterial communities in surface soil rather than subsoil [2,3,4,24,25]. Surface soil has higher bacterial activity and biomass [2,3,6,26] and higher density of plant roots [25]. Studies show that land-use change influences bacterial taxonomic and functional composition, a trend that has been observed in grasslands [14], forests [27,28], and wetlands [22,29], but has not been yet been fully explored in Australian riparian ecosystems.
Revegetation is often conducted with the purpose of increasing functionality and reverting composition to the pre-disturbance state [28]. However, revegetation success is often determined by evaluating changes aboveground rather than belowground [30]. Due to the complexity and variability observed in the taxonomic and functional distribution belowground, the changes to microbial communities (including fungi, bacteria and archaea) are often called ‘the black box of microbial diversity’ [31]. For instance, in grasslands revegetated sites had higher bacterial taxonomic and functional diversity compared to agricultural sites, and bacterial taxonomic and functional diversity was positively correlated with plant diversity [14]. However, in coastal forest revegetation, soil bacterial taxonomic diversity was related to soil physicochemical properties, and revegetated ex-agricultural pastures were comparable to the remnant sites after 15 years [32]. Yan, et al. [32] also showed a clear effect of soil depth at phyla level, where soil depth had a significant effect on many taxa, most showed greater diversity and abundance in surface soils. In riparian ecosystems it is currently unclear what vegetation or edaphic properties are related to bacterial (taxonomic and functional) composition, how conversion to pasture and subsequent revegetation may impact soil bacterial community composition and function, or if recently revegetated sites (<10 years post planting) are similar to remnant sites.
Riparian ecosystems have high plant diversity, complex stream geomorphology, and are centers of anthropogenic activity. They are often referred to as a complex mosaic of habitats that differ in soil properties, moisture availability, and plant community composition [33,34,35,36,37,38]. Due to the high fertility of riparian soils and easy access to watercourses, these ecosystems were often converted to productive agricultural areas for cultivation and livestock [10,39]. This conversion has affected many waterways worldwide, resulting in the loss of capacity to provide valuable ecosystem services in riparian areas [40]. In recent decades, a greater understanding of the relevance that riparian forests play in landscape hydrology has made revegetation of this area a high priority [41].
It is assumed that since soil bacterial communities are significantly influenced by vegetation complexity [42,43,44], bacterial communities would also be recovered through revegetation. However, to what extent vegetation clearance affects soil abiotic and biotic factors and whether that effect is reverted through revegetation with native riparian forest species is unknown. To address this gap, we investigated bacterial community composition (taxonomic and functional) in riparian ecosystems that have been converted to agricultural pasture and later revegetated. We aimed to address the following questions: (a) Does soil bacterial community composition (taxonomic and functional) change with the conversion of riparian forest to agricultural pasture? (b) To what extent does revegetation of pasture restore the soil bacterial community composition (taxonomic and functional) to its original state? (c) Do land-use change effects on bacterial composition differ in the surface soil (0–10 cm) relative to subsoil (20–30 cm)? (d) Which vegetation properties and/or soil physicochemical properties better predict the soil bacterial community composition (taxonomic and functional)? We hypothesized that bacterial community composition (taxonomic and functional) would change with land use and soil depth. Furthermore, we predicted that composition in revegetated areas would be most similar to remnant riparian vegetation, particularly in the surface soil.
2. Materials and Methods
2.1. Study Sites
Six study sites were located around the city of Melbourne in south-eastern Australia near different rivers and creeks. Sites encompassed a range of climate, topographies, and soil types (Figure 1, Table 1).
Figure 1.
Map of the six study sites, showing the location of the state of in south-eastern Australia (top left), the state of Victoria (top right), and the six study locations (bottom).

Table 1.
Characteristics of the six study sites around the city of Melbourne in south-eastern Australia.
Each site supported three land-use types representative of the conversion of riparian forest to cleared pasture, and the subsequent revegetation of pasture with species native to remnant riparian forest. Remnant riparian forest was dominated by Eucalyptus viminalis Labill., Eucalyptus ovata Labill., and Acacia dealbata Link; pasture was dominated by exotic grasses including Holcus lanatus L. and Anthoxanthum odoratum L. and periodically grazed by sheep or cattle. Revegetated areas were formerly pasture that had been planted with nursery grown indigenous trees and shrubs typical of the riparian forest and were three to nine years of age at time of sampling (Table 1, Supplementary Table S1).
2.2. Vegetation Surveys
One 25 m × 25 m quadrat (625 m2) was set out within each land-use type (remnant, revegetated, and pasture) at each site for sampling vegetation and soils. Quadrats were located 50–2057 m apart across all sites. Vegetation surveys were conducted in three strata defined as: canopy (trees with diameter at breast height (DBH) ≥15 cm), sub-canopy (trees and tall shrubs with DBH <15 cm), and ground cover (grasses and forbs). Cover and basal area of canopy species were assessed using the point-centered quarter method [47], at four randomly selected points within each quadrat. For each quadrant we measured diameter and distance from each point to the nearest tree with a DBH ≥15 cm. Average point to plant distance (APPD) was calculated by summing total distance measured from point to tree and dividing by the number of plants. For pasture quadrats with sparse canopy, apart from the occasional canopy tree, a maximum value for APPD of 50 m was applied. Basal area was calculated across all plants and for Eucalyptus species. Relative density of canopy species was calculated by dividing the number of individuals found at each land-use type by the total number possible for a quadrat (4 points × 4 quadrants = 16).
Cover of the sub-canopy was assessed via the line-intercept method, with three 20 m transects established within each quadrat [48]. Transects were placed 2 m inside the north boundary of each quadrat at 7 m, 14 m, and 21 m. For each transect, estimates of percentage vegetation cover were recorded at 1 m intervals for vegetation 1.5 m aboveground. We used the line intercept data to determine the total number of species and total number of Acacia spp. in the sub-canopy. For ground cover, assessments were made using a small 1 m2 quadrat placed every 5 m along each transect (5 quadrats per transect). In small quadrats we determined; cover of litter, bare ground, cover of each species and total cover as a percentage, estimated to the nearest 5%.
2.3. Soil Sampling and Analyses
2.3.1. Soil Sampling and Physicochemical Analyses
Soils were sampled from within each quadrat according to protocols established by the Biomes of Australian Soil Environments (BASE) project [49] and our data is available online in the BASE database. Samples were collected in November 2017, Austral spring. Each soil sample was a composite of 27 cores, taken within a 1 m radius of the nine points regularly spaced within each quadrat, at two depths (0–10 cm and 20–30 cm) using a soil auger (27 mm inner diameter). Soils were kept on ice until homogenized in the laboratory when anything larger than 5 mm was removed (litter, roots, and stones). Two 50 mL tubes were each filled with approximately 20 g of field moist homogenized soil which was frozen at −80 °C for later genetic analysis and community-level physiological profiling (CLPP). The remaining soil was weighed, then dried in an oven at 40 °C and weighed again for soil moisture (%). Approximately 500 g of soil was sent to CSBP Soil and Plant Analysis Laboratory (Bibra Lake, Western Australia) for soil physical and chemical analyses including: pH, electrical conductivity, available nitrogen, available phosphorus, sulfur, organic carbon, total nitrogen, exchangeable cations (Mg, K, Na, Ca), extractable trace elements (Cu, Fe, Mn, Zn, Al, B), and soil particle size [50]. Soil bulk density was measured by the volumetric method for surface soil (0–10 cm depth) in one core per quadrat. Samples were not adjusted for stone content and dried at 80 °C.
2.3.2. Soil DNA Extraction, Sequencing, Operational Taxonomic Unit (zOTU) Identification, and Filtering
DNA extraction and sequence analyses were conducted according to the methods described in Bissett et al. [49], with amplicon sequence variants produced instead of 97% zOTUs. Briefly, soil DNA was extracted and then pooled from 3 × 0.25 g soil replicates from each sample at the Australian Genome Research Facility (AGRF, Adelaide, Australia) using MoBio powersoil DNA extraction kits (MoBio, Carlsbad, CA, USA) according to manufacturer’s instructions. Bacterial 16S ribosomal DNA was PCR-amplified using primers 27F and 519R [51]. Sequences (300 bp PE) were then produced using the Illumina MiSEQ platform. Sequenced 16S rRNA gene amplicons were merged with FLASH [52]. Merged reads <400 bp and those containing Ns or homopolymer runs >8 bp were removed using MOTHUR v1.34.1 [53]. Sequences were denoised using the UNOISE3 algorithm [54] and USEARCH v11 [55] (–minsize 4) with abundance profiles built by mapping all data to identified zOTU (usearch–otutab–maxaccepts 0). Amplicon sequence variants were classified using the RDP classifier [56] in MOTHUR at a 60% probability cut-off, against SILVA132 [57].
Across all samples we found a total of 127,662 bacterial zOTUs across 40 phyla. Data were then filtered to include only phyla with 2% or more of the total number of zOTUs found, leaving 10 phyla. The reduced dataset had 120,863 zOTUs. The number of zOTUs present in samples ranged from 62,959 to 143,581, with a mean of 101,756. Samples were not rarefied as preliminary analysis showed no significant difference in the sequence numbers per sample among land use and depth, and observed richness was strongly correlated with rarefied richness ([58] Supplementary Figures S1 and S2). Singletons at sample level were removed from dataset and the number of observed zOTUs per sample was used for further analyses. No significant differences in the number of singletons removed were observed based on land use or depth (data not shown). The filtered dataset was used to investigate differences in abundance, richness and Shannon’s diversity index [59] with land use, and depth overall, and by phyla. Relative abundance was calculated by dividing the total sequence abundance for each phylum by the sum total sequence abundance for that land use and depth.
2.3.3. Bacterial Functional Composition (Community-Level Physiological Profiles—CLPP)
Community-level physiological profiling (CLPP) was used to characterize soil bacterial community carbon use as a measure of activity and functional composition. CLPP was measured using Biolog EcoPlates™, which have tetrazolium violet dye and 31 different carbon compounds that have a redox reaction when a carbon source is used [60]. The redox reaction makes the well change color and measurements of the intensity of bacterial activity for specific carbon sources can be collected using a spectrometer. Color development only occurs once the total number of cells in a well reaches approximately 108 cells m−1 [61,62], triggering the color change.
A solution of 5 g of field-moist frozen soil from each sample was suspended in 45 mL of 0.87% NaCl solution, shaken for 30 min and left to settle for a further 30 min in accordance with Garland [60]. A 130 µL aliquot of the soil solution was added to each EcoPlate well (one EcoPlate per soil sample). The EcoPlates were dark incubated at 30 °C for 168 h during which time the optical density (OD) of plates was read every 24 h at 590 nm using a photometric microplate reader with an incubator (Multiscan GO, Thermo Scientific, Ratastie, Finland). Plates were shaken for 30 s to mix the solution after each read. OD was corrected by subtracting the control well OD (only contained reverse osmosis water) from carbon source OD, for each well from each plate. Values <0.25 and negative values were set to zero. Each EcoPlate had 96 wells comprising of three replicates of 31 carbon sources (Supplementary Table S2).
Average well color development (AWCD) for each carbon source was calculated as the average OD of the three replicate wells containing the same carbon source in each plate [60]. The optimal range for OD values was reached at 120 h, after this point the curve began to flatten. Area under the curve (AUC) used AWCD at a range of time points () in the upwards slope of the curve (0, 24, 48, 72, 96, 120 h, respectively). AUC is an integral rate of increased activity overtime [63,64] which captures the differences in carbon source utilization profiles by integrating all the information contained in the kinetic variables [65,66]:
AUC has been used to compare changes over time with those observed at one time point (AWCD). However, methods for CLPP statistical analysis have not been standardized yet [64], and many studies use more than one measure of CLPP to quantify changes (e.g., Jałowiecki, et al. [63], Poyraz and Mutlu [67], and Schmitt, et al. [68]).
Further analyses used AWCD and AUC to investigate differences in catabolism (abundance), number of different carbon sources used (richness), and evenness of response (Shannon’s diversity index [59]) based on land-use, and depth overall. Then similar carbon compounds were grouped into guilds (carbohydrates, carboxylic and ketonic acid, amino acids, amines/amides, or polymers, Supplementary Table S2). AWCD was used to investigate overarching patterns in catabolism, richness, and evenness within carbon guilds for land use and depth. Relative catabolism was calculated by dividing the total AWCD for each carbon guild by the sum total catabolism for that land use and depth.
2.4. Statistical Analyses
Prior to analyses, data was either log10 or square-root transformed as necessary to meet assumptions of normality and homogeneity of variance. Significance of land use and depth (soils only), effects on vegetation, soil physicochemical properties, and bacterial community (taxonomic and functional composition) were assessed using generalized linear models (GLMs). Land use and depth were fixed factors, and site (river/creek) was a random factor. Models used the Gaussian method and were implemented using R package lme4 [69]. Significant pair-wise differences were assessed using post-hoc Tukey HSD tests using R package emmeans [70].
Non-parametric permutational multivariate analysis of variance (PERMANOVA) was used to test the overall effects of land use and depth on environmental variables (vegetation, soil properties) and bacterial community taxonomic and functional composition. Environmental variables (vegetation properties and soil physicochemical properties) had significance testing of the Euclidean dissimilarity measures and post-hoc comparisons were made using 999 permutations in the R package vegan version 2.4-6 [71]. Bacterial community taxonomic (within class) and functional composition (within guild) were visualized using non-metric multidimensional scaling (NMDS). The ordination used Bray–Curtis distances from Hellinger transformed data, with 999 permutations. Dispersion was assessed by calculating the average distance between data points and the centroid in multivariate space with the significance of differences (by land use or depth) analyzed using ANOVAs, with post-hoc Tukey HSD tests. Analyses were conducted using vegan version 2.4-6 R package [71].
Principal component analysis (PCA) was used to identify differences in variables between land-use types and depth. PCA was conducted on vegetation and soil properties separately. Visualization of PCA used the function prcomp in R core stats package [72], and packages factoextra [73] and FactoMineR [74]. Dimensions for variables with eigenvalues ≥1 that cumulatively explained 80% or more of the variation were used in redundancy analysis (Supplementary Table S3).
Redundancy analysis (RDA) was used to assess the relationship between environmental properties and soil bacterial community composition (taxonomic and functional). Variables included in the RDA are dimensions from the vegetation and soil PCAs that were fitted using the envfit function in vegan package [71]. Vegetation properties have been duplicated for both depths; bulk density and soil moisture have been removed as they were only sampled in surface soil. All statistical analyses were conducted in R version 1.0.153 [72].
3. Results
3.1. Environmental Factors
3.1.1. Vegetation
Vegetation diversity differed among land-use types. Species richness of canopy, sub-canopy, and native ground cover in remnant and revegetated land-uses was significantly greater than in pasture (Table 2). Pasture lacked canopy and sub-canopy vegetation, had larger APPD than other land uses, and had a greater richness of exotic ground cover (Table 2). Average tree basal area and total density of canopy species were similar between revegetated and remnant land uses; however, remnant quadrats had greater sub-canopy cover, greater native ground cover richness, and lower exotic ground cover richness than revegetated quadrats (Table 2). Consistent with most univariate analyses, multivariate PERMANOVA indicated significant differences in vegetation properties between pasture and other land uses, but no differences between remnant and revegetated land use (Table 2).

Table 2.
Vegetation properties of the three land-use types (P, pasture; RV, revegetated; RM, remnant) across six sites. Values represent means (±SE) with p values from general linear models (GLM) testing for the effect of land use (L). Significant differences (Tukey HSD test) between land uses are indicated by superscript letters and are bold for p values < 0.05. Study site was included as a random factor. PERMANOVA tested for differences in the Euclidean distance matrix among land-uses across all variables.
Principal component analysis (PCA) of vegetation properties clearly separated quadrats based on land use (Figure 2a for PC1 and 2, all other variations in Supplementary Figure S3). The first four components explained 84% of the total variance with each subsequent component explaining less than 10% of the variance (Supplementary Table S3). PC1 (45% variance explained) reflected the results of the GLM and PERMANOVA (Table 2) and separated pasture from revegetated and remnant quadrats. Accordingly, along PC1, the separation of quadrats by land use was most strongly correlated with canopy richness, average basal area, abundance of eucalypts, and total richness of sub-canopy (contribution >10%, Supplementary Figure S5). None of the other PC axes showed a clear separation of quadrats based on land use, instead reflecting the variability in ground cover characteristics (bare ground, litter, total ground cover, Figure 2a, Supplementary Figure S3).
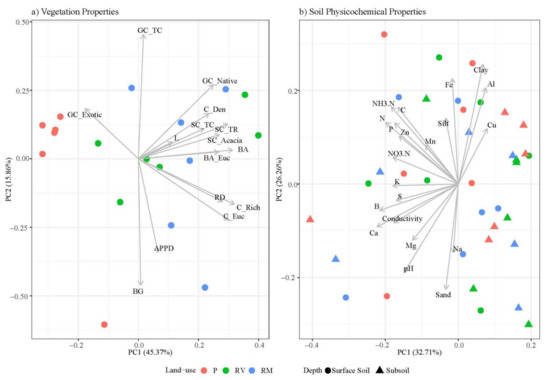
Figure 2.
PCA of (a) vegetation properties and (b) soil physicochemical properties. Points are colored based on land use (red = pasture (P), green = revegetated (RV), and blue = remnant (RM)) with symbols distinguishing soil depth (circle = surface soil and triangle = subsoil). Abbreviations for vegetation are described in Table 2 and those for soil in Table 3.
3.1.2. Soil
A limited number of soil physicochemical properties differed with land use, which included significantly greater available P, clay, and silt in pasture relative to the other land uses and significantly lower pH in remnant relative to other land uses (Table 3). Soil nutrients consistently decreased with depth, although these differences were not always significant (Table 3). C and N decreased significantly with depth but did not differ with land use (Table 3).

Table 3.
Soil physicochemical properties of the three land-use types (P, pasture; RV, revegetated; RM, remnant) across six sites. Values represent means (±SE) with p values from general linear models (GLM) testing for the effects of land use (L) and depth (D).
In accordance with the univariate results, multivariate PERMANOVA indicated no overall effects of land use or sampling depth on soil physicochemical properties (Table 3) and the PCA of soil physicochemical properties showed no clear separation of quadrats according to land-use or depth (Figure 2b for PC1, Supplementary Figure S4). The first five PCs explained 83% of the total variance with each subsequent component, explaining less than 10% of the variance (Supplementary Table S3). PC1 (33%) separated the quadrats along an overall gradient of fertility whereas separation along PC2 (26%) was largely driven by differences in soil texture (Supplementary Figure S6). PC3 (11%) and PC4 (7%) separated quadrats based on Mn and Na, respectively.
3.2. Bacterial Community Taxonomic Composition
Bacterial community taxonomic composition at the phylum level differed significantly among land uses (PERMANOVA, p = 0.01, F = 3.25), where remnant soils were significantly different to pasture (Figure 3a, Table 4). Revegetated soil was similar to pasture (p = 0.52) and remnant (p = 0.09) soil (Table 4). There were no significant differences in dispersion (p = 0.91, F = 0.09).
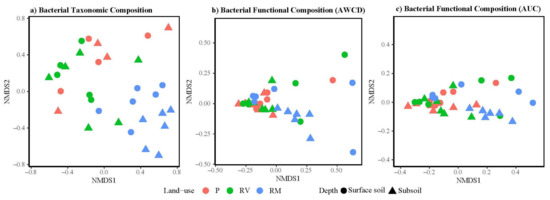
Figure 3.
Non-metric multidimensional scaling (NMDS) ordination of (a) bacterial taxonomic composition (stress 0.15, non-metric R2 0.87); (b) bacterial functional composition (community-level physiological profiling (CLPP)—represented as average well color development (AWCD), stress 0.07, linear fit R2 0.99); and (c) bacterial functional composition (CLPP—represented as area under the curve (AUC), stress 0.04, linear fit R2 0.99). Points are colored based on land use (red = pasture (P), green = revegetated (RV), and blue = remnant (RM)) with symbols distinguishing soil depth (circle = surface soil and triangle = subsoil).

Table 4.
PERMANOVA pairwise comparisons for each land-use combination, p values < 0.05 are in bold.
Bacterial richness and Shannon’s diversity index (diversity) were significantly greater in remnant and revegetated soils than in pasture, a trend that was evident both in top and subsoil (Figure 4b,c). Across selected phyla there were relatively few significant differences in abundance, richness, and diversity with land use, and the direction of any land-use effects was not consistent among phyla. For example, remnant soils were characterized by a significantly higher abundance, richness, and diversity of Acidobacteria than pasture and revegetated quadrats. Whereas Actinobacteria showed the opposite trend, with significantly higher abundance, richness, and diversity in pasture and revegetated quadrats than remnant quadrats (Supplementary Figures S7–S9).
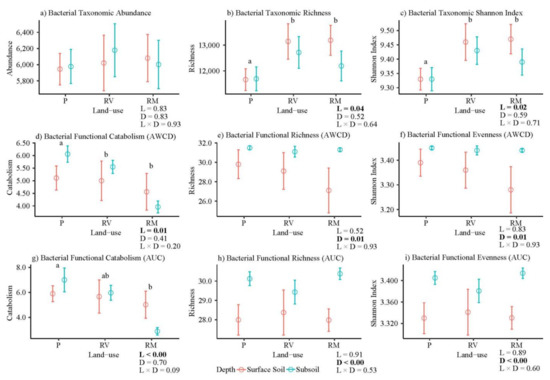
Figure 4.
Overall bacterial taxonomic and functional diversity (abundance/catabolism (a,d,g), richness (b,e,h), Shannon’s diversity index (c,f,i)). Values represent means according to land use (P = pasture, RV = revegetated, and RM = remnant) and colored by soil depth. Bars represent standard error of the mean, p values for GLM shown below graphs (L = land-use, D = depth, and L × D = interaction), bold indicates significant difference (p values < 0.05). Letters indicate significant pairwise differences among land uses (across depth) as determined by Tukey HSD tests.
Overall, bacterial community taxonomic composition did not differ with depth (p = 0.22, F = 1.16); however, richness, relative abundance, and diversity of Proteobacteria and Bacteriodetes were greater in surface soil than in subsoil. Furthermore, Chloroflexi had higher species richness, relative abundance, and diversity in subsoil than surface soil (Supplementary Figures S7–S9).
Comparisons of relative abundances of sequences by phylum showed that the most abundant bacterial phylum found in the study was Proteobacteria (~32%), followed by Acidobacteria (~23%, Figure 5a). Surface soil of pasture was characterized by a higher proportion of Firmicutes (15%), with all other land uses or depths having 7% or less (Figure 5a).
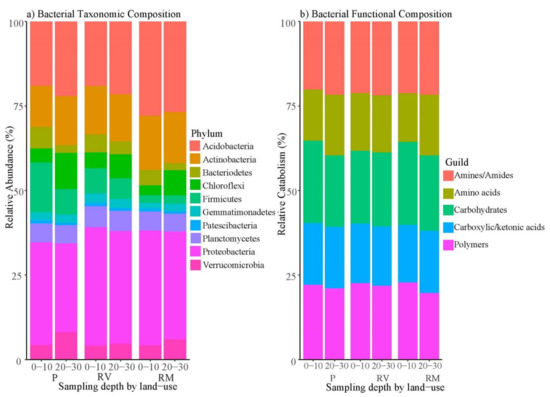
Figure 5.
Relative abundance of bacterial community showing (a) bacterial taxonomic composition by phylum and (b) bacterial functional composition (CLPP—represented as AWCD) by carbon guild for each by land-use type; pasture (P), revegetated (RV), and remnant (RF), indicating proportions of each phylum or guild by land use across both depths (0–10 cm = surface soil and 20–30 cm = subsoil).
3.3. Bacterial Functional Composition (Community-Level Physiological Profiles—CLPP)
Bacterial functional composition measured as the average well color development (AWCD—at 120 h) or area under the curve (AUC—incorporates all time intervals 0–120 h) are indicative of the intensity of the bacterial activity. Bacterial functional composition varied marginally for carbon compounds with land use (AWCD p = 0.07 F = 2.36, and AUC p = 0.07 F = 2.45) and no difference was observed with sampling depth (AWCD p = 0.55 F = 0.53, and AUC p = 0.37 F = 0.89, Figure 3b,c). Pairwise PERMANOVA showed no differences for any land-use combinations for AWCD (Table 4). However, AUC showed that revegetated quadrats were different to remnant and pasture quadrats, with no differences between remnant and pasture (Table 4). The functional composition as depicted by AWCD in pasture quadrats had less variability between quadrats than in other land-use types (PERMANOVA dispersion, p = 0.01 F = 5.35, Figure 3b). Variability in functional composition of revegetated quadrats measured as AUC was less than for other land-use types (PERMANOVA dispersion, p = 0.01 F = 5.94, Figure 3c).
Bacterial functional catabolism differed among land uses, but the variation was mostly due to differences in the subsoil (AWCD and AUC, Figure 4d,g). Both AWCD and AUC showed a similar trend where remnant soil had the lowest bacterial catabolism, pasture had the highest, and revegetated soil was in between (Figure 4d,g). Among functional guilds, there was significantly higher catabolism of amines/amides in pasture quadrats than revegetated or remnant quadrats (AWCD, Supplementary Figure S10a). Remnant quadrats had significantly lower catabolism of all carbon guilds (amino acids, carbohydrates, and carboxylic and ketonic acids) than pasture and revegetated quadrats, polymers were the exception where no differences were found (AWCD, Supplementary Figure S10b–d).
Bacterial functional richness and evenness of response (diversity—Shannon’s diversity index) were significantly greater in subsoil than in surface soil with no overall land-use effect detected, although both measures tended to be greater in pasture soils and lower in remnant quadrats (Figure 4e,f,h,i). Notably, bacterial functional richness and diversity of carbon source usage varied among land uses in the surface soil (0–10 cm) but remained similar in the deeper soil (20–30 cm, Figure 4e,f,h,i). Remnant quadrats showed larger increases in bacterial functional richness with soil depth than other land uses, catabolizing more of the 31 carbon sources in subsoil (Figure 4e,f,h,i). Across all land uses, there was greater variability, both overall and within guilds for bacterial functional diversity measures, in surface soil than subsoil (Figure 4 and Supplementary Figures S10–S12).
Among carbon substrates, pasture soils harbored a greater bacterial functional richness within carbohydrates, carboxylic and ketonic acids than remnant soils, with revegetated soils intermediate (AWCD, Supplementary Figure S11). Only carboxylic and ketonic acids showed this difference in diversity (AWCD, Supplementary Figure S12). In regard to catabolism of particular carbon substrates, revegetated and remnant soils shared similar lower levels of amine/amide catabolism than pasture soil, whereas revegetated and pasture soils shared similar higher levels of carboxylic and ketonic acid catabolism than remnant soils (Supplementary Figure S10). Little variation in relative catabolism was observed for land use or depth (Figure 5b). Carbohydrates had the highest relative catabolism at 23%, closely followed by polymers (22%, Figure 5b).
3.4. Relationships between Environmental Factors and Bacterial Community Composition
Environmental variables explained 56% of the constrained variation in bacterial community taxonomic composition (p < 0.00, Figure 6a,b). RDA1 and RDA2 explained 15% and 7.8% of the total variation. No clear groups were observed based on land use or depth (Figure 6a). Bacterial community taxonomic composition was not related to any vegetation principal components (Supplementary Table S4) but was related to two different soil physicochemical principal components: the overall gradient of fertility (Soil PC1, p < 0.00), and the soil texture (Soil PC2, p < 0.00, Supplementary Figure S4). RDA1 was positively correlated with both Soil PC1 and Soil PC2, with RDA2 positively correlated with Soil PC2 but not Soil PC1 (Figure 6a). The positioning of bacterial phyla on the RDA illustrates that high abundance of Proteobacteria and Bacteriodetes was related to nutrient-poor soils (particularly low in Ca), with high abundance of Chloroflexi and Verrucomicrobia found in higher fertility soils (Figure 6a). Soils with high proportions of clay and sand had high abundance of Acidobacteria, although more phyla preferred soil with low proportions of clay and sand (Firmicutes, Planctomycetes, Gemmatimonadetes, Patescibacteria, and Actinobacteria—Figure 6a).
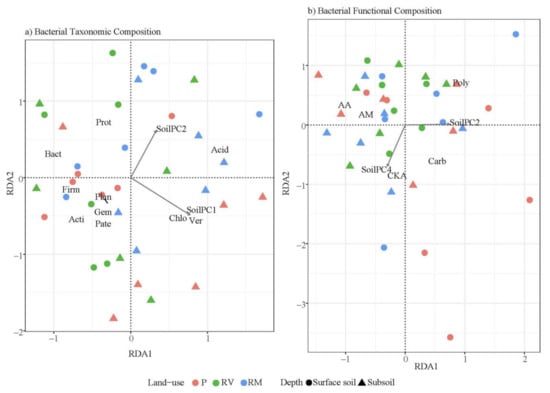
Figure 6.
Redundancy analysis using environmental variables from PCA dimensions as seen in Figure 1 and Supplementary Figures S1 and S2 to explain variation in (a) bacterial taxonomic composition by phyla and direction of constraints from principal components, (b) bacterial functional composition showing the direction of constraints from principal components (CLPP—represented as AWCD). Points are colored based on land use (red = pasture (P), green = revegetated (RV), and blue = remnant (RM)) with symbols distinguishing soil depth (circle = surface soil and triangle = subsoil). Only significant environmental variables are shown. Bacterial taxonomic composition had a p value of <0.00 and R2 of 0.42, and bacterial functional composition had a p value of 0.01 and R2 of 0.17. Names for phyla have been abbreviated in Figure 6a; abbreviations are as follows: Proteobacteria (Prot), Actinobacteria (Acti), Acidobacteria (Acid), Planctomycetes (Plan), Chloroflexi (Chlo), Firmicutes (Firm), Bacteroidetes (Bact), Verrucomicrobia (Ver), Gemmatimonadetes (Gem), and Patescibacteria (Pate). Names for carbon guilds have been abbreviated in Figure 6b; abbreviations are as follows: Amines and amides (AM), amino acids (AA), carbohydrates (Carb), carboxylic and ketonic acids (CKA), and polymers (Poly).
Environmental variables explained 38% of the constrained variation in bacterial functional composition (p = 0.02, R2 = 0.17, Figure 6b). RDA1 and RDA2 explained 7.6% and 4.8% of the total variation, respectively. Similar to bacterial community taxonomic composition, no clear groups were observed based on land use or depth for bacterial functional composition, and no relationships with vegetation properties were found (Figure 6b and Supplementary Table S4). Bacterial functional composition was related to two different soil physicochemical principal components: a gradient in soil texture (Soil PC2, p < 0.00) and in salinity (Na) (Soil PC4, p = 0.01 in Supplementary Figure S4). Soil PC2 was positively correlated with RDA1 and RDA2, whereas Soil PC4 was negatively correlated with both RDA1 and 2 (Figure 6b). The positioning of carbon guilds on the RDA shows that higher carbohydrate catabolism was related to greater proportions of clay and sand, whereas increased catabolism of amines/amides and amino acids was related to lower proportions of clay and sand (Figure 6b). Carboxylic acids were catabolized more in soils with high Na; conversely, high carbohydrate catabolism is related to low Na (Figure 6b).
4. Discussion
Our study investigated the response of bacterial taxonomic and functional composition to land-use change, specifically the conversion of riparian forest to cleared pasture, and subsequent revegetation of that pasture with species native to local riparian forest. We found pasture was characterized by a lack of tree canopy cover, higher soil P and Ca levels and more acidic pH (~4), higher proportions of clay and silt, with lower bacterial taxonomic richness and diversity, but higher bacterial functional activity than revegetated or remnant areas. Soils in revegetated areas were intermediate in overall bacterial taxonomic and functional composition. Consistent patterns were found for bacterial taxonomic and functional composition with soil depth regardless of land use. Contrary to our hypotheses, there was no relationship between vegetation properties and bacterial community composition (taxonomic or functional). Instead, we found that bacterial community composition was related to soil texture and chemistry.
4.1. Changes in Bacterial Taxonomic Composition with Land Use and Depth
Remnant and revegetated soils had significantly greater bacterial taxonomic diversity despite having a similar level of abundance to pasture as observed in GLMs. Pasture had the lowest richness and diversity across most phyla studied. Species composition in remnant riparian forest soils was different to that in pastures, with revegetated soils showing intermediary composition. The intermediary composition of revegetated soils could be indicative that soil bacterial communities are on a trajectory of recovery [75]. No differences were found overall with depth, yet richness, abundance, and diversity of Proteobacteria and Bacteriodetes were greater in surface soil than in subsoil and Chloroflexi had higher species richness, abundance, and diversity in subsoil. Our results suggest that revegetation has contributed to the return of some components of bacterial taxonomic diversity, but recovery is lacking in some phyla (e.g., Acidobacteria, Actinobacteria, Gemmatimonadetes, and Firmicutes). Despite bacterial taxonomic composition showing differences with land use, these patterns could not be associated with differences in vegetation properties.
Bacterial community composition (taxonomic and functional) has been linked to changes in vegetation [16,17,18,19], but we found no relationships with measures of vegetation properties and diversity. In our study, revegetated and remnant quadrats shared many vegetation properties with the exception of exotic ground cover. A study by Yang et al. [17] showed that invasion of an exotic cordgrass in wetlands significantly increased soil bacterial abundance, species richness, and diversity compared to three other native dominated vegetation communities. Our study contrasts Yang et al. [17], as quadrats with exotic ground cover (grasses and forbs) had less bacterial taxonomic diversity but higher functional catabolism and showed no relationship with vegetation properties. Previous research has shown plant diversity influences soil microbial (including fungi, bacteria, archaea) abundance [18]. Vegetation showed no significant relationship to bacterial community composition (taxonomic or functional) despite the inclusion of richness of native and exotic species as vegetation properties. The inclusion of alternative vegetation properties could have changed the lack of relationship found between soil bacterial communities and vegetation properties. Our results clearly point to soil physicochemical properties as the best predictors of soil bacterial communities.
Soil chemistry emerged as one of the most important predictors for soil bacterial taxonomic composition. Pasture soils had greater available P, clay, and silt relative to the other land-uses, with higher pH. Lauber et al. [76], found that the abundance of the largely oligotrophic phylum Acidobacteria was highest in acidic soils, whereas the largely copiotrophic phylum Actinobacteria had higher abundance in basic soils (pH 7–9). Our findings support Lauber et al. [76], as we observed greater proportions of Acidobacteria in remnant soils and Actinobacteria in pastures. Acidobacteria responds to changes in soil pH, potentially due to increased cell specialization and enzyme stability at more acidic pH [77]. This is consistent with our remnant soils being more acidic (~4) than revegetated and pasture (~5), and harboring a greater proportion of Acidobacteria. Higher abundance of Acidobacteria, a phylum generally more abundant in soils with low resource availability [77], has been associated with less disturbed areas [78]. This is in agreement with the poor nutrient status of Australian soils [79], and was observed in soils of remnant riparian forests. We found pasture soils were characterized as high nutrient sites with a fast turnover of nutrients [7,21], and had greater soil fertility. Our results contrast with research by Guo et al. [14] who found that agricultural pastures had higher proportions of Acidobacteria compared to revegetated pastures (5, 15, and 30 years post planting). This contrasting result might be related to nutrient differences since Guo et al. [14] measured total nitrogen 2–3 times higher than what we have observed.
Soil texture was the most important attribute underpinning changes in soil bacterial taxonomic diversity and composition. Soil texture shapes the microhabitats for bacteria and other members of the soil microbial community [80]. Seaton et al. [81] found that clay and silt particles were more important for soil bacterial composition than sand in a range of habitats including arable, improved, and unimproved grassland, broadleaved and coniferous woodland, and heathland. Similarly, Kim et al. [12] reported that soil texture was related to bacterial taxonomic composition, noting that plant community and drainage conditions were of secondary importance. However, previous research by Waymouth et al. [82] showed that variability in soil chemical properties, but not soil texture, was related to variability in soil bacterial taxonomic composition in different vegetation communities. The relationship between soil bacterial composition and soil texture found in this study may be due to differences between sites (river/creek) and thus reflect geographical variation in the taxonomic composition.
Our results showed some differences in the composition of soil bacterial communities with soil depth. Surface soil was characterized as having higher C and N, greater abundance, richness, and diversity of Proteobacteria and Bacteriodetes, but lower richness, abundance, and diversity of Chloroflexi. A study by Cong et al. [83] showed that soil bacterial abundance was lower in subsoil which had lower C content. Additionally, Upton et al. [24] found that, deeper in the soil (depths 30–100 cm), there was a decrease in abundance of Bacteroidetes and Proteobacteria which was likely due to decrease in C input. However, variation in Chloroflexi with depth may be related to changes in soil N along the soil profile. Research by Pedrinho et al. [84] showed a link between high abundance of Chloroflexi and nitrogen stress, which is in agreement with our results.
4.2. Changes in Bacterial Functional Composition with Land Use and Depth
Pasture had the highest catabolism of carbon sources followed by revegetated and remnant soils. The few differences found in soil nutrient content (i.e., P and Ca) followed the same trend, potentially due to agricultural nutrient inputs. Specific edaphic and anthropic factors such as pH, phosphorus availability and concentration, are important for the function of microbial communities [1]. Zhao et al. [29] found that, in wetlands, phosphorus was a determining factor for microbial activity, with higher catabolism of carbon sources in areas with higher phosphorus and denser vegetation (30–60% vegetation cover). This is consistent with the covariation of microbial activity and soil phosphorus found in our study. The intermediate levels of soil fertility and microbial activity of revegetated soil between remnant and revegetated soils found are in accordance with our findings from taxonomic composition. These findings further suggest that revegetation is reverting changes resulting from the conversion of riparian forest to agricultural pastures.
Our results indicate that bacterial functional composition was related to the proportion of clay and sand and the soil sodium content, a mineral related to salinity that affects soil texture (i.e., dispersiveness). Salinity has been previously found to influence the size and activity of soil microbial biomass [85,86,87]. Clay content is also known to influence soil microbial biomass [88,89], as it creates microsites and increases soil organic matter stabilization and water retention, thus changing soil conditions for microbial development [90]. Our findings are consistent with results from Zhao et al. [29] who showed a relationship between soil salinity, soil texture, and bacterial functional composition. Whilst we did not find a difference in salinity with land use, Main Creek was close to the ocean and had a slightly higher salinity, introducing potential site (river/creek) differences.
Richness and diversity of function was greater in subsoil (20–30 cm) than surface soil (0–10 cm). In subsoil, bacteria may have a starvation lifestyle [91], so diversity of activity or carbon use is important to maximize use of scarce resources. In our study, subsoil had lower carbon and nitrogen and could be related to the observed higher richness and evenness of soil bacterial functional composition. However, our finding contrasts with Tian et al. [92], who also found carbon and nitrogen decreased with increasing soil depth, but evenness decreased with depth in subtropical forests. Goberna et al. [93] found similar differences to Tian et al. [92], with bacterial evenness decreasing with depth, attributing differences to disturbances from agricultural practices as subsoil is less affected by these practices in Mediterranean semiarid forests. The contrasting results between our study and those of Tian et al. [92] and Goberna et al. [93] could be related to the distinct climate and rainfall in the study sites. The relationship between bacterial community composition and depth should be further explored in different climates to determine global patterns.
5. Conclusions
Soil bacterial community taxonomic and functional composition varied with land use and soil depth. Former riparian forests converted into agricultural pastures showed lower bacterial taxonomic diversity and higher bacterial functional activity than remnant riparian forests. Pastures and remnant forests also differed in their soil bacterial taxonomic composition. Revegetated soils harbored bacterial communities that were intermediate to those in pastures and remnant forests. Contrary to our expectations, soil bacterial community composition was strongly related to soil physicochemical properties, but not vegetation properties. In particular, soil texture (clay content) and soil fertility (P and Ca) had strong relationships with bacterial taxonomic and functional composition. Our results suggest that the conversion of riparian forest to agricultural land is associated with a significant change in the composition and diversity of the soil bacterial community, with a loss of taxonomic diversity and an increase in bacterial activity. This reflects the land-use history and changes in soil nutrient content as riparian forests had oligotrophic soils and pasture had copiotrophic soil. Revegetation of pastures with native species tended to reverse those changes, where soil bacterial communities under revegetation were intermediary between remnant riparian forest and pasture soils.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/12/2/157/s1, Table S1. Species composition of three strata at each site, listed alphabetically by family. Average relative density (RD, stems ha-1) is shown for species in canopy and average percentage cover is shown for species in sub-canopy and ground cover. Table S2: Carbon sources tested in EcoPlates and the corresponding guild group. Table S3: Eigenvalues and percent variance for principal component analysis for environmental factors. Table S4: Redundancy analysis fit of environmental variables. p values < 0.05 are in bold and have been included in Figure 5. Figure S1: Observed richness compared to rarefied richness. Figure S2: Rarefaction curve showing richness of each sample. Figure S3: Principal component analysis (PCA) of vegetation factors (a) PC1 and 2, (b) PC1 and 3, (c) PC1 and 4, (d) PC2 and 3, (e) PC2 and 4, (f) PC3 and 4. Points are colored based on land-use. Abbreviations of predictor variables are provided in Table 2. Figure S4: Principal component analysis (PCA) of soil physicochemical factors (a) PC1 and 2, (b) PC1 and 3, (c) PC1 and 4, (d) PC2 and 3, (e) PC2 and 4, (f) PC3 and 4, (g) PC2 and PC5, (h) PC3 and PC4 (i) PC3 and 5, and (j) PC4 and 5. Points are colored based on land use and shape is based on depth. Abbreviations of predictor variables are provided in Table 3. Figure S5: Dimensions for principal components of vegetation characteristics showing the contribution of each variable to that component. Abbreviations of predictor variables are provided in Table 2. Figure S6: Dimensions for soil physicochemical principal components showing the contribution of each variable to that component. Abbreviations for predictor variables are provided in Table 3. Figure S7: Bacterial taxonomic abundance by phylum showing mean land use (P = pasture, RV = Revegetated and RM = Remnant) grouped by soil depth, error bars represent standard error of the mean, p values for GLM shown on graphs (L = Land-use, D = Depth and L × D = Interaction), bold indicates significant difference (<0.05). Figure S8: Bacterial taxonomic richness by phylum showing mean land use (P = pasture, RV = Revegetated and RM = Remnant) grouped by soil depth, error bars represent standard error of the mean, p values for GLM shown on graphs (L = Land-use, D = Depth and L × D = Interaction), bold indicates significant difference (<0.05). Figure S9: Bacterial taxonomic Shannon Index by phylum showing mean land use (P = pasture, RV = Revegetated and RM = Remnant) grouped by soil depth, error bars represent standard error of the mean, p values for GLM shown on graphs (L = Land-use, D = Depth and L × D = Interaction), bold indicates significant difference (<0.05). Figure S10: Bacterial Functional catabolism (abundance) calculated using AWCD shown by carbon guild, points indicate mean land use (P = pasture, RV = Revegetated and RM = Remnant) grouped by soil depth, error bars represent standard error of the mean, p values for GLM shown on graphs (L = Land-use, D = Depth and L × D = Interaction), bold indicates significant difference (<0.05). Figure S11: Bacterial Functional richness calculated using AWCD shown by carbon guild, points indicate mean land use (P = pasture, RV = Revegetated and RM = Remnant) grouped by soil depth, error bars represent standard error of the mean, p values for GLM shown on graphs (L = Land-use, D = Depth and L × D Interaction), bold indicates significant difference (<0.05). Figure S12: Bacterial Functional Shannon’s Index of diversity calculated using AWCD shown by carbon guild, points indicate mean land-use (P = pasture, RV = Revegetated and RM = Remnant) grouped by soil depth, error bars represent standard error of the mean, p values for GLM shown on graphs (L = Land-use, D = Depth and L × D = Interaction), bold indicates significant difference (<0.05).
Author Contributions
Conceptualization, V.W., R.E.M., F.E., and C.A.; methodology, V.W., R.E.M., F.E., and C.A.; software, V.W., A.B., and C.A.; formal analysis, V.W., S.K., A.B., R.E.M., and C.A.; investigation, V.W., R.E.M., S.K., F.E., and C.A.; resources, V.W., F.E., A.B., R.E.M., and C.A.; data curation, V.W., and A.B.; writing—original draft preparation, V.W., R.E.M., S.K., F.E., and C.A.; writing—review and editing, V.W., R.E.M., S.K., F.E., and C.A.; validation, V.W., R.E.M., S.K., F.E., and C.A.; project administration, V.W., R.E.M., S.K., F.E., and C.A.; funding acquisition, V.W., R.E.M., A.B., F.E., and C.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Holsworth Wildlife Research Endowment from the Ecological Society of Australia, Melbourne Water, and the Madeleine Selwyn Smith Memorial Scholarship. Vicky Waymouth was a recipient of a Research Training Program Scholarship. Cybec Foundation supported Rebecca Miller during her lectureship at Melbourne University. We would like to acknowledge the contributions of the Biomes of Australian Soil Environments (BASE) and Australian Microbiome consortiums to the generation of genetic data for this study. The Australian Microbiome initiative is supported by funding from Bioplatforms Australia and the Integrated Marine Observing System (IMOS) through the Australian Government’s National Collaborative Research Infrastructure Strategy (NCRIS), Parks Australia through the Bush Blitz program funded by the Australian Government and BHP, and CSIRO. Soil collection and field sampling was conducted under Parks Victoria permit number 10008494.
Data Availability Statement
Data is available on Bioplatforms Australia data portal: https://data.bioplatforms.com/organization/about/australian-microbiome.
Acknowledgments
We thank Lisa Wittick, Sascha Andrusiak, Nicholas Osborne for technical support, and field assistants Cordula Gutekunst, Ana Bermudez Contreras, Sarah Fischer, Tony Lovell, Kathy Russell, Robert Dabal, Anu Singh and Scott McKendrick; and Jess Gardner, Kathryn Russell, Fred Cherqui, Sarah Fischer, Joe Greet and Joanne McGimpsey for reviewing early drafts.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lopes, L.D.; Fernandes, M.F. Changes in microbial community structure and physiological profile in a kaolinitic tropical soil under different conservation agricultural practices. Appl. Soil Ecol. 2020, 152, 103545. [Google Scholar] [CrossRef]
- Potthoff, M.; Steenwerth, K.L.; Jackson, L.E.; Drenovsky, R.E.; Scow, K.M.; Joergensen, R.G. Soil microbial community composition as affected by restoration practices in California grassland. Soil Biol. Biochem. 2006, 38, 1851–1860. [Google Scholar] [CrossRef]
- Yan, D.; Mills, J.G.; Gellie, N.J.; Bissett, A.; Lowe, A.J.; Breed, M.F. High-throughput eDNA monitoring of fungi to track functional recovery in ecological restoration. Biol. Conserv. 2018, 217, 113–120. [Google Scholar] [CrossRef]
- Eilers, K.G.; Debenport, S.; Anderson, S.; Fierer, N. Digging deeper to find unique microbial communities: The strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol. Biochem. 2012, 50, 58–65. [Google Scholar] [CrossRef]
- McKinley, V. Microbial biomass and activity in soils from virgin prairies compared with prairie restoration, forest and agricultural sites in Illinois. In Proceedings of the Seventeenth North American Prairie Conference: Seeds for the Future, Roots of the Past North Iowa Area Community College, Mason City, IA, USA, 16–20 July 2000; pp. 107–117. [Google Scholar]
- Horwath, W.R. The role of the soil microbial biomass in cycling nutrients. In Microbial Biomass: A Paradigm Shift in Terrestrial Biochemistry; Tate, K.R., Ed.; World Scientific: Hackensack, NJ, USA, 2017; pp. 41–66. [Google Scholar]
- Duchicela, J.; Sullivan, T.S.; Bontti, E.; Bever, J.D. Soil aggregate stability increase is strongly related to fungal community succession along an abandoned agricultural field chronosequence in the Bolivian Altiplano. J. Appl. Ecol. 2013, 50, 1266–1273. [Google Scholar] [CrossRef]
- Rillig, M.C.; Mummey, D.L. Mycorrhizas and soil structure. New Phytol. 2006, 171, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Schlatter, D.C.; Bakker, M.G.; Bradeen, J.M.; Kinkel, L.L. Plant community richness and microbial interactions structure bacterial communities in soil. Ecology 2015, 96, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.A.; Zwart, A.B.; Broadhurst, L.M.; Young, A.G.; Richardson, A.E. The influence of sampling strategies and spatial variation on the detected soil bacterial communities under three different land-use types. FEMS Microbiol. Ecol. 2011, 78, 70–79. [Google Scholar] [CrossRef]
- Ren, C.; Liu, W.; Zhao, F.; Zhong, Z.; Deng, J.; Han, X.; Yang, G.; Feng, Y.; Ren, G. Soil bacterial and fungal diversity and compositions respond differently to forest development. Catena 2019, 181, 104071. [Google Scholar] [CrossRef]
- Kim, M.; Boldgiv, B.; Singh, D.; Chun, J.; Lkhagva, A.; Adams, J.M. Structure of soil bacterial communities in relation to environmental variables in a semi-arid region of Mongolia. J. Arid Environ. 2013, 89, 38–44. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, C.; Cui, Z.; Qian, W.; Liang, C.; Wang, C. Soil bacterial community restoration along a chronosequence of sand-fixing plantations on moving sand dunes in the Horqin sandy land in northeast China. J. Arid Environ. 2019, 165, 81–87. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, X.; Wu, Y.; Zhang, L.; Cheng, J.; Wei, G.; Lin, Y. Natural revegetation of a semiarid habitat alters taxonomic and functional diversity of soil microbial communities. Sci. Total Environ. 2018, 635, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.P.d.A.; Andrade, P.A.M.d.; Bini, D.; Durrer, A.; Robin, A.; Bouillet, J.P.; Andreote, F.D.; Cardoso, E.J.B.N. Shifts in the bacterial community composition along deep soil profiles in monospecific and mixed stands of Eucalyptus grandis and Acacia mangium. PLoS ONE 2017, 12, e0180371. [Google Scholar] [CrossRef] [PubMed]
- Banning, N.C.; Gleeson, D.B.; Grigg, A.H.; Grant, C.D.; Andersen, G.L.; Brodie, E.L.; Murphy, D. Soil microbial community successional patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 2011, 77, 6158–6164. [Google Scholar] [CrossRef]
- Yang, W.; Cai, A.; Wang, J.; Luo, Y.; Cheng, X.; An, S. Exotic Spartina alterniflora Loisel. Invasion significantly shifts soil bacterial communities with the successional gradient of saltmarsh in eastern China. Plant Soil 2020, 449, 97–115. [Google Scholar] [CrossRef]
- Lange, M.; Eisenhauer, N.; Sierra, C.A.; Bessler, H.; Engels, C.; Griffiths, R.I.; Mellado-Vázquez, P.G.; Malik, A.A.; Roy, J.; Scheu, S. Plant diversity increases soil microbial activity and soil carbon storage. Nat. Commun. 2015, 6, 6707. [Google Scholar] [CrossRef]
- Koo, B.J.; Adriano, D.C.; Bolan, N.S.; Barton, C.D. Root Exudates and Microorganisms. In Encyclopedia of Soils in the Environment; Hillel, D., Ed.; Elsevier: Oxford, UK, 2005; pp. 421–428. [Google Scholar] [CrossRef]
- Card, S.; Quideau, S. Microbial community structure in restored riparian soils of the Canadian prairie pothole region. Soil Biol. Biochem. 2010, 42, 1463–1471. [Google Scholar] [CrossRef]
- Nielsen, U.N.; Wall, D.H.; Six, J. Soil biodiversity and the environment. Annu. Rev. Environ. Resour. 2015, 40, 63–90. [Google Scholar] [CrossRef]
- Lu, M.; Ren, Y.; Wang, S.; Kun, T.; Xiangyang, S.; Shuxian, P. Contribution of soil variables to bacterial community composition following land use change in Napahai plateau wetlands. J. Environ. Manag. 2019, 246, 77–84. [Google Scholar] [CrossRef]
- Rodrigues, J.L.; Pellizari, V.H.; Mueller, R.; Baek, K.; Jesus, E.d.C.; Paula, F.S.; Mirza, B.; Hamaoui, G.S.; Tsai, S.M.; Feigl, B. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2013, 110, 988–993. [Google Scholar] [CrossRef]
- Upton, R.N.; Checinska Sielaff, A.; Hofmockel, K.S.; Xu, X.; Polley, H.W.; Wilsey, B.J. Soil depth and grassland origin cooperatively shape microbial community co-occurrence and function. Ecosphere 2020, 11, e02973. [Google Scholar] [CrossRef]
- Gregory, P.J. Plant Roots: Growth, Activity and Interactions with the Soil; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Churchland, C.; Grayston, S.J. Specificity of plant-microbe interactions in the tree mycorrhizosphere biome and consequences for soil C cycling. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Cheng, X.; Shu, X.; Liu, Y.; Zhang, Q. Linking soil bacterial and fungal communities to vegetation succession following agricultural abandonment. Plant Soil 2018, 431, 19–36. [Google Scholar] [CrossRef]
- Standards Reference Group SERA. National Standards for the Practice of Ecological Restoration in Australia, 2nd ed.; Society for Ecological Restoration Australasia: Hamilton, New Zealand, 2018. [Google Scholar]
- Zhao, M.; Yin, C.; Tao, Y.; Li, C.; Fang, S. Diversity of soil microbial community identified by Biolog method and the associated soil characteristics on reclaimed Scirpus mariqueter wetlands. SN Appl. Sci. 2019, 1, 1408. [Google Scholar] [CrossRef]
- González, E.; Sher, A.A.; Tabacchi, E.; Masip, A.; Poulin, M. Restoration of riparian vegetation: A global review of implementation and evaluation approaches in the international, peer-reviewed literature. J. Environ. Manag. 2015, 158, 85–94. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Asuming-Brempong, S.; Nüsslein, K.; Marsh, T.L.; Flynn, S.J. Opening the black box of soil microbial diversity. Appl. Soil Ecol. 1999, 13, 109–122. [Google Scholar] [CrossRef]
- Yan, D.; Bissett, A.; Gellie, N.; Mills, J.G.; Lowe, A.J.; Breed, M.F. Soil bacterial community differences along a coastal restoration chronosequence. Plant Ecol. 2020, 221, 795–811. [Google Scholar] [CrossRef]
- Whited, D.C.; Lorang, M.S.; Harner, M.J.; Hauer, F.R.; Kimball, J.S.; Stanford, J.A. Climate, hydrologic disturbance, and succession: Drivers of floodplain pattern. Ecology 2007, 88, 940–953. [Google Scholar] [CrossRef]
- Naiman, R.J.; Decamps, H.; McClain, M.E. Riparia: Ecology, Conservation, and Management of Streamside Communities; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Harner, M.J.; Opitz, N.; Geluso, K.; Tockner, K.; Rillig, M.C. Arbuscular mycorrhizal fungi on developing islands within a dynamic river floodplain: An investigation across successional gradients and soil depth. Aquat. Sci. 2011, 73, 35–42. [Google Scholar] [CrossRef]
- Van Der Nat, D.; Tockner, K.; Edwards, P.J.; Ward, J.; Gurnell, A.M. Habitat change in braided flood plains (Tagliamento, NE-Italy). Freshw. Biol. 2003, 48, 1799–1812. [Google Scholar] [CrossRef]
- Stanford, J.A.; Lorang, M.; Hauer, F. The shifting habitat mosaic of river ecosystems. Int. Ver. Für Theor. Und Angew. Limnol. Verh. 2005, 29, 123–136. [Google Scholar] [CrossRef]
- Nakamura, F.; Shin, N.; Inahara, S. Shifting mosaic in maintaining diversity of floodplain tree species in the northern temperate zone of Japan. For. Ecol. Manag. 2007, 241, 28–38. [Google Scholar] [CrossRef]
- McClain, C.D.; Holl, K.D.; Wood, D.M. Successional models as guides for restoration of riparian forest understory. Restor. Ecol. 2011, 19, 280–289. [Google Scholar] [CrossRef]
- Hansen, B.D.; Reich, P.; Cavagnaro, T.R.; Lake, P. Challenges in applying scientific evidence to width recommendations for riparian management in agricultural Australia. Ecol. Manag. Restor. 2015, 16, 50–57. [Google Scholar] [CrossRef]
- Naiman, R.J.; Decamps, H.; Pollock, M. The role of riparian corridors in maintaining regional biodiversity. Ecol. Appl. 1993, 3, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Cantrell, S.A.; Molina, M.; Lodge, D.J.; Rivera-Figueroa, F.J.; Ortiz-Hernández, M.L.; Marchetti, A.A.; Cyterski, M.J.; Pérez-Jiménez, J.R. Effects of a simulated hurricane disturbance on forest floor microbial communities. For. Ecol. Manag. 2014, 332, 22–31. [Google Scholar] [CrossRef]
- González, G.; Lodge, D.J.; Richardson, B.A.; Richardson, M.J. A canopy trimming experiment in Puerto Rico: The response of litter decomposition and nutrient release to canopy opening and debris deposition in a subtropical wet forest. For. Ecol. Manag. 2014, 332, 32–46. [Google Scholar] [CrossRef]
- Ossola, A.; Aponte, C.; Hahs, A.K.; Livesley, S.J. Contrasting effects of urban habitat complexity on metabolic functional diversity and composition of litter and soil bacterial communities. Urban Ecosyst. 2017, 20, 595–607. [Google Scholar] [CrossRef]
- CSIRO Land and Water. Australian Soil Resource Information System; CSIRO Land and Water: Collingwood, Australia, 2013. [Google Scholar]
- Bureau of Meterology. Climate Statistics for Australian locations; Bureau of Meterology: Melbourne, Australia, 2020. [Google Scholar]
- Cottam, G.; Curtis, J.T. The use of distance measures in phytosociological sampling. Ecology 1956, 37, 451–460. [Google Scholar] [CrossRef]
- Canfield, R.H. Application of the line interception method in sampling range vegetation. J. For. 1941, 39, 388–394. [Google Scholar] [CrossRef]
- Bissett, A.; Fitzgerald, A.; Meintjes, T.; Mele, P.M.; Reith, F.; Dennis, P.G.; Breed, M.F.; Brown, B.; Brown, M.V.; Brugger, J.; et al. Introducing BASE: The biomes of Australian soil environments soil microbial diversity database. GigaScience 2016, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- CSBP Lab. CSBP Lab Methods. CSBP Lab: 2 Altona Street, Bibra Lake, WA 6163. 2019. Available online: https://www.csbp.com.au/docs/default-source/csbp-lab/csbp-lab-methods-1118.pdf (accessed on 22 January 2020).
- Lane, D. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; Volume 5, pp. 125–175. [Google Scholar]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Peet, R.K. The measurement of species diversity. Annu. Rev. Ecol. Syst. 1974, 5, 285–307. [Google Scholar] [CrossRef]
- Garland, J.L. Analysis and interpretation of community-level physiological profiles in microbial ecology. Fems Microbiol. Ecol. 1997, 24, 289–300. [Google Scholar] [CrossRef]
- Hill, G.; Mitkowski, N.; Aldrich-Wolfe, L.; Emele, L.; Jurkonie, D.; Ficke, A.; Maldonado-Ramirez, S.; Lynch, S.; Nelson, E. Methods for assessing the composition and diversity of soil microbial communities. Appl. Soil Ecol. 2000, 15, 25–36. [Google Scholar] [CrossRef]
- Haack, S.K.; Garchow, H.; Odelson, D.A.; Forney, L.J.; Klug, M.J. Accuracy, reproducibility, and interpretation of fatty acid methyl ester profiles of model bacterial communities. Appl. Environ. Microbiol. 1994, 60, 2483–2493. [Google Scholar] [CrossRef] [PubMed]
- Jałowiecki, Ł.; Chojniak, J.M.; Dorgeloh, E.; Hegedusova, B.; Ejhed, H.; Magnér, J.; Płaza, G.A. Microbial community profiles in wastewaters from onsite wastewater treatment systems technology. PLoS ONE 2016, 11, e0147725. [Google Scholar] [CrossRef]
- Miki, T.; Yokokawa, T.; Ke, P.J.; Hsieh, I.F.; Hsieh, C.h.; Kume, T.; Yoneya, K.; Matsui, K. Statistical recipe for quantifying microbial functional diversity from EcoPlate metabolic profiling. Ecol. Res. 2018, 33, 249–260. [Google Scholar] [CrossRef]
- Garland, J. Potential and limitations of BIOLOG for microbial community analysis. In Microbial Biosystems: New Frontiers, Proceedings of the 8th International Symposium on Microbial Ecology, Halifax, NS, Canada, 9–14 August 1998; Atlantic Canada Society for Microbial Ecology: Halifax, NS, Canada, 1999; pp. 1–7. [Google Scholar]
- Perujo, N.; Romaní, A.M.; Martín-Fernández, J.A. Microbial community-level physiological profiles: Considering whole data set and integrating dynamics of colour development. Ecol. Indic. 2020, 117, 106628. [Google Scholar] [CrossRef]
- Poyraz, N.; Mutlu, M.B. Assessment of Changes in Microbial Communities in Different Operational Units from a Wastewater Treatment Plant. Pol. J. Environ. Stud. 2017, 26. [Google Scholar] [CrossRef]
- Schmitt, H.; Haapakangas, H.; van Beelen, P. Effects of antibiotics on soil microorganisms: Time and nutrients influence pollution-induced community tolerance. Soil Biol. Biochem. 2005, 37, 1882–1892. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. arXiv 2014, arXiv:0.18637/jss.v067.i01. [Google Scholar]
- Lenth, R. Estimated Marginal Means, Aka Least-Squares Means., 1.4.8; R Package. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 30 December 2020).
- Oksanen, J. Multivariate Analysis of Ecological Communities in R: Vegan Tutorial., 43; R Doc. Available online: https://john-quensen.com/wp-content/uploads/2018/10/Oksanen-Jari-vegantutor.pdf (accessed on 30 December 2020).
- RStudio Team. RStudio: Integrated Development Environment for R 1.0.153; RStudio, Inc.: Boston, MA, USA, 2019; Available online: http://www.r-project.org/ (accessed on 30 December 2020).
- Kassambara, A.; Mundt, F. Extract and Visualize the Results of Multivariate Data Analyses., 1.0.5. R Package. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 30 December 2020).
- Husson, F.; Josse, J.; Le, S.; Mazet, J. Multivariate Exploratory Data Analysis and Data Mining, 2.2; R Package; RStudio, Inc.: Boston, MA, USA, 2020. [Google Scholar]
- Barber, N.A.; Chantos-Davidson, K.M.; Amel Peralta, R.; Sherwood, J.P.; Swingley, W.D. Soil microbial community composition in tallgrass prairie restorations converge with remnants across a 27-year chronosequence. Environ. Microbiol. 2017, 19, 3118–3131. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The ecology of Acidobacteria: Moving beyond genes and genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef] [PubMed]
- Tin, H.S.; Palaniveloo, K.; Anilik, J.; Vickneswaran, M.; Tashiro, Y.; Vairappan, C.S.; Sakai, K. Impact of land-use change on vertical soil bacterial communities in Sabah. Microb. Ecol. 2018, 75, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Brundrett, M.C.; Grierson, P.F.; Bennett, L.T.; Weston, C.J. Soils and the below-ground interactions that shape Australian vegetation. In Australian Vegetation, 3rd ed.; Keith, D.A., Ed.; Cambridge University Press: Cambridge, UK, 2017; pp. 156–181. [Google Scholar]
- Hemkemeyer, M.; Dohrmann, A.B.; Christensen, B.T.; Tebbe, C.C. Bacterial preferences for specific soil particle size fractions revealed by community analyses. Front. Microbiol. 2018, 9, 149. [Google Scholar] [CrossRef] [PubMed]
- Seaton, F.M.; George, P.B.; Lebron, I.; Jones, D.L.; Creer, S.; Robinson, D.A. Soil textural heterogeneity impacts bacterial but not fungal diversity. Soil Biol. Biochem. 2020, 144, 107766. [Google Scholar] [CrossRef]
- Waymouth, V.; Miller, R.E.; Ede, F.; Bissett, A.; Aponte, C. Variation in soil microbial communities: Elucidating relationships with vegetation and soil properties, and testing sampling effectiveness. Plant Ecol. 2020, 221, 837–851. [Google Scholar] [CrossRef]
- Cong, P.; Wang, J.; Li, Y.; Liu, N.; Dong, J.; Pang, H.; Zhang, L.; Gao, Z. Changes in soil organic carbon and microbial community under varying straw incorporation strategies. Soil Tillage Res. 2020, 204, 104735. [Google Scholar] [CrossRef]
- Pedrinho, A.; Mendes, L.W.; Merloti, L.F.; Andreote, F.D.; Tsai, S.M. The natural recovery of soil microbial community and nitrogen functions after pasture abandonment in the Amazon region. Fems. Microbiol. Ecol. 2020, 96, fiaa149. [Google Scholar] [CrossRef]
- Canfora, L.; Bacci, G.; Pinzari, F.; Papa, G.L.; Dazzi, C.; Benedetti, A. Salinity and bacterial diversity: To what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 2014, 9, e106662. [Google Scholar] [CrossRef]
- Rietz, D.; Haynes, R. Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biol. Biochem. 2003, 35, 845–854. [Google Scholar] [CrossRef]
- Tripathi, S.; Kumari, S.; Chakraborty, A.; Gupta, A.; Chakrabarti, K.; Bandyapadhyay, B.K. Microbial biomass and its activities in salt-affected coastal soils. Biol. Fertil. Soils 2006, 42, 273–277. [Google Scholar] [CrossRef]
- Aponte, C.; Marañón, T.; García, L.V. Microbial C, N and P in soils of Mediterranean oak forests: Influence of season, canopy cover and soil depth. Biogeochemistry 2010, 101, 77–92. [Google Scholar] [CrossRef]
- Aponte, C.; Matías, L.; González-Rodríguez, V.; Castro, J.; García, L.V.; Villar, R.; Marañón, T. Soil nutrients and microbial biomass in three contrasting Mediterranean forests. Plant Soil 2014, 380, 57–72. [Google Scholar] [CrossRef]
- Sparling, G. Ratio of microbial biomass carbon to soil organic carbon as a sensitive indicator of changes in soil organic matter. Soil Res. 1992, 30, 195–207. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Tian, Q.; Wang, X.; Wang, D.; Wang, M.; Liao, C.; Yang, X.; Liu, F. Decoupled linkage between soil carbon and nitrogen mineralization among soil depths in a subtropical mixed forest. Soil Biol. Biochem. 2017, 109, 135–144. [Google Scholar] [CrossRef]
- Goberna, M.; Insam, H.; Klammer, S.; Pascual, J.; Sanchez, J. Microbial community structure at different depths in disturbed and undisturbed semiarid Mediterranean forest soils. Microb. Ecol. 2005, 50, 315–326. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).