Individual Tree Crown Segmentation Directly from UAV-Borne LiDAR Data Using the PointNet of Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Laser Data Acquisition
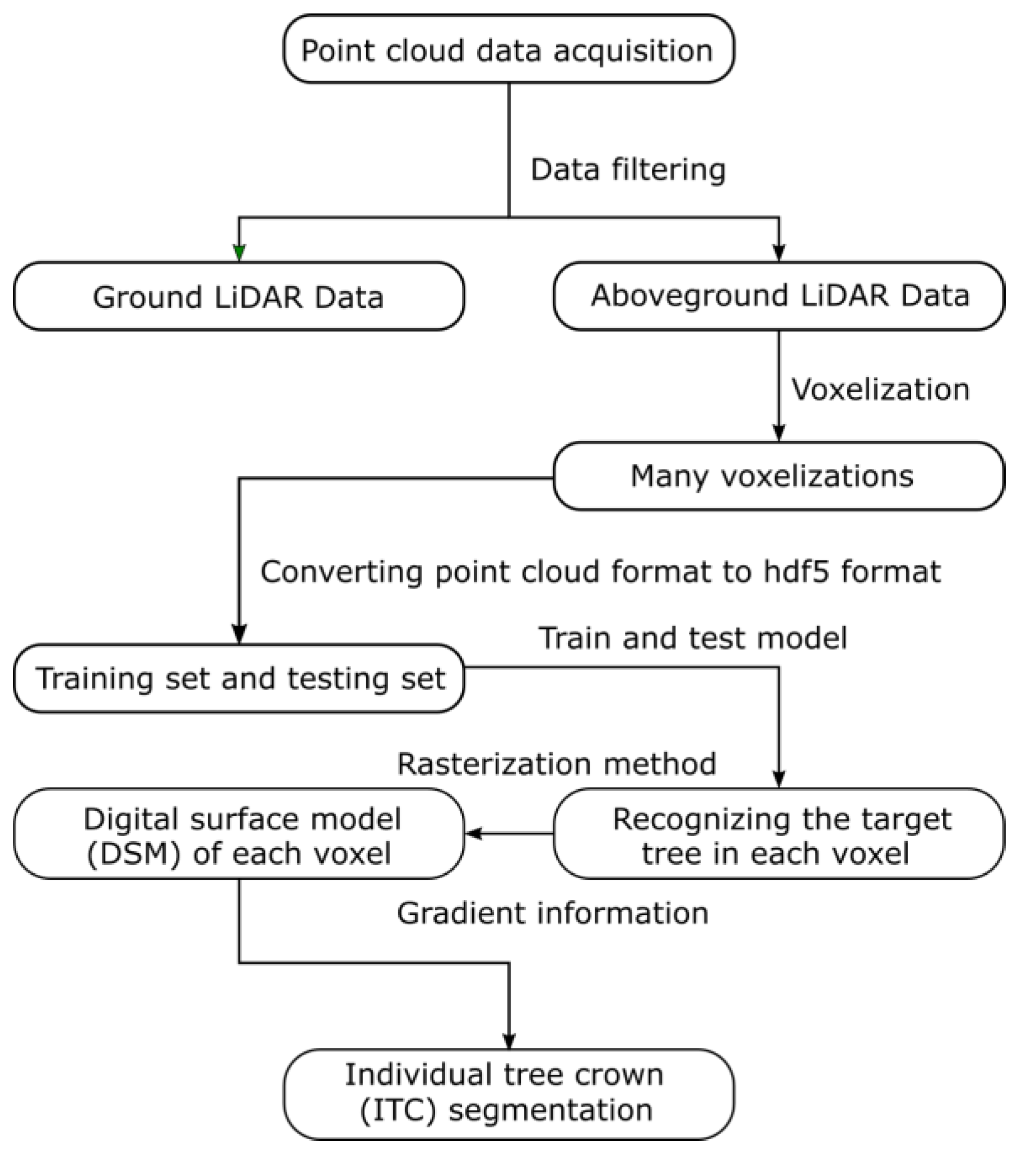
2.3. Data Pre-Processing
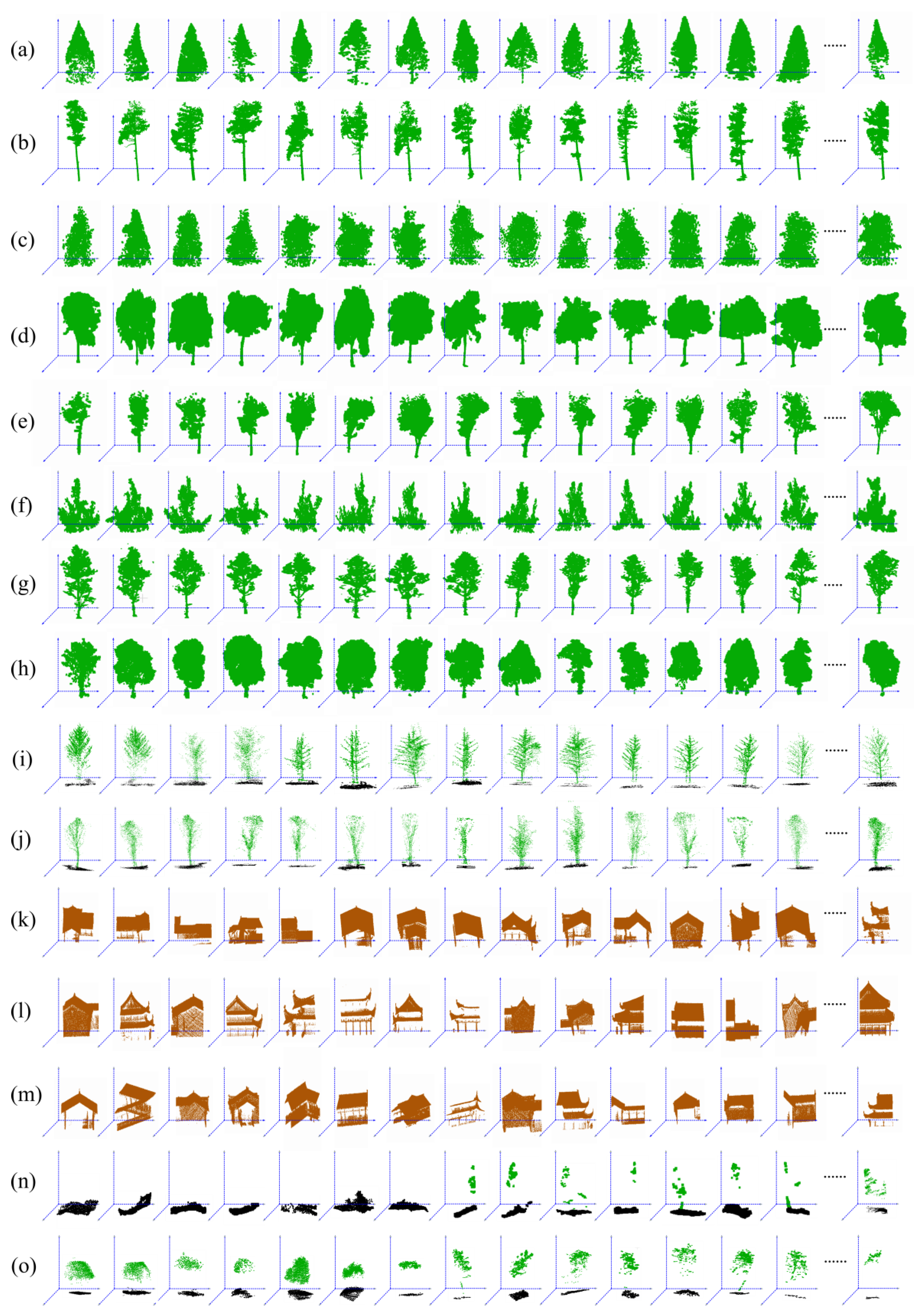
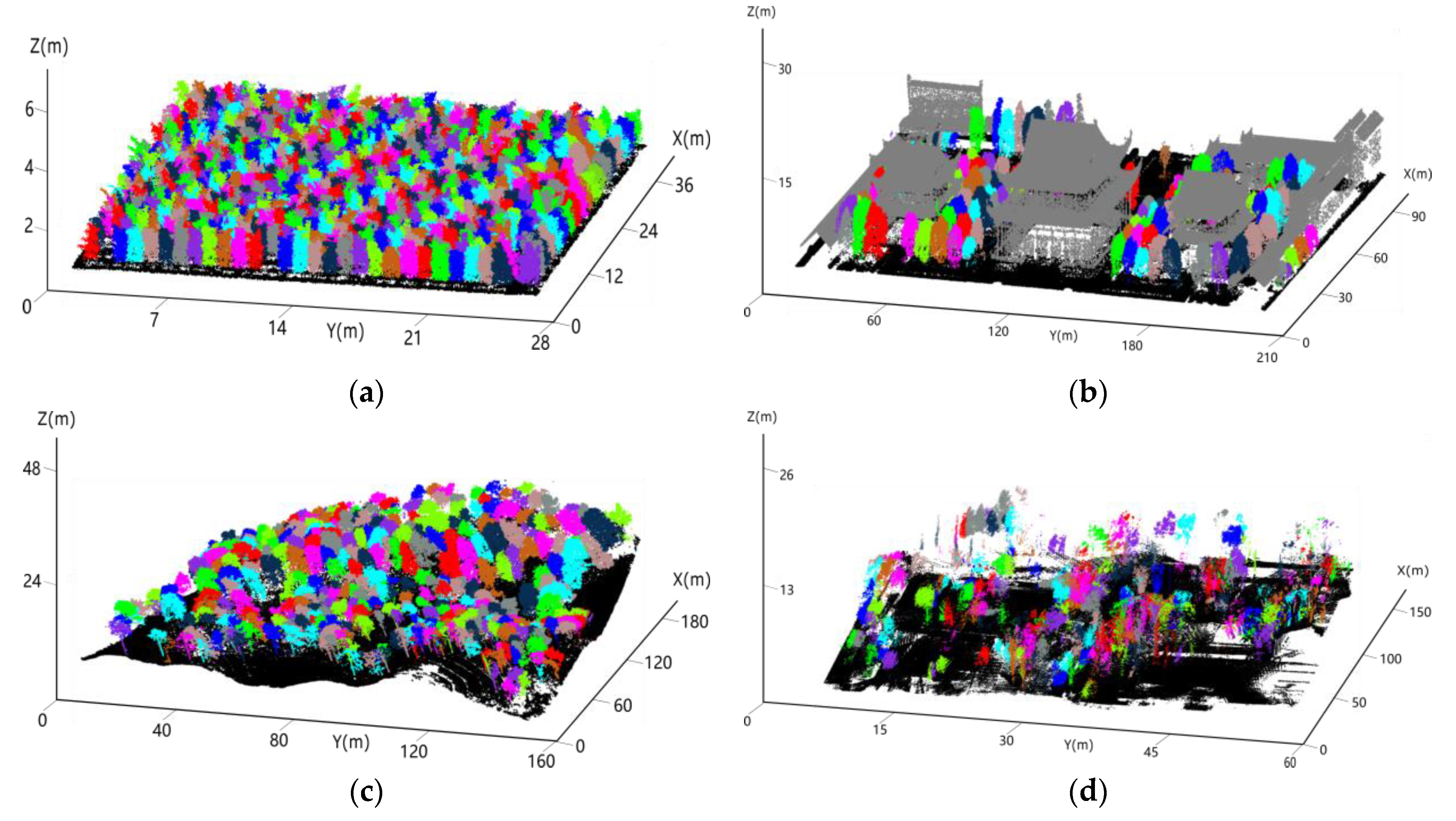
2.3.1. Training Data
2.3.2. Testing Data
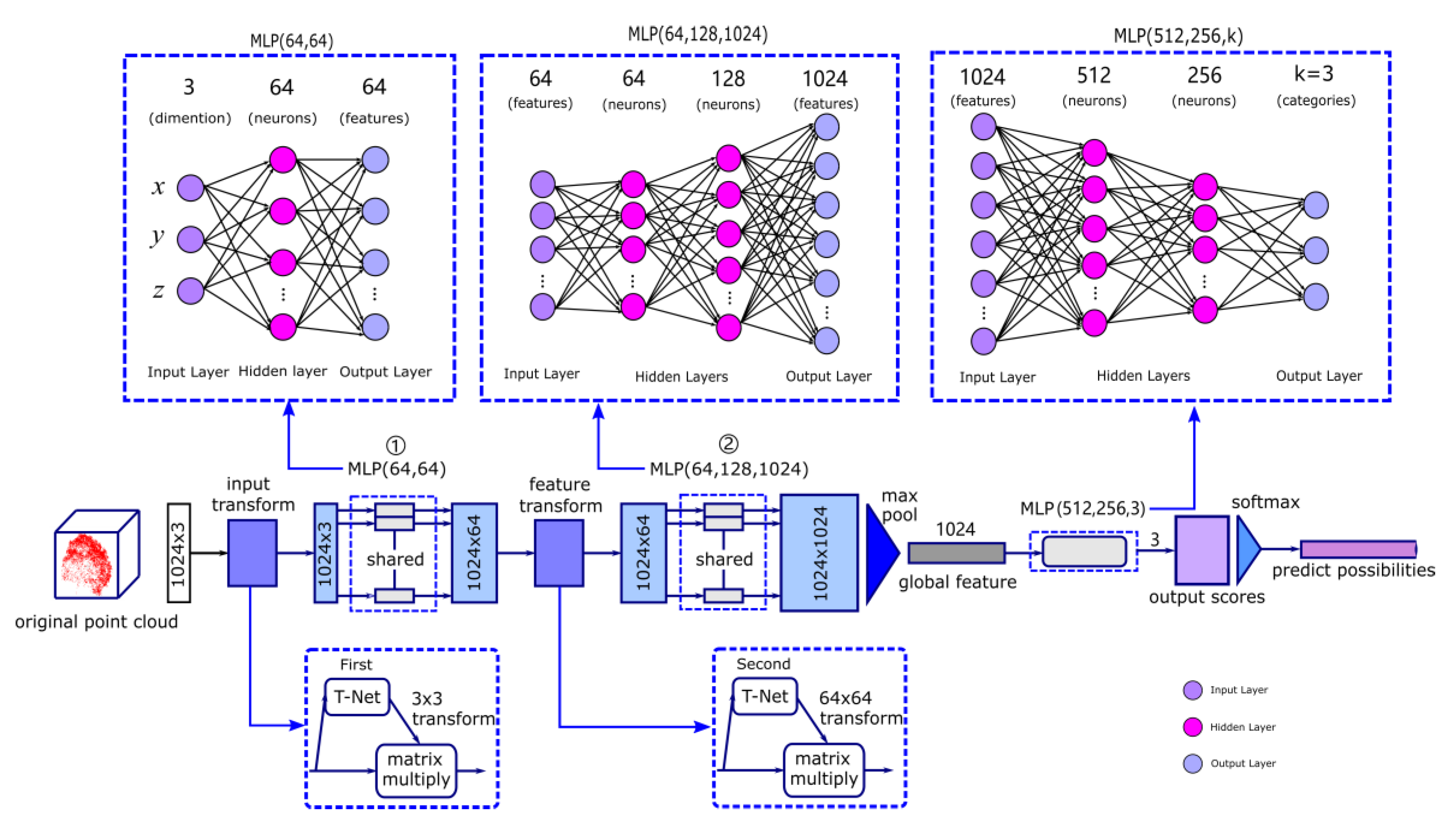
2.4. Training by PointNet
2.5. The loss Function of the Training Process
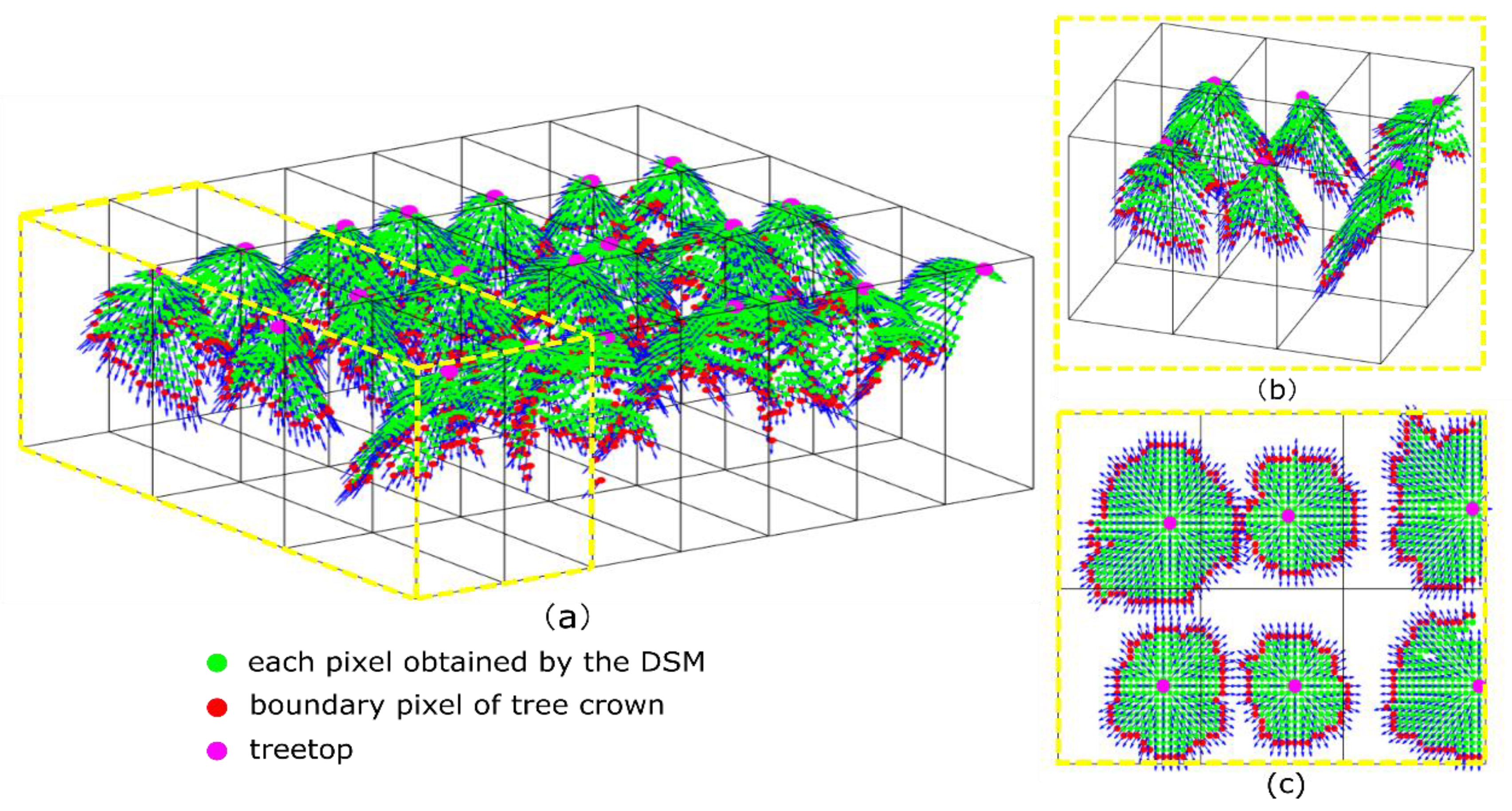
2.6. Individual Tree Segmentation
3. Results
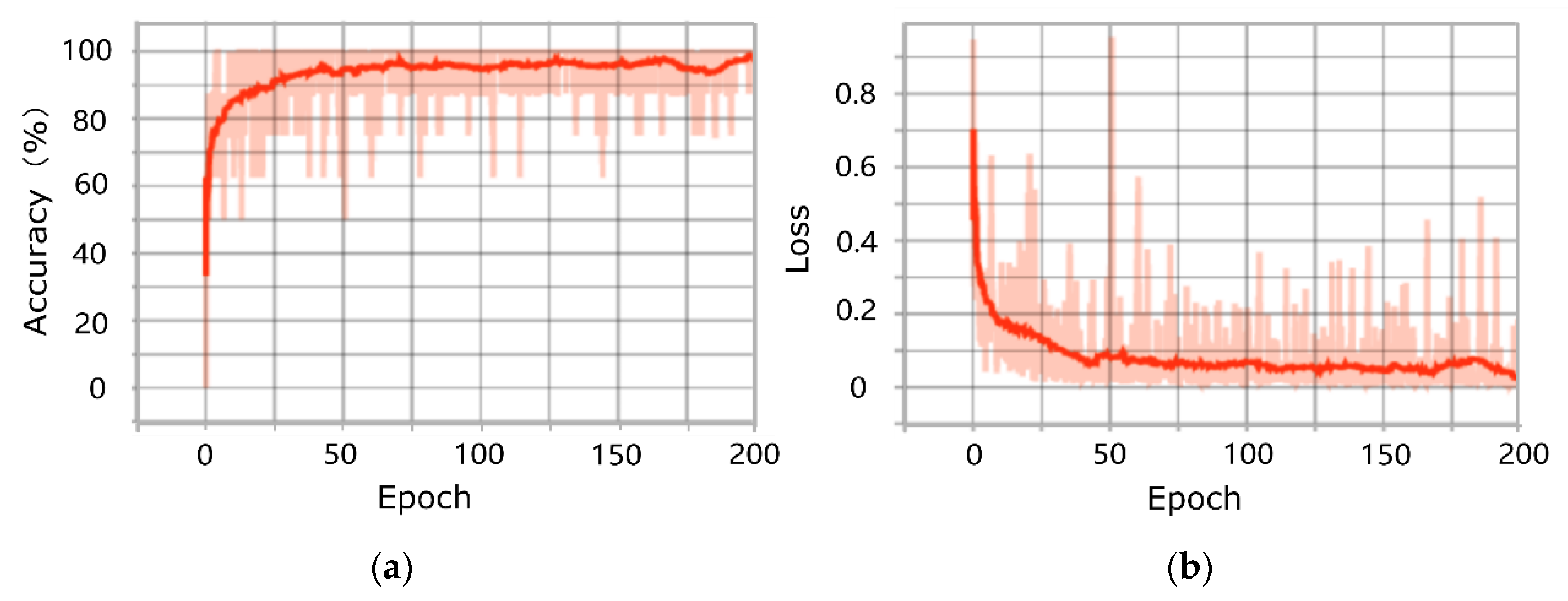
3.1. Results of Training and Testing of the PointNet Model
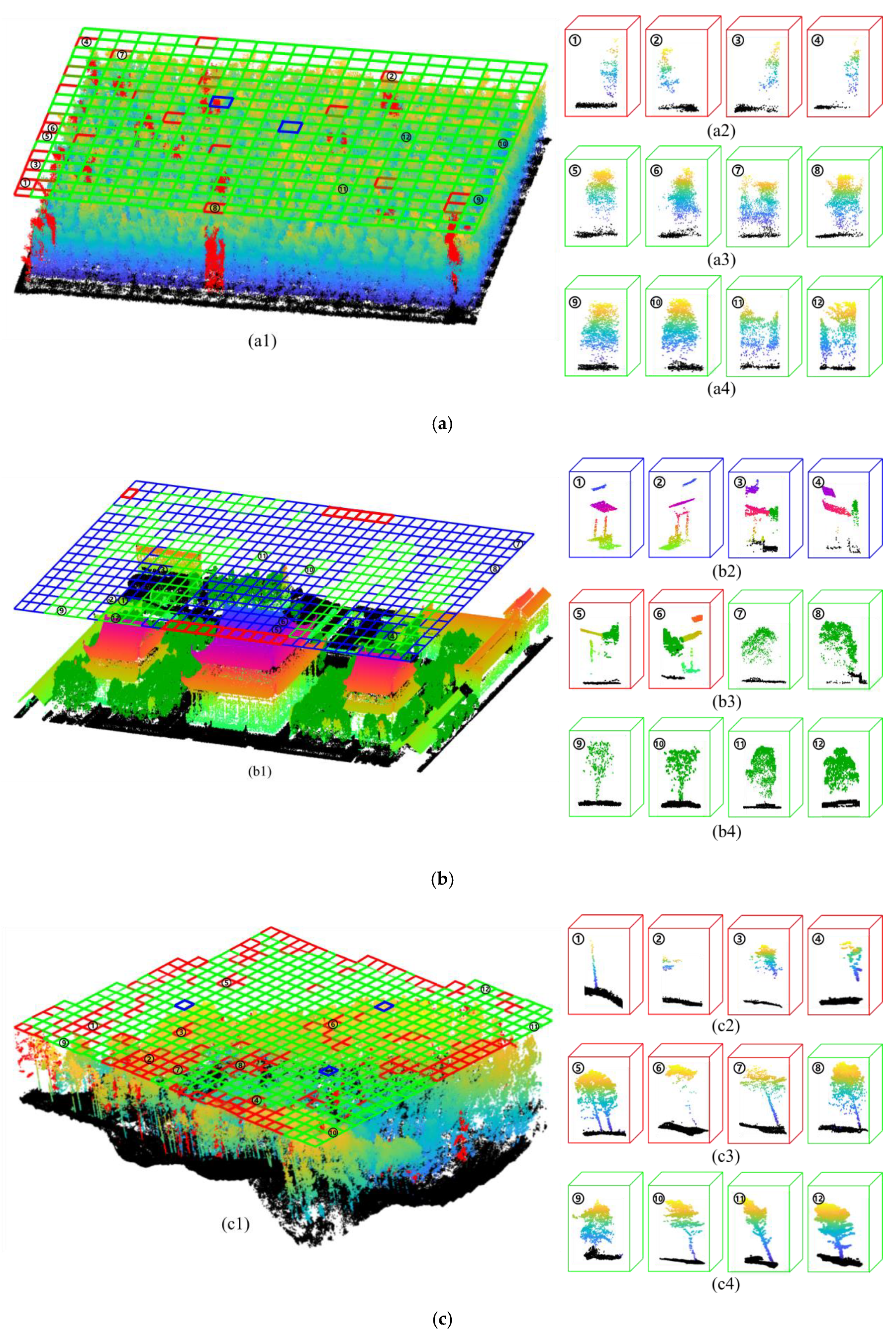
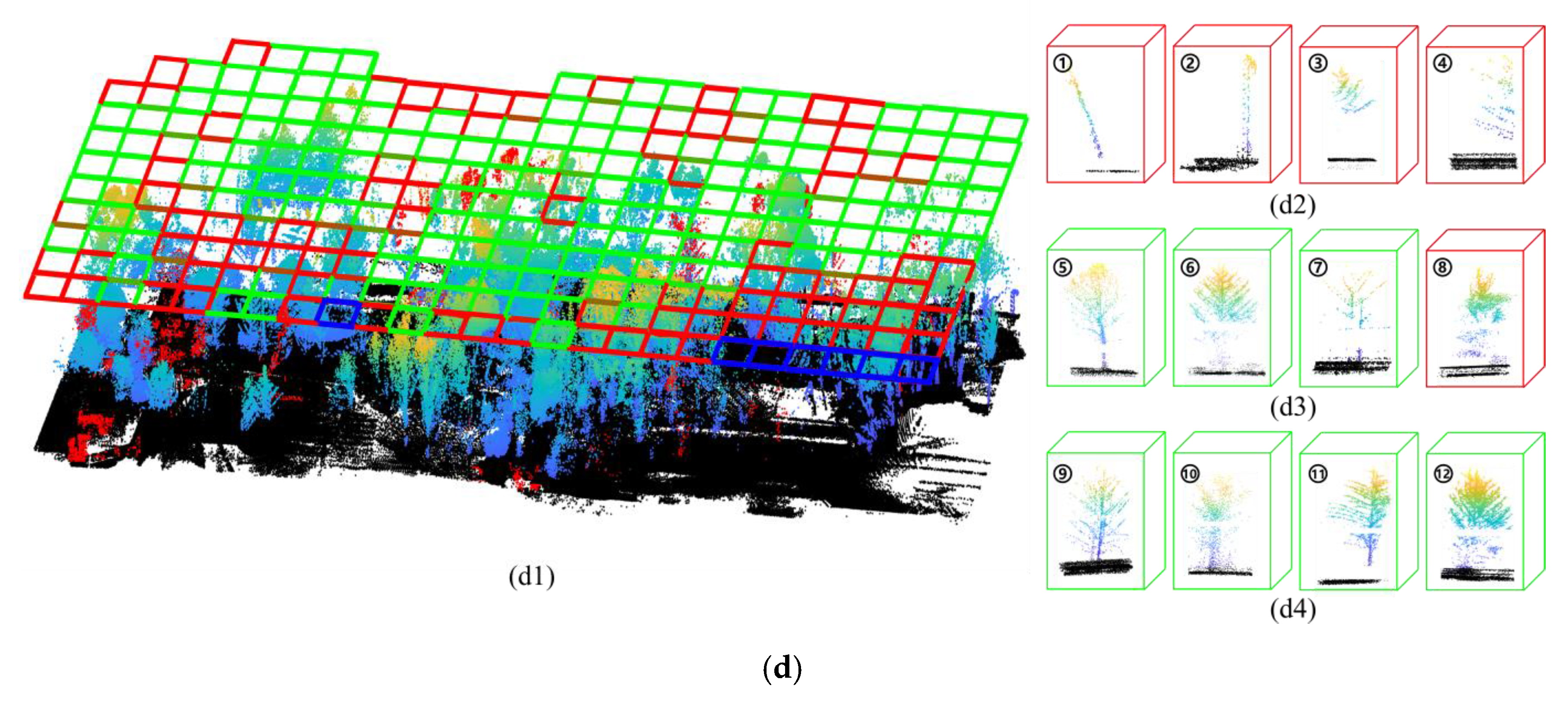
3.2. Accuracy of Tree Crown Width Estimation
4. Discussion
4.1. The Advantages of Our Approach
4.2. Comparison with Existing Methods
4.3. Potential Improvements
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sun, Y.; Liang, X.; Liang, Z.; Welham, C.; Li, W. Deriving Merchantable Volume in Poplar through a Localized Tapering Function from Non-Destructive Terrestrial Laser Scanning. Forests 2016, 7, 87. [Google Scholar] [CrossRef]
- Xu, S.; Xu, S.; Ye, N.; Zhu, F. Individual stem detection in residential environments with MLS data. Remote. Sens. Lett. 2017, 9, 51–60. [Google Scholar] [CrossRef]
- Lee, H.; Slatton, K.C.; Roth, B.E.; Cropper, W.P. Adaptive clustering of airborne LiDAR data to segment individual tree crowns in managed pine forests. Int. J. Remote. Sens. 2010, 31, 117–139. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppa, J.; Vastaranta, M.; Holopainen, M.; Viitala, R. Predicting individual tree attributes from airborne laser point clouds based on the random forests technique. ISPRS J. Photogramm. Remote. Sens. 2011, 66, 28–37. [Google Scholar] [CrossRef]
- Goodbody, T.R.H. Four steps to extend drone use in research. Nature 2019, 7–9. [Google Scholar] [CrossRef]
- Kato, A.; Moskal, L.M.; Batchelor, J.L.; Thau, D.; Hudak, A.T. Relationships between Satellite-Based Spectral Burned Ratios and Terrestrial Laser Scanning. Forests 2019, 10, 444. [Google Scholar] [CrossRef]
- Xu, S.; Xu, S.; Ye, N.; Zhu, F. Automatic extraction of street trees’ nonphotosynthetic components from MLS data. Int. J. Appl. Earth Obs. Geoinf. 2018, 69, 64–77. [Google Scholar] [CrossRef]
- Hu, C.; Pan, Z.; Zhong, T. Leaf and wood separation of poplar seedlings combining locally convex connected patches and K-means++ clustering from terrestrial laser scanning data. J. Appl. Remote. Sens. 2020, 14, 018502. [Google Scholar] [CrossRef]
- Peng, X.; Li, X.; Wang, C.; Zhu, J.; Liang, L.; Fu, H.; Du, Y.; Yang, Z.; Xie, Q. SPICE-based SAR tomography over forest areas using a small number of P-band airborne F-SAR images characterized by non-uniformly distributed baselines. Remote Sens. 2019, 11, 975. [Google Scholar] [CrossRef]
- Dai, W.; Yang, B.; Dong, Z.; Shaker, A. A new method for 3D individual tree extraction using multispectral airborne LiDAR point clouds. ISPRS J. Photogramm. Remote. Sens. 2018, 144, 400–411. [Google Scholar] [CrossRef]
- Hyyppa, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote. Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Duncanson, L.; Cook, B.; Hurtt, G.C.; O Dubayah, R. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote. Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Strîmbu, V.F.; Strimbu, B.M. A graph-based segmentation algorithm for tree crown extraction using airborne LiDAR data. ISPRS J. Photogramm. Remote. Sens. 2015, 104, 30–43. [Google Scholar] [CrossRef]
- Wu, B.; Yu, B.; Wu, Q.; Huang, Y.; Chen, Z.; Wu, J. Individual tree crown delineation using localized contour tree method and airborne LiDAR data in coniferous forests. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 82–94. [Google Scholar] [CrossRef]
- Shen, X.; Cao, L.; Chen, D.; Sun, Y.; Wang, G.; Ruan, H. Prediction of Forest Structural Parameters Using Airborne Full-Waveform LiDAR and Hyperspectral Data in Subtropical Forests. Remote. Sens. 2018, 10, 1729. [Google Scholar] [CrossRef]
- Li, Q.; Yuan, P.; Liu, X.; Zhou, H. Street tree segmentation from mobile laser scanning data. Int. J. Remote. Sens. 2020, 41, 7145–7162. [Google Scholar] [CrossRef]
- Morsdorf, F.; Meier, E.; Allgöwer, B.; Uesch, D. Clustering in airborne laser scanning raw data for segmentation of single trees. Remote Sens. Spat. Inf. Sci. 2003, 34, W13. [Google Scholar]
- Hu, X.; Chen, W.; Xu, W. Adaptive Mean Shift-Based Identification of Individual Trees Using Airborne LiDAR Data. Remote. Sens. 2017, 9, 148. [Google Scholar] [CrossRef]
- Wang, Y.; Weinacker, H.; Koch, B.; Stereńczak, K. Lidar point cloud based fully automatic 3D single tree modelling in forest and evaluations of the procedure. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, 8, 1682–1750. [Google Scholar]
- Tao, S.; Wu, F.; Guo, Q.; Wang, Y.; Li, W.; Xue, B.; Hu, X.; Li, P.; Tian, D.; Li, C.; et al. Segmenting tree crowns from terrestrial and mobile LiDAR data by exploring ecological theories. ISPRS J. Photogramm. Remote. Sens. 2015, 110, 66–76. [Google Scholar] [CrossRef]
- Yang, X.; Yang, H.; Zhang, F.; Fan, X.; Ye, Q.; Feng, Z. A random-weighted plane-Gaussian artificial neural network. Neural Comput. Appl. 2019, 31, 8681–8692. [Google Scholar] [CrossRef]
- Fan, X.; Yang, X.; Ye, Q.; Yang, Y. A discriminative dynamic framework for facial expression recognition in video sequences. J. Vis. Commun. Image Represent. 2018, 56, 182–187. [Google Scholar] [CrossRef]
- Fan, X.; Tjahjadi, T. Fusing dynamic deep learned features and handcrafted features for facial expression recognition. J. Vis. Commun. Image Represent. 2019, 65, 102659. [Google Scholar] [CrossRef]
- Fujiyoshi, H.; Hirakawa, T.; Yamashita, T. Deep learning-based image recognition for autonomous driving. IATSS Res. 2019, 43, 244–252. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, M.; Liu, M.; Zhang, D. A Survey on Deep Learning for Neuroimaging-Based Brain Disorder Analysis. Front. Neurosci. 2020, 14, 779. [Google Scholar] [CrossRef]
- Liu, X.; Hu, C.; Li, P. Automatic segmentation of overlapped poplar seedling leaves combining Mask R-CNN and DBSCAN. Comput. Electron. Agric. 2020, 178, 105753. [Google Scholar] [CrossRef]
- Hu, C.; Liu, X.; Pan, Z.; Li, P. Automatic Detection of Single Ripe Tomato on Plant Combining Faster R-CNN and Intuitionistic Fuzzy Set. IEEE Access 2019, 7, 154683–154696. [Google Scholar] [CrossRef]
- Jin, S.; Su, Y.; Gao, S.; Wu, F.; Hu, T.; Liu, J.; Li, W.; Wang, D.; Chen, S.; Jiang, Y.; et al. Deep Learning: Individual Maize Segmentation from Terrestrial Lidar Data Using Faster R-CNN and Regional Growth Algorithms. Front. Plant Sci. 2018, 9, 866. [Google Scholar] [CrossRef]
- Fang, Y.; Xie, J.; Dai, G.; Wang, M.; Zhu, F.; Xu, T.; Wong, E. 3D deep shape descriptor. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 2319–2328. [Google Scholar] [CrossRef]
- Pittaras, N.; Markatopoulou, F.; Mezaris, V.; Patras, I. Comparison of Fine-Tuning and Extension Strategies for Deep Convolutional Neural Networks. Lect. Notes Comput. Sci. 2016, 10132, 102–114. [Google Scholar] [CrossRef]
- Savva, M.; Yu, F.; Su, H.; Kanezaki, A.; Furuya, T.; Ohbuchi, R.; Zhou, Z.; Yu, R.; Bai, S.; Bai, X.; et al. SHREC’17 Track Large-Scale 3D Shape Retrieval from ShapeNet Core55. In Proceedings of the Workshop on 3D Object Retrieval, Lyon, France, 24–28 April 2017. [Google Scholar] [CrossRef]
- Wu, Z.; Song, S.; Khosla, A.; Yu, F.; Zhang, L.; Tang, X.; Xiao, J. 3D ShapeNets: A deep representation for volumetric shapes. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1912–1920. [Google Scholar] [CrossRef]
- Kang, Z.; Yang, J.; Zhong, R.; Wu, Y.; Shi, Z.; Lindenbergh, R. Voxel-Based Extraction and Classification of 3-D Pole-Like Objects From Mobile LiDAR Point Cloud Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2018, 11, 4287–4298. [Google Scholar] [CrossRef]
- Charles, R.Q.; Su, H.; Kaichun, M.; Guibas, L.J. PointNet: Deep Learning on Point Sets for 3D Classification and Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 77–85. [Google Scholar]
- Klokov, R.; Lempitsky, V. Escape from Cells: Deep Kd-Networks for the Recognition of 3D Point Cloud Models. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 863–872. [Google Scholar] [CrossRef]
- Li, Y.; Bu, R.; Sun, M.; Wu, W.; Di, X.; Chen, B. PointCNN: Convolution on X-transformed points. arXiv 2018, arXiv:1801.07791. [Google Scholar]
- Fan, S.-Y.; Tsai, M.-F.; Chou, L.-H.; Lai, Y.-T.; Lee, M.-T. Application of UVA in Soil and Water Conservation: Preparation of Planting Materials for UVA; IEEE: Piscataway, NJ, USA, 2018; pp. 163–165. [Google Scholar]
- Zong, C.; Zhang, D.; Ding, Z.; Liu, Y. Mixed Propagation Modes in Three Bragg Propagation Periods of Variable Chain Structures. IEEE Trans. Antennas Propag. 2020, 68, 311–318. [Google Scholar] [CrossRef]
- Zhang, W.; Qi, J.; Wan, P.; Wang, H.; Xie, D.; Wang, X.; Yan, G. An Easy-to-Use Airborne LiDAR Data Filtering Method Based on Cloth Simulation. Remote. Sens. 2016, 8, 501. [Google Scholar] [CrossRef]
- Manduchi, G. Commonalities and differences between MDSplus and HDF5 data systems. Fusion Eng. Des. 2010, 85, 583–590. [Google Scholar] [CrossRef]
- Ohno, H. Training data augmentation: An empirical study using generative adversarial net-based approach with normalizing flow models for materials informatics. Appl. Soft Comput. 2020, 86, 105932. [Google Scholar] [CrossRef]
- Liu, W.; Sun, J.; Li, W.; Hu, T.; Wang, P. Deep Learning on Point Clouds and Its Application: A Survey. Sensors 2019, 19, 4188. [Google Scholar] [CrossRef]
- Peng, X.; Li, L.; Wang, F.-Y. Accelerating Minibatch Stochastic Gradient Descent Using Typicality Sampling. IEEE Trans. Neural Netw. Learn. Syst. 2019, 31, 1–11. [Google Scholar] [CrossRef]
- Xu, Z.; Li, W.; Li, Y.; Shen, X.; Ruan, H. Estimation of secondary forest parameters by integrating image and point cloud-based metrics acquired from unmanned aerial vehicle. J. Appl. Remote. Sens. 2019, 14, 022204. [Google Scholar] [CrossRef][Green Version]
- Zhang, F.; Zhao, P.; Xu, S.; Wu, Y.; Yang, X.; Zhang, Y. Integrating multiple factors to optimize watchtower deployment for wildfire detection. Sci. Total. Environ. 2020, 737, 139561. [Google Scholar] [CrossRef]
- Ai, T. The drainage network extraction from contour lines for contour line generalization. ISPRS J. Photogramm. Remote. Sens. 2007, 62, 93–103. [Google Scholar] [CrossRef]
- Lin, H.; Bai, D.; Liu, Y. Maximum data collection rate routing for data gather trees with data aggregation in rechargeable wireless sensor networks. Clust. Comput. 2017, 22, 597–607. [Google Scholar] [CrossRef]
- Parkan, M.; Lausanne, C.; Zürich, C. Individual tree segmentation in deciduous forests using geodesic voting geographic. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium, Milan, Italy, 31 July 2015; Volume 2012, pp. 637–640. [Google Scholar]
- Mongus, D.; Žalik, B. An efficient approach to 3D single tree-crown delineation in LiDAR data. ISPRS J. Photogramm. Remote. Sens. 2015, 108, 219–233. [Google Scholar] [CrossRef]
- Sylvain, J.-D.; Drolet, G.; Brown, N. Mapping dead forest cover using a deep convolutional neural network and digital aerial photography. ISPRS J. Photogramm. Remote. Sens. 2019, 156, 14–26. [Google Scholar] [CrossRef]
- Hu, B.; Li, J.; Jing, L.; Judah, A. Improving the efficiency and accuracy of individual tree crown delineation from high-density LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 145–155. [Google Scholar] [CrossRef]
- Qi, C.; Yi, L.; Su, H.; Guibas, L. PointNet++: Deep Hierarchical Feature Learning on point sets in a metric space. In Proceedings of the NIPS’17, 31st Conference on Neural Information Processing Systems (NIPS), Long Beach, CA, USA, 4–9 December 2017; pp. 5105–5114. [Google Scholar]
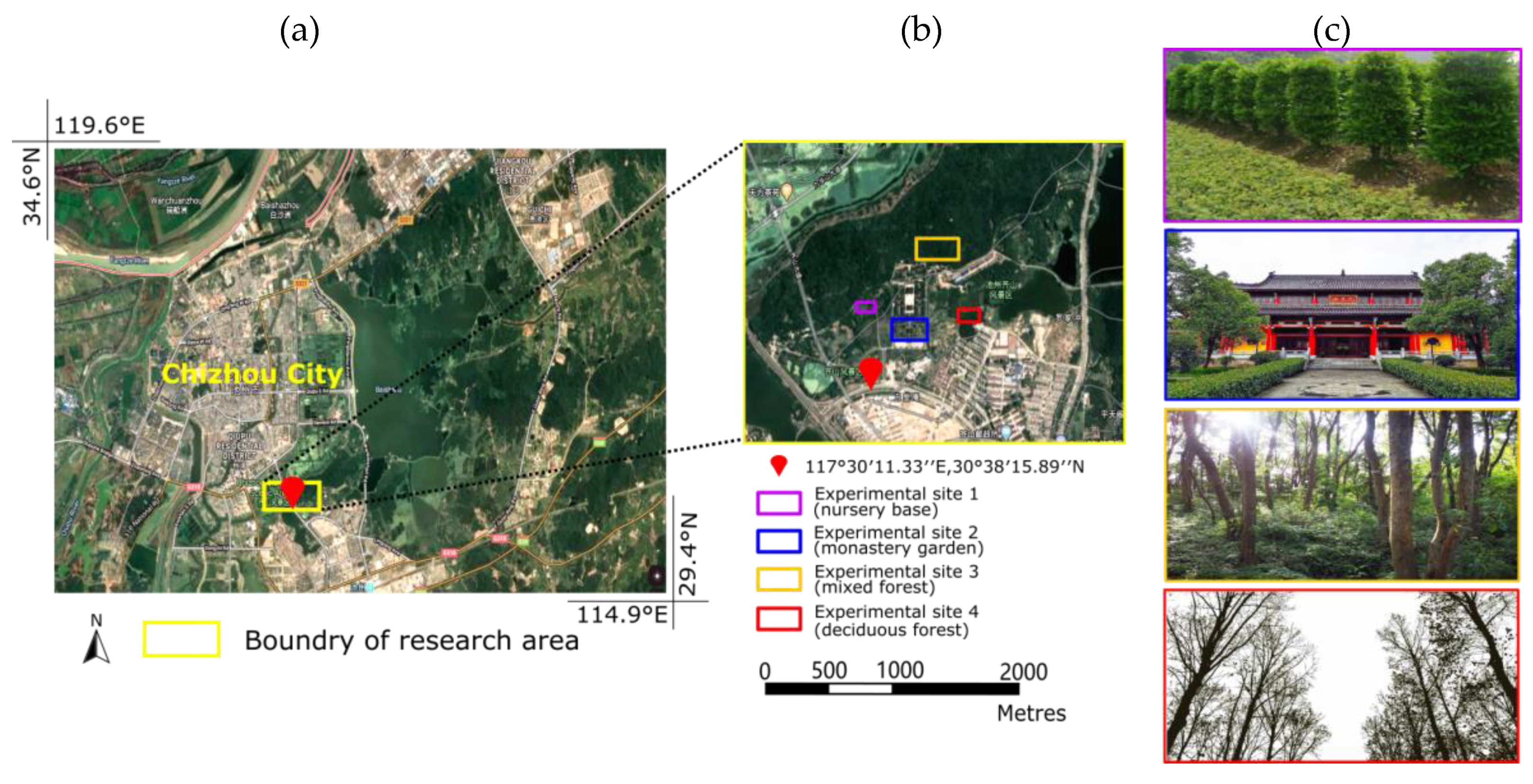

| Nursery Base | Monastery Garden | Mixed Forest | Defoliated Forest | ||
|---|---|---|---|---|---|
| NT | 1059 | 336 | 921 | 338 | |
| NP | 2,942,740 | 44,693,237 | 43,773,108 | 7,424,662 | |
| NPPT | 2779 | 31,020 | 47,528 | 21,966 | |
| Area (m2) | 1947.16 | 44,596.64 | 60,601.78 | 14,780.11 | |
| Training sites | NT | 537 | 176 | 465 | 171 |
| NP | 1,485,416 | 22,432,004 | 22,044,103 | 3,722,314 | |
| Area (m2) | 984.13 | 22,377.18 | 30,461.12 | 7439.57 | |
| Testing sites | NT | 522 | 160 | 456 | 167 |
| NP | 1,457,324 | 22,261,233 | 21,729,005 | 3,702,348 | |
| Area (m2) | 963.03 | 22,219.46 | 30,140.66 | 7340.54 | |
| Nursery Base | Monastery Garden | Mixed Forest | Defoliated Forest | |
|---|---|---|---|---|
| Average tree crown length/width/heights (m) | 1.35/1.36/3.29 | 6.46/5.81/6.34 | 7.08/6.59/13.71 | 5.23/5.2/14.95 |
| Length/width/height of voxels (m) | 1.35/1.36/4.92 | 6.46/5.81/26.96 | 7.08/6.59/48.06 | 5.23/5.2/20.96 |
| Total number of voxels after voxelization | 528 | 592 | 646 | 270 |
| Identification results | T:497; B:5; O:26 | T:168; B:331; O:93 | T:424; B:30; O:192 | T:165; B:19; O:86 |
| NT | NS | TP | FP | FN | r | P | F | |
|---|---|---|---|---|---|---|---|---|
| Nursery base | 522 | 511 | 470 | 41 | 52 | 0.90 | 0.92 | 0.91 |
| Monastery garden | 160 | 151 | 136 | 15 | 24 | 0.85 | 0.90 | 0.87 |
| Mixed forest | 456 | 445 | 365 | 80 | 91 | 0.80 | 0.82 | 0.81 |
| Defoliatedforest | 167 | 163 | 137 | 26 | 30 | 0.82 | 0.84 | 0.83 |
| Overall | 1305 | 1270 | 1108 | 162 | 197 | 0.85 | 0.87 | 0.86 |
| Method | Experimental Forest Plots | NT/NS | TP | FP | FN | r | P | F |
|---|---|---|---|---|---|---|---|---|
| Watershed algorithm | Nursery base | 522/534 | 470 | 64 | 52 | 0.90 | 0.88 | 0.89 |
| Monastery garden | 160/156 | 134 | 22 | 26 | 0.84 | 0.86 | 0.85 | |
| Mixed forest | 456/451 | 365 | 86 | 91 | 0.80 | 0.81 | 0.80 | |
| Defoliated forest | 167/165 | 127 | 38 | 40 | 0.76 | 0.77 | 0.76 | |
| Overall | 1305/1306 | 1096 | 210 | 209 | 0.84 | 0.84 | 0.84 | |
| Point cloud-based cluster segmentation algorithm | Nursery base | 522/517 | 465 | 52 | 57 | 0.89 | 0.90 | 0.89 |
| Monastery garden | 160/147 | 134 | 13 | 26 | 0.84 | 0.91 | 0.87 | |
| Mixed forest | 456/439 | 351 | 88 | 105 | 0.77 | 0.80 | 0.78 | |
| Defoliated forest | 167/169 | 127 | 42 | 40 | 0.76 | 0.75 | 0.75 | |
| Overall | 1305/1272 | 1077 | 195 | 228 | 0.83 | 0.85 | 0.84 | |
| Our method | Nursery base | 522/511 | 470 | 41 | 52 | 0.90 | 0.92 | 0.91 |
| Monastery garden | 160/151 | 136 | 15 | 24 | 0.85 | 0.90 | 0.87 | |
| Mixed forest | 456/445 | 365 | 80 | 91 | 0.80 | 0.82 | 0.81 | |
| Defoliated forest | 167/163 | 137 | 26 | 30 | 0.82 | 0.84 | 0.83 | |
| Overall | 1305/1270 | 1108 | 162 | 197 | 0.85 | 0.87 | 0.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Jiang, K.; Zhu, Y.; Wang, X.; Yun, T. Individual Tree Crown Segmentation Directly from UAV-Borne LiDAR Data Using the PointNet of Deep Learning. Forests 2021, 12, 131. https://doi.org/10.3390/f12020131
Chen X, Jiang K, Zhu Y, Wang X, Yun T. Individual Tree Crown Segmentation Directly from UAV-Borne LiDAR Data Using the PointNet of Deep Learning. Forests. 2021; 12(2):131. https://doi.org/10.3390/f12020131
Chicago/Turabian StyleChen, Xinxin, Kang Jiang, Yushi Zhu, Xiangjun Wang, and Ting Yun. 2021. "Individual Tree Crown Segmentation Directly from UAV-Borne LiDAR Data Using the PointNet of Deep Learning" Forests 12, no. 2: 131. https://doi.org/10.3390/f12020131
APA StyleChen, X., Jiang, K., Zhu, Y., Wang, X., & Yun, T. (2021). Individual Tree Crown Segmentation Directly from UAV-Borne LiDAR Data Using the PointNet of Deep Learning. Forests, 12(2), 131. https://doi.org/10.3390/f12020131






