Soil Microbial Community Succession Based on PhoD and Gcd Genes along a Chronosequence of Sand-Fixation Forest
Abstract
:1. Introduction
2. Materials and Methods
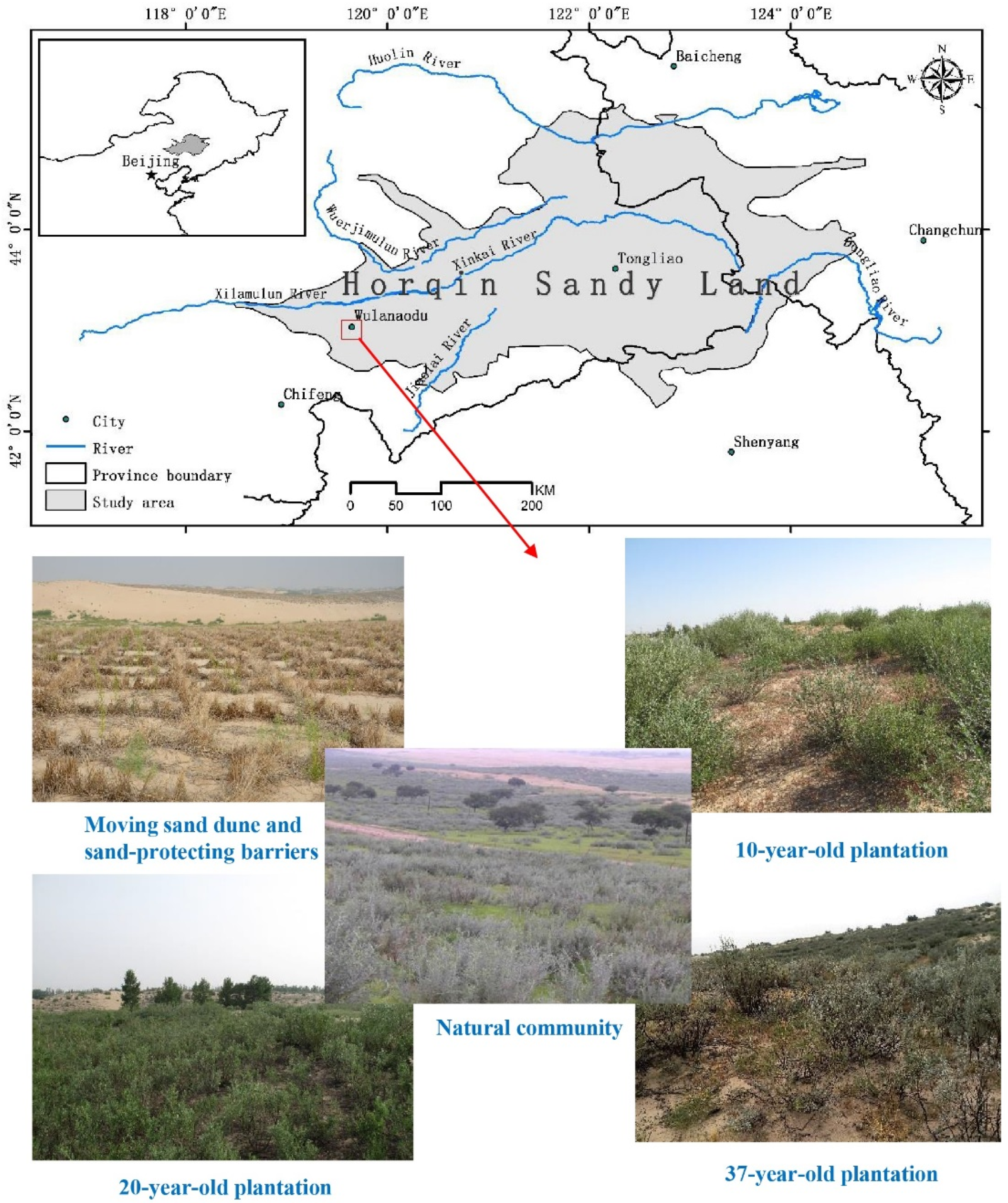
2.1. Study Location and Site Description
2.2. Experimental Design and Soil Sampling
2.3. Soil Physicochemical Properties
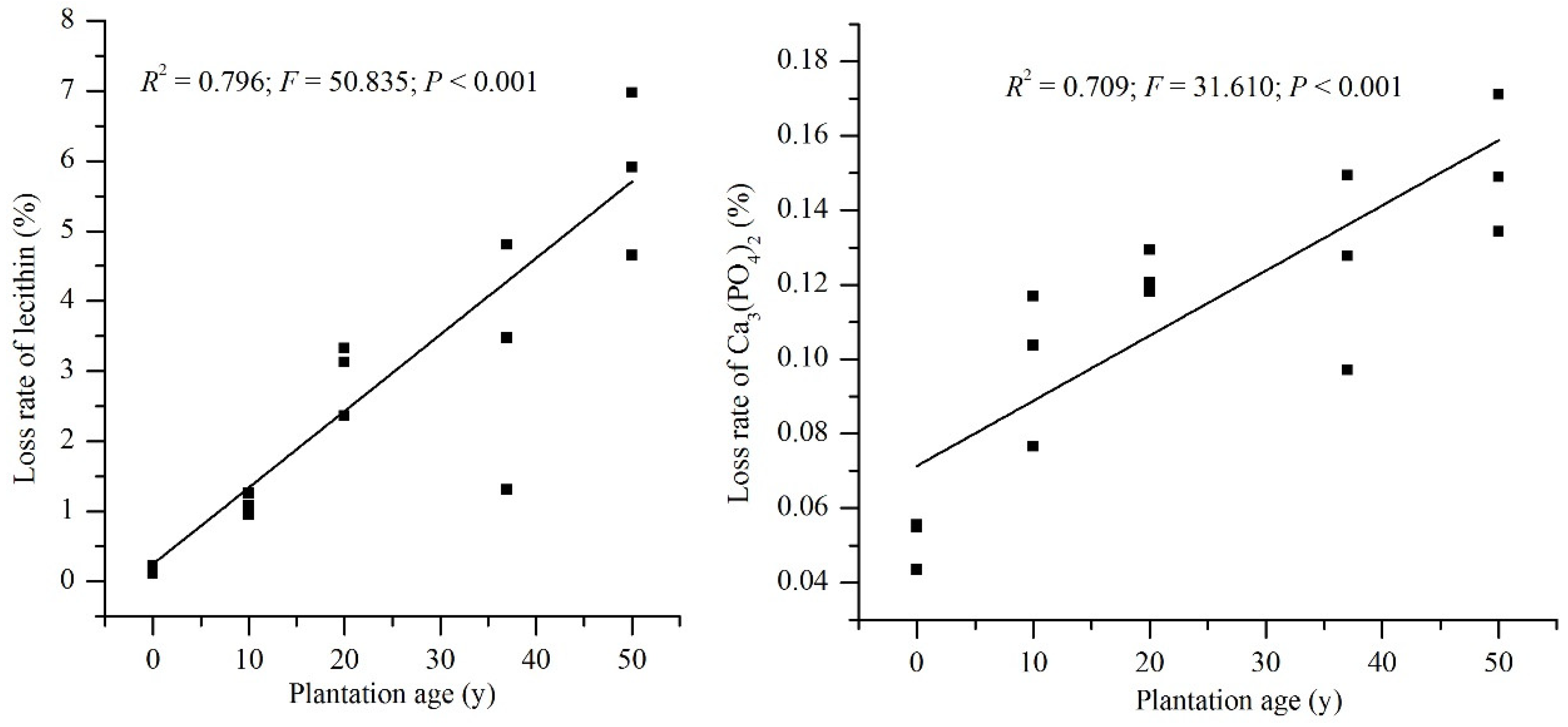
2.4. Potential of Mineralization of OP and Solubilization of IOP
2.5. Soil Enzymatic Activity
2.6. Soil P Fraction
2.7. Amplification, Quantification, and Sequencing of PhoD and Gcd Genes
2.8. Data Analysis
3. Results
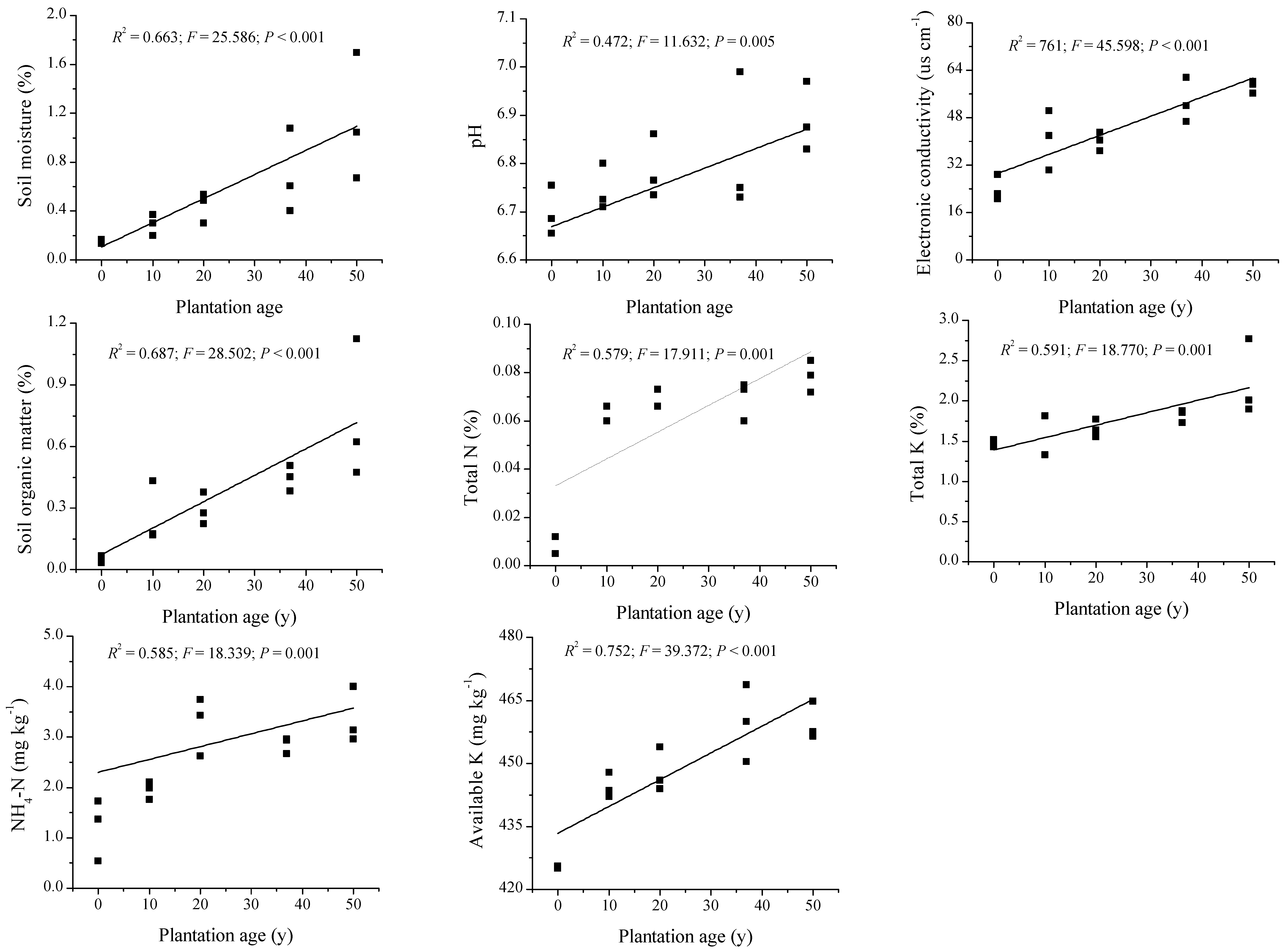
3.1. Soil Properties and P Fractions along the Plantation Development
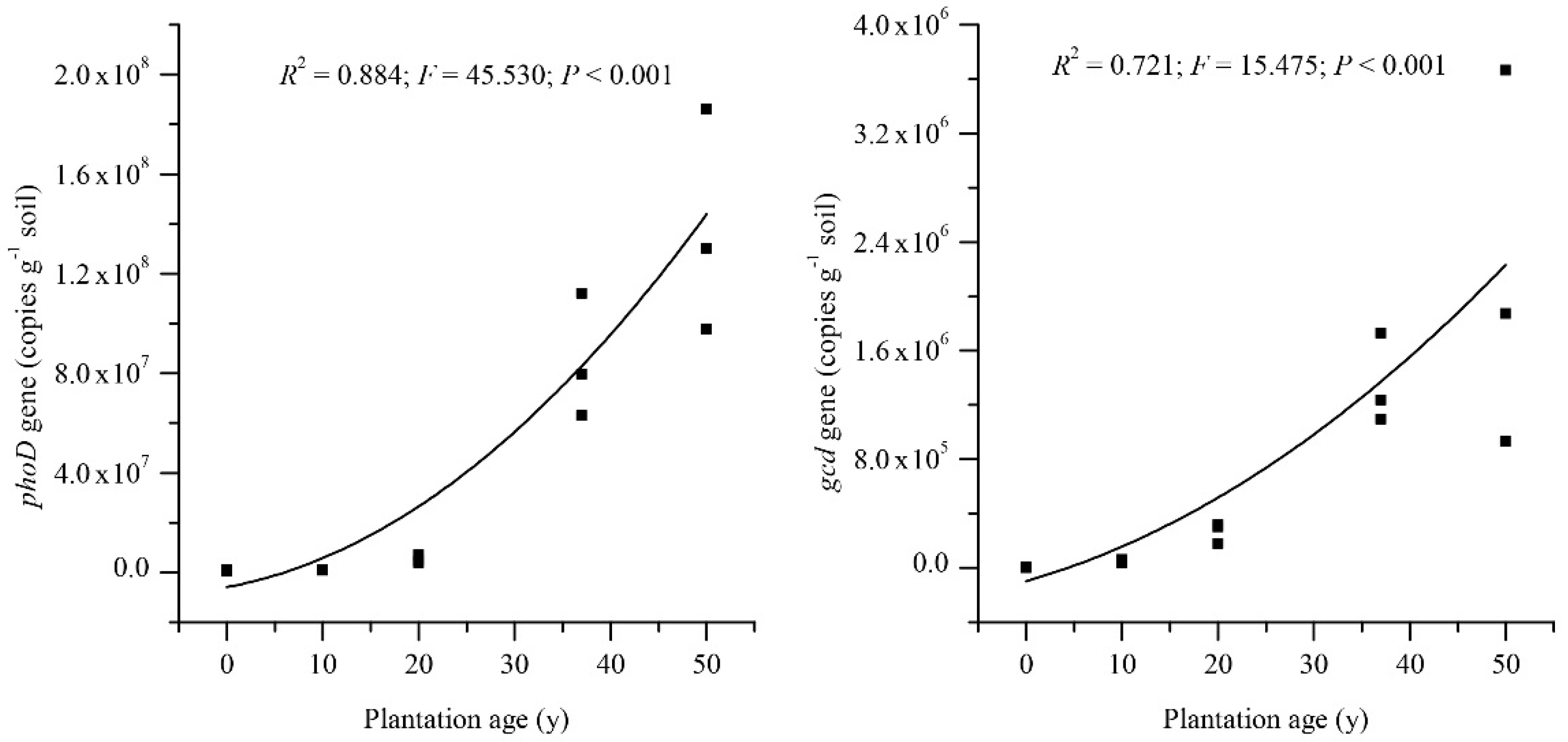
3.2. Abundance of PhoD and Gcd Genes
3.3. Structures of Soil PhoD and Gcd Communities
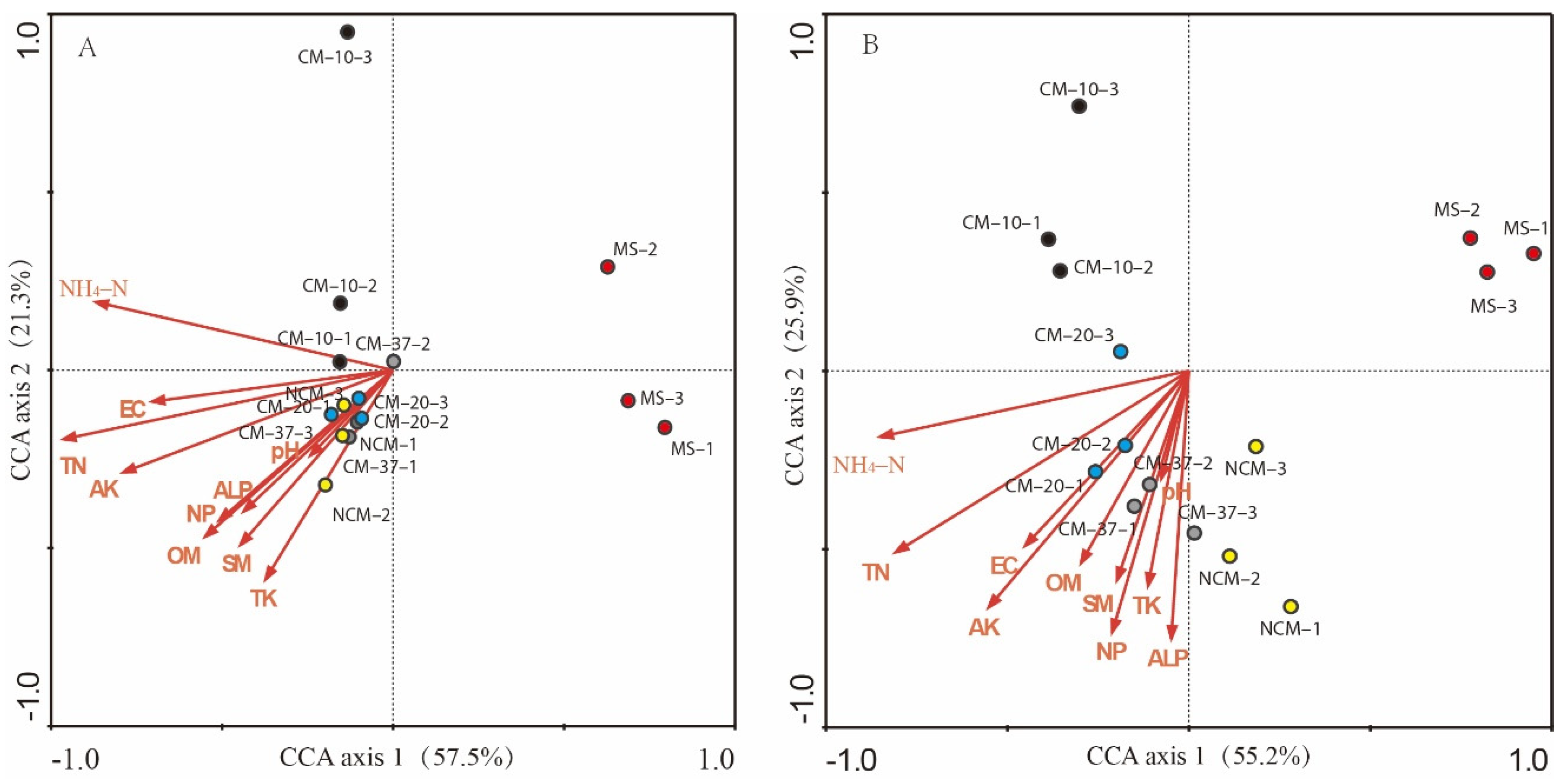
3.4. Dependence of PhoD and Gcd Communities on Soil Properties
4. Discussion
4.1. Improvement of Soil Properties through the Revegetation of C. microphylla on Sand Dunes
4.2. Relationship of PhoD Gene Abundance to Phosphomonoesterase Activity
4.3. Structures of PhoD and Gcd Microbial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Westheimer, F.H. Why nature chose phosphates. Science 1987, 235, 1173–1178. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Porder, S.; Houlton, B.Z.; Chadwick, O.A. Terrestrial phosphorus limitation: Mechanisms, implications, and nitrogen phosphorus interactions. Ecol. Appl. 2010, 20, 5–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragot, S.A.; Kertesz, M.A.; Bünemann, E.K. phoD alkaline phosphatase gene diversity in soil. Appl. Environ. Microbiol. 2015, 81, 7281–7289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, H.; Park, M.; Madhaiyan, M.; Seshadri, S.; Song, J.; Cho, H.; Sa, T. Isolation and characterization of phosphate solubilizing bacteria from the rhizosphere of crop plants of Korea. Soil Biol. Biochem. 2005, 37, 1970–1974. [Google Scholar] [CrossRef]
- Fraser, T.; Lynch, D.H.; Entz, M.H.; Dunfield, K.E. Linking alkaline phosphatase activity with bacterial phoD gene abundance in soil from a long-term management trial. Geoderma 2015, 257, 115–122. [Google Scholar] [CrossRef]
- Tanuwidjaja, I.; Vogel, C.; Pronk, G.J.; Scholer, A.; Kublik, S.; Vestergaard, G.; Kogel-Knabner, I.; Fuka, M.M.; Schloter, M.; Schulz, S. Microbial key players involved in P turnover differ in artificial soil mixtures depending on clay mineral composition. Microb. Ecol. 2021, 81, 897–907. [Google Scholar] [CrossRef]
- Chen, W.M.; Yang, F.; Zhang, L.; Wang, J.M. Organic acid secretion and phosphate solubilizing efficiency of pseudomonas sp. PSB12: Effects of phosphorus forms and carbon sources. Geomicrobiol. J. 2016, 33, 870–877. [Google Scholar] [CrossRef]
- Tan, H.; Barret, M.; Mooij, M.J.; Rice, O.; Morrissey, J.P.; Dobson, A.; Griffiths, B.; O’Gara, F. Long-term phosphorus fertilization increased the diversity of the total bacterial community and the phoD phosphorus mineraliser group in pasture soils. Biol. Fertil. Soils 2013, 49, 661–672. [Google Scholar] [CrossRef] [Green Version]
- Vershinina, O.A.; Znamenskaya, L.V. The Pho regulons of bacteria. Microbiology 2002, 71, 497–511. [Google Scholar] [CrossRef]
- Wu, J.R.; Shien, J.H.; Shieh, H.K.; Hu, C.C.; Gong, S.R.; Chen, L.Y.; Chang, P.C. Cloning of the gene and characterization of the enzymatic properties of the monomeric alkaline phosphatase (PhoX) from Pasteurella multocida strain X-73. FEMS Microbiol. Lett. 2007, 267, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Zappa, B.; Rolland, J.; Flament, D.; Gueguen, Y.; Boudrant, J.; Dietrich, J. Characterization of a highly thermostable alkaline phosphatase from the euryarchaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 2001, 67, 4504–4511. [Google Scholar] [CrossRef] [Green Version]
- Gomez, P.F.; Ingram, L.O. Cloning, sequencing and characterization of the alkaline-phosphatase gene (phoD) from Zymomonas-mobilis. FEMS Microbiol. Lett. 1995, 125, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.E.; Martiny, A.; Allison, S.D. Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J. 2013, 7, 1187–1199. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.W.; Benner, R.; Long, R.A.; Hu, J.J. Subcellular localization of marine bacterial alkaline phosphatases. Proc. Natl. Acad. Sci. USA 2009, 106, 21219–21223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kageyama, H.; Tripathi, K.; Rai, A.K.; Cha-um, S.; Waditee-Sirisattha, R.; Takabe, T. An alkaline phosphatase/phosphodiesterase, phoD, induced by salt stress and secreted out of the cells of aphanothece halophytica, a halotolerant Cyanobacterium. Appl. Environ. Microbiol. 2011, 77, 5178–5183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, M.J.; Penuelas, J.; Sardans, J.; Tong, C.; Chang, C.T.; Cao, W.Z. Dynamics of phosphorus speciation and the phoD phosphatase gene community in the rhizosphere and bulk soil along an estuarine freshwater-oligohaline gradient. Geoderma 2020, 365, 114236. [Google Scholar] [CrossRef]
- Chen, X.D.; Jiang, N.; Condron, L.M.; Dunfield, K.E.; Chen, Z.H.; Wang, J.K.; Chen, L.J. Impact of long-term phosphorus fertilizer inputs on bacterial phoD gene community in a maize field, Northeast China. Sci. Total Environ. 2019, 669, 1011–1018. [Google Scholar] [CrossRef]
- Hu, Y.J.; Xia, Y.H.; Sun, Q.; Liu, K.P.; Chen, X.B.; Ge, T.D.; Zhu, B.L.; Zhu, Z.K.; Zhang, Z.H.; Su, Y.R. Effects of long-term fertilization on phoD-harboring bacterial community in Karst soils. Sci. Total Environ. 2018, 628–629, 53–63. [Google Scholar] [CrossRef]
- Ragot, S.A.; Kertesz, M.A.; Meszaros, E.; Frossard, E.; Bunemann, E.K. Soil phoD and phoX alkaline phosphatase gene diversity responds to multiple environmental factors. FEMS Microbiol. Ecol. 2017, 93, fiw212. [Google Scholar] [CrossRef] [Green Version]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.W.; Wu, X.Q.; Wen, X.Y. Effects of soluble phosphate on phosphate-solubilizing characteristics and expression of gcd gene in Pseudomonas frederiksbergensis JW-SD2. Curr. Microbiol. 2016, 72, 198–206. [Google Scholar] [CrossRef]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-solubilizing microorganisms: Mechanism and their role in phosphate solubilization and uptake. J. Soil Sci. Plant Nutr. 2021, 21, 49–68. [Google Scholar] [CrossRef]
- An, R.; Moe, L.A. Regulation of pyrroloquinoline quinone-dependent glucose dehydrogenase activity in the model rhizosphere-dwelling bacterium Pseudomonas putida KT2440. Appl. Environ. Microbiol. 2016, 82, 4955–4964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, H.; Rossolini, G.M.; Gonzalez, T.; Li, J.; Glick, B.R. Isolation of a gene from Burkholderia cepacia IS-16 encoding a protein that facilitates phosphatase activity. Curr. Microbiol. 2000, 40, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.L.; Zhou, R.L.; Su, Y.Z.; Zhang, H.; Zhao, L.Y.; Drake, S. Shrub facilitation of desert land restoration in the Horqin Sand Land of Inner Mongolia. Ecol. Eng. 2007, 31, 1–8. [Google Scholar] [CrossRef]
- Li, S.G.; Harazono, Y.; Zhao, H.L.; He, Z.Y.; Chang, X.L.; Zhao, X.Y.; Zhang, T.H.; Oikawa, T. Micrometeorological changes following establishment of artificially established Artemisia vegetation on desertified sandy land in the Horqin sandy land, China and their implication on regional environmental change. J. Arid Environ. 2002, 52, 101–119. [Google Scholar] [CrossRef]
- Cao, C.Y.; Jiang, D.M.; Alamsa; Luo, Y.M.; Kou, Z.W. Ecological processes of vegetation restoration of Caragana mirophylla in the sand fixing area. Chin. J. Appl. Ecol. 2000, 11, 349–354. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, C.Y.; Cui, Z.B.; Qian, W.; Liang, C.P.; Wang, C.M. Soil bacterial community restoration along a chronosequence of sand-fixing plantations on moving sand dunes in the Horqin sandy land in northeast China. J. Arid Environ. 2019, 165, 81–87. [Google Scholar] [CrossRef]
- Cao, C.Y.; Jiang, D.M.; Teng, X.H.; Jiang, Y.; Liang, W.J.; Cui, Z.B. Soil chemical and microbiological properties along a chronosequence of Caragana microphylla Lam. plantations in the Horqin Sandy Land of Northeast China. Appl. Soil Ecol. 2008, 40, 78–85. [Google Scholar] [CrossRef]
- Li, H.L.; Zhang, Y.; Wang, T.T.; Feng, S.W.; Ren, Q.; Cui, Z.B.; Cao, C.Y. Responses of soil denitrifying bacterial communities carrying nirS, nirK, and nosZ genes to revegetation of moving sand dunes. Ecol. Indic. 2019, 107, 105541. [Google Scholar] [CrossRef]
- Cao, C.Y.; Zhang, Y.; Cui, Z.B.; Feng, S.W.; Wang, T.T.; Ren, Q. Soil bacterial community responses to revegetation of moving sand dune in semi-arid grassland. Appl. Environ. Microbiol. 2017, 101, 6217–6228. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, C.Y.; Peng, M.; Xu, X.J.; Zhang, P.; Yu, Q.J.; Sun, T. Diversity of nitrogen-fixing, ammonia-oxidizing, and denitrifying bacteria in biological soil crusts of a revegetation area in Horqin Sandy Land, Northeast China. Ecol. Eng. 2014, 71, 71–79. [Google Scholar] [CrossRef]
- Lin, D.Y. Guidance of Soil Science Experiment; China Forestry Publishing House: Beijing, China, 2004; pp. 45–70. [Google Scholar]
- Zheng, H.Y.; Zhang, D.S. Methods on Dynamics of Soil Biochemistry Study; Science Press: Beijing, China, 1982; pp. 167–169. [Google Scholar]
- Kandeler, E.; Gerber, H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils 1988, 6, 68–72. [Google Scholar] [CrossRef]
- Institute of Soil Science, Chinese Academy of Sciences (ISSCAS). Methods on Soil Microorganism Study; Science Press: Beijing, China, 1985; pp. 260–275. [Google Scholar]
- Tabatabai, M.A. Soil enzymes. In Methods of Soil Analysis; SSSA; Page, A.L., Millar, E.M., Keeney, D.R., Eds.; ASA: Madison, WI, USA, 1982; pp. 501–538. [Google Scholar]
- Schinner, F.; Öhlinger, R.; Kandeler, E.; Margesin, R. Methods in Soil Biology; Springer: Berlin, Germany, 1996. [Google Scholar]
- Sardans, J.; Peñuelas, J. Drought decreases soil enzyme activity in a Mediterranean Quercus ilex L. forest. Soil Biol. Biochem. 2005, 37, 455–461. [Google Scholar] [CrossRef]
- Ladd, J.N.; Butler, J.H.A. Short-term assays of soil proteolytic enzymeactivities using proteins and dipeptide derivatives as substrates. Soil Biol. Biochem. 1972, 4, 19–30. [Google Scholar] [CrossRef]
- Perucci, P.; Casucc, C.; Dumontet, S. An improved method to evaluate the o-diphenol oxidase activity of soil. Soil Biol. Biochem. 2000, 32, 1927–1933. [Google Scholar] [CrossRef]
- Xu, G.H.; Zheng, H.Y. Manual of Analytical Methods of Soil Microorganism; China Agriculture Press: Beijing, China, 1986; pp. 266–269. [Google Scholar]
- Hedley, M.J.; Stewart, J.W.B.; Chauhan, B.S. Changes in inorganic and organic soil phosphorus fractions induced by cultivation practices and by laboratory incubations. Soil Sci. Soc. Am. J. 1982, 46, 970–976. [Google Scholar] [CrossRef]
- da Silva, V.M.; Antoniolli, Z.I.; Jacques, R.J.S.; Ott, R.; Rodrigues, P.E.D.; Andrade, F.V.; Passos, R.R.; Mendonca, E.D. Influence of the tropical millipede, Glyphiulus granulatus (Gervais, 1847), on aggregation, enzymatic activity, and phosphorus fractions in the soil. Geoderma 2017, 289, 135–141. [Google Scholar] [CrossRef]
- Bergkemper, F.; Kublik, S.; Lang, F.; Kruger, J.; Vestergaard, G.; Schloter, M.; Schulz, S. Novel oligonucleotide primers reveal a high diversity of microbes which drive phosphorous turnover in soil. J. Microbiol. Methods 2016, 125, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinsinger, P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 2001, 237, 173–195. [Google Scholar] [CrossRef]
- Wang, R.Z.; Liu, H.Y.; Sardans, J.; Feng, X.; Xu, Z.W.; Penuelas, J. Interacting effects of urea and water addition on soil mineral-bound phosphorus dynamics in semi-arid grasslands with different land-use history. Eur. J. Soil Sci. 2021, 72, 946–962. [Google Scholar] [CrossRef]
- Alamgir, M.; McNeill, A.; Tang, C.X.; Marschner, P. Changes in soil P pools during legume residue decomposition. Soil Biol. Biochem. 2012, 49, 70–77. [Google Scholar] [CrossRef]
- Turner, B.L.; McKelvie, I.D.; Haygarth, P.M. Characterisation of water-extractable soil organic phosphorus by phosphatase hydrolysis. Soil Biol. Biochem. 2002, 34, 27–35. [Google Scholar] [CrossRef]
- Brookes, P.C.; Powlson, D.S.; Jenkinson, D.S. Phosphorus in the soil microbial biomass. Soil Biol. Biochem. 1984, 16, 169–175. [Google Scholar] [CrossRef]
- Cui, H.; Zhou, Y.; Gu, Z.H.; Zhu, H.H.; Fu, S.L.; Yao, Q. The combined effects of cover crops and symbiotic microbes on phosphatase gene and organic phosphorus hydrolysis in subtropical orchard soils. Soil Biol. Biochem. 2015, 82, 119–126. [Google Scholar] [CrossRef]
- Turner, B.L.; Haygarth, P.M. Phosphatase activity in temperate pasture soils: Potential regulation of labile organic phosphorus turnover by phosphodiesterase activity. Sci. Total Environ. 2005, 344, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.W.; Ling, N.; Nannipieri, P.; Chen, H.; Raza, W.; Wang, M.; Guo, S.W.; Shen, Q.R. Long-term fertilisation regimes affect the composition of the alkaline phosphomonoesterase encoding microbial community of a vertisol and its derivative soil fractions. Biol. Fertil. Soils 2017, 53, 375–388. [Google Scholar] [CrossRef]
- Hemkemeyer, M.; Christensen, B.T.; Martens, R.; Tebbe, C.C. Soil particle size fractions harbour distinct microbial communities and differ in potential for microbial mineralisation of organic pollutants. Soil Biol. Biochem. 2015, 90, 255–265. [Google Scholar] [CrossRef]
- Wang, C.Y.; Wang, R.Y.; Zhang, S.J.; Zhang, X.L.; Tian, L.J. Soil type and soil forming process in Wulanaodu Area. In Study on Integrated Control of Windy-Sandy Arid Zone in Northeast Inner Mongolia (I); Cao, X.S., Ed.; Inner Mongolia People Publishing House: Huhehaote, China, 1984; pp. 45–59. [Google Scholar]
- Lagos, L.M.; Acuna, J.J.; Maruyama, F.; Ogram, A.; Mora, M.D.; Jorquera, M.A. Effect of phosphorus addition on total and alkaline phosphomonoesterase-harboring bacterial populations in ryegrass rhizosphere microsites. Biol. Fertil. Soils 2016, 52, 1007–1019. [Google Scholar] [CrossRef]
- Yang, P.X.; Ma, L.; Chen, M.H.; Xi, J.Q.; He, F.; Duan, C.Q.; Mo, M.H.; Fang, D.H.; Duan, Y.Q.; Yang, F.X. Phosphate solubilizing ability and phylogenetic diversity of bacteria from P-rich soils around Dianchi Lake drainage area of China. Pedosphere 2012, 22, 707–716. [Google Scholar] [CrossRef]
- Jiang, D.M.; Cao, C.Y.; Zhang, Y.; Cui, Z.B.; Han, X.S. Plantations of native shrub species restore soil microbial diversity in the Horqin Sandy Land, northeastern China. J. Arid Land 2014, 6, 445–453. [Google Scholar] [CrossRef] [Green Version]
- Suleiman, A.K.A.; Manoeli, L.; Boldo, J.T.; Pereira, M.G.; Roesch, L.F.W. Shifts in soil bacterial community after eight years of land-use change. Syst. Appl. Microbiol. 2013, 36, 137–144. [Google Scholar] [CrossRef] [PubMed]


| Fractions | MS | CM-10 | CM-20 | CM-37 | NCM | ANOVA in Response to Plantation Age | ||
|---|---|---|---|---|---|---|---|---|
| R2 | F | p | ||||||
| H2O-Pi | 1.35 ± 0.09 | 1.64 ± 0.46 | 2.18 ± 0.74 | 2.90 ± 0.77 | 4.24 ± 1.56 | 0.667 | 26.066 | <0.001 |
| NaHCO3-Pi (0.5 mol L−1) | 5.82 ± 0.32 | 7.11 ± 0.77 | 10.69 ± 1.63 | 10.16 ± 1.02 | 15.17 ± 3.01 | 0.784 | 47.267 | <0.001 |
| NaHCO3-Po(0.5 mol L−1) | 43.18 ± 4.51 | 37.89 ± 2.37 | 44.31 ± 3.45 | 41.34 ± 10.19 | 32.83 ± 8.19 | 0.169 | 2.651 | 0.127 |
| HCl-Pi (1.0 mol L−1) | 4.22 ± 0.98 | 6.66 ± 1.14 | 12.70 ± 1.28 | 15.05 ± 2.44 | 19.36 ± 0.67 | 0.918 | 145.00 | <0.001 |
| HCl-Po (1.0 mol L−1) | 11.44 ± 0.61 | 13.34 ± 2.72 | 12.64 ± 4.01 | 17.61 ± 6.42 | 13.97 ± 1.78 | 0.105 | 1.519 | 0.240 |
| NaOH-Pi (0.5 mol L−1) | 6.82 ± 1.77 | 7.56 ± 0.84 | 8.06 ± 1.98 | 7.5 ± 1.98 | 9.19 ± 2.06 | 0.168 | 2.624 | 0.129 |
| NaOH-Po (0.5 mol L−1) | 44.68 ± 2.07 | 49.44 ± 3.37 | 51.44 ± 5.45 | 52.50 ± 4.06 | 52.81 ± 4.18 | 0.169 | 2.641 | 0.128 |
| NaOH-Pi (0.1 mol L−1) | 9.21 ± 0.98 | 9.66 ± 0.77 | 11.68 ± 0.83 | 11.85 ± 2.00 | 14.13 ± 1.95 | 0.662 | 25.47 | <0.001 |
| NaOH-Po (0.1 mol L−1) | 40.29 ± 5.25 | 48.84 ± 9.90 | 40.32 ± 4.77 | 54.15 ± 8.85 | 61.87 ± 14.62 | 0.413 | 9.158 | 0.009 |
| Residual-P | 67.50 ± 26.15 | 91.00 ± 27.10 | 135.0 ± 15.80 | 258.5 ± 62.93 | 518.0 ± 32.32 | 0.897 | 113.1 | <0.001 |
| Total-P | 234.5 ± 16.09 | 273.1 ± 24.94 | 329.0 ± 22.31 | 471.6 ± 70.48 | 741.6 ± 49.90 | 0.917 | 143.6 | <0.001 |
| Enzyme | MS | CM-10 | CM-20 | CM-37 | NCM | ANOVA in Response to Plantation Age | ||
|---|---|---|---|---|---|---|---|---|
| R2 | F | p | ||||||
| DHA (mg TPF kg−1 24 h−1) | 46.21 ± 1.33 | 48.86 ± 2.40 | 64.92 ± 11.35 | 97.65 ± 48.42 | 255.9 ± 71.29 | 0.711 | 31.97 | <0.001 |
| Urease (mg 100 g−1 24 h−1) | 0.51 ± 0.07 | 3.81 ± 0.05 | 9.81 ± 2.17 | 18.31 ± 11.33 | 32.28 ± 5.80 | 0.844 | 70.12 | <0.001 |
| PHA (mg g−1 h−1) | 2.28 ± 0.95 | 17.58 ± 8.11 | 45.99 ± 15.20 | 81.44 ± 10.40 | 159.4 ± 41.07 | 0.896 | 112.3 | <0.001 |
| POA (µmol g−1 10 min−1) | 1.99 ± 0.38 | 2.73 ± 0.68 | 3.25 ± 0.38 | 4.79 ± 1.00 | 4.66 ± 0.84 | 0.682 | 27.88 | <0.001 |
| Protease (mg Tyr g−1 2 h−1) | 8.17 ± 0.85 | 26.97 ± 11.02 | 55.54 ± 4.93 | 91.52 ± 14.89 | 96.03 ± 19.32 | 0.843 | 69.55 | <0.001 |
| GLA (μg g−1 h−1) | 0.14 ± 0.01 | 0.56 ± 0.04 | 0.58 ± 0.10 | 1.04 ± 0.03 | 1.372 ± 0.05 | 0.950 | 249.1 | <0.001 |
| Index | MS | CM-10 | CM-20 | CM-37 | NCM | One-Way ANOVA | ||
|---|---|---|---|---|---|---|---|---|
| F | p | |||||||
| Simpson | phoD | 0.939 ± 0.006 a | 0.986 ± 0.004 a | 0.973 ± 0.018 a | 0.985 ± 0.006 a | 0.960 ± 0.038 a | 3.078 | 0.068 |
| gcd | 0.940 ± 0.011 a | 0.878 ± 0.038 b | 0.948 ± 0.025 b | 0.967 ± 0.013 b | 0.941 ± 0.026 b | 5.550 | 0.013 | |
| Shannon-Wiener | phoD | 6.068 ± 0.143 a | 7.916 ± 0.157 b | 7.592 ± 0.362 b | 7.836 ± 0.306 b | 7.356 ± 0.959 b | 7.111 | 0.006 |
| gcd | 4.906 ± 0.315 a | 4.592 ± 0.404 a | 5.799 ± 0.497 b | 6.346 ± 0.414 b | 5.961 ± 0.471 b | 9.131 | 0.002 | |
| Pielou | phoD | 0.667 ± 0.015 a | 0.791 ± 0.015 b | 0.760 ± 0.035 b | 0.783 ± 0.027 b | 0.733 ± 0.078 a | 4.465 | 0.025 |
| gcd | 0.666 ± 0.041 b | 0.581 ± 0.057 a | 0.688 ± 0.050 b | 0.744 ± 0.017 b | 0.689 ± 0.039 b | 5.754 | 0.011 | |
| Chao1 | phoD | 765 ± 9 a | 1371 ± 78 b | 1382 ± 70 b | 1427 ± 67 b | 1450 ± 280 b | 13.35 | 0.001 |
| gcd | 239 ± 59 a | 289 ± 58 a | 413 ± 27 b | 431 ± 124 b | 472 ± 73 b | 5.260 | 0.015 | |
| Observed species | phoD | 548 ± 15 a | 1028 ± 51 b | 1014 ± 37 b | 1027 ± 48 b | 1048 ± 186 b | 17.08 | <0.001 |
| gcd | 168 ± 33 a | 243 ± 40 a | 345 ± 37 b | 377 ± 90 b | 404 ± 58 b | 5.572 | 0.002 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Zhang, Y.; Xia, Y.; Cui, Z.; Cao, C. Soil Microbial Community Succession Based on PhoD and Gcd Genes along a Chronosequence of Sand-Fixation Forest. Forests 2021, 12, 1707. https://doi.org/10.3390/f12121707
Wang F, Zhang Y, Xia Y, Cui Z, Cao C. Soil Microbial Community Succession Based on PhoD and Gcd Genes along a Chronosequence of Sand-Fixation Forest. Forests. 2021; 12(12):1707. https://doi.org/10.3390/f12121707
Chicago/Turabian StyleWang, Fei, Ying Zhang, Yong Xia, Zhenbo Cui, and Chengyou Cao. 2021. "Soil Microbial Community Succession Based on PhoD and Gcd Genes along a Chronosequence of Sand-Fixation Forest" Forests 12, no. 12: 1707. https://doi.org/10.3390/f12121707
APA StyleWang, F., Zhang, Y., Xia, Y., Cui, Z., & Cao, C. (2021). Soil Microbial Community Succession Based on PhoD and Gcd Genes along a Chronosequence of Sand-Fixation Forest. Forests, 12(12), 1707. https://doi.org/10.3390/f12121707





