Improving Plot-Level Model of Forest Biomass: A Combined Approach Using Machine Learning with Spatial Statistics
Abstract
:1. Introduction
2. Materials and Methods
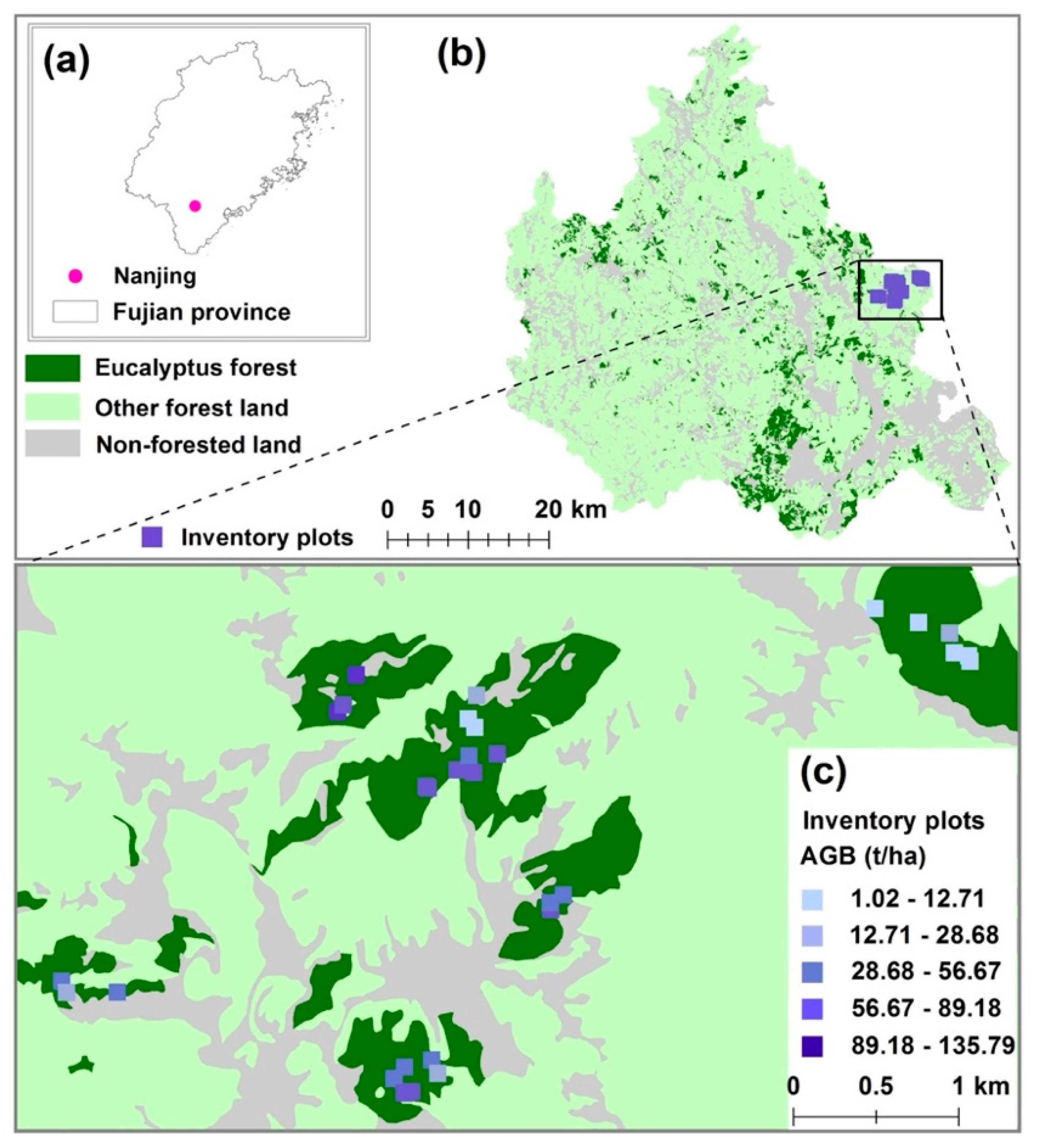
2.1. Study Area
2.2. Reference AGB Data
2.2.1. Destructive Sampling in Inventory Plots: Tree Harvest
2.2.2. Construction of Tree-Level Allometric Models
2.2.3. Calculating Reference AGB of Inventory Plots
2.2.4. Spatial Characteristic Test of Forest Reference AGB
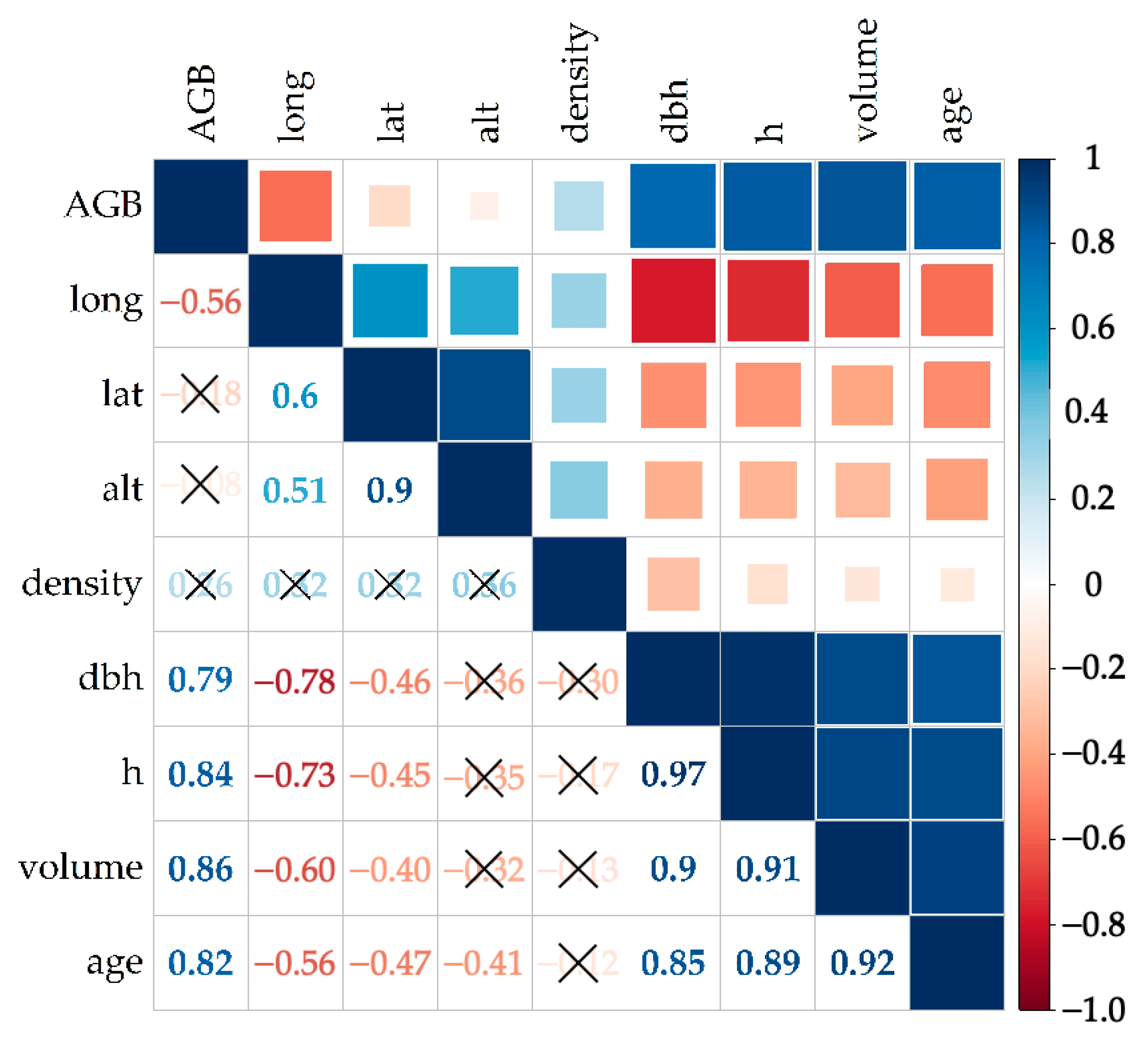
2.3. Selection of Variables
2.4. Model Development
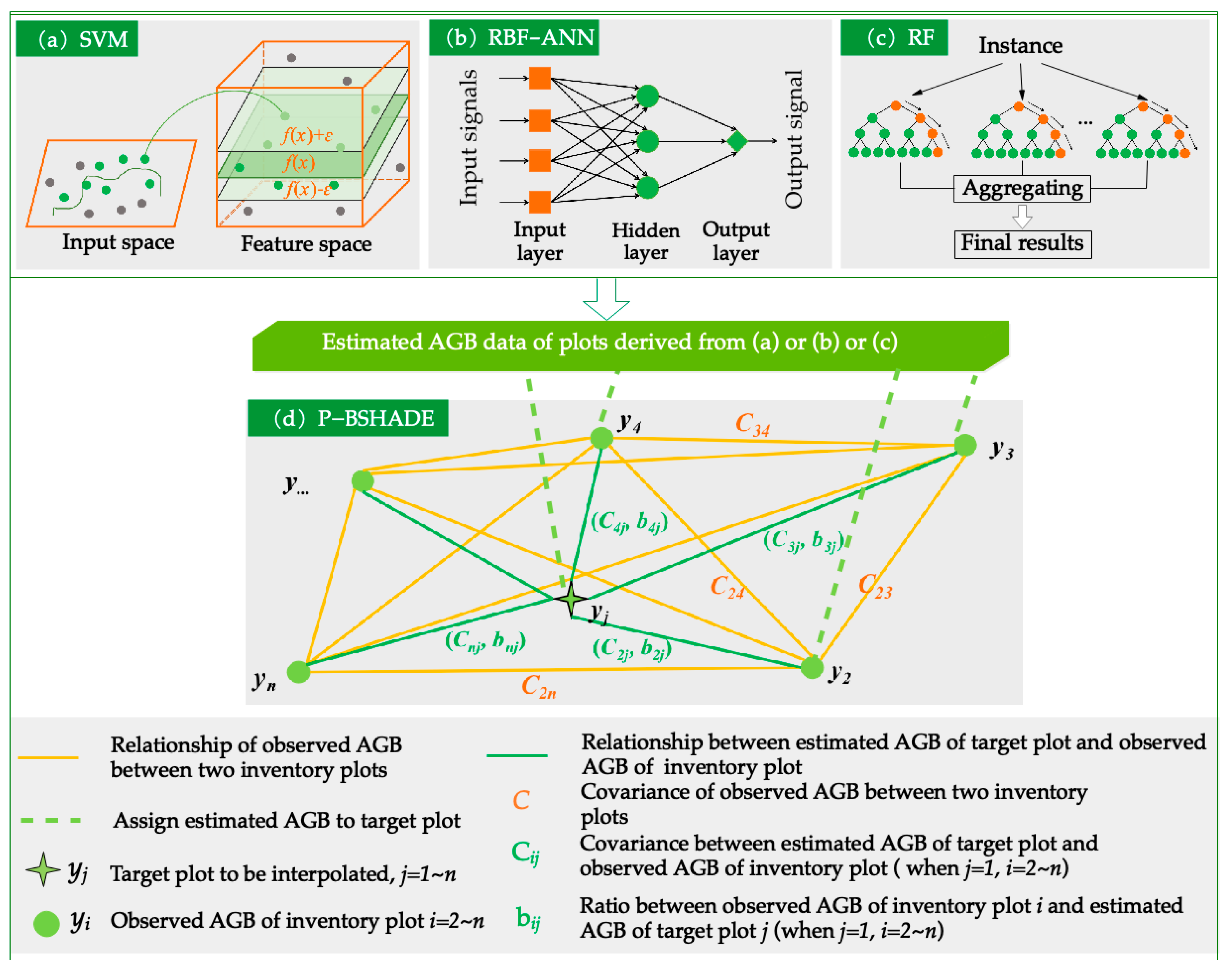
2.4.1. Machine Learning
2.4.2. Spatial Statistical Model: P-BSHADE
2.4.3. Combination of Machine Learning and Spatial Statistical Models
2.5. Model Evaluation and Comparison
2.5.1. Split Datasets
2.5.2. Calculate Indicators
2.5.3. Robustness of Combined Models
3. Results
3.1. Spatial Characteristic Test of Forest Reference AGB
3.2. Selection of Variables
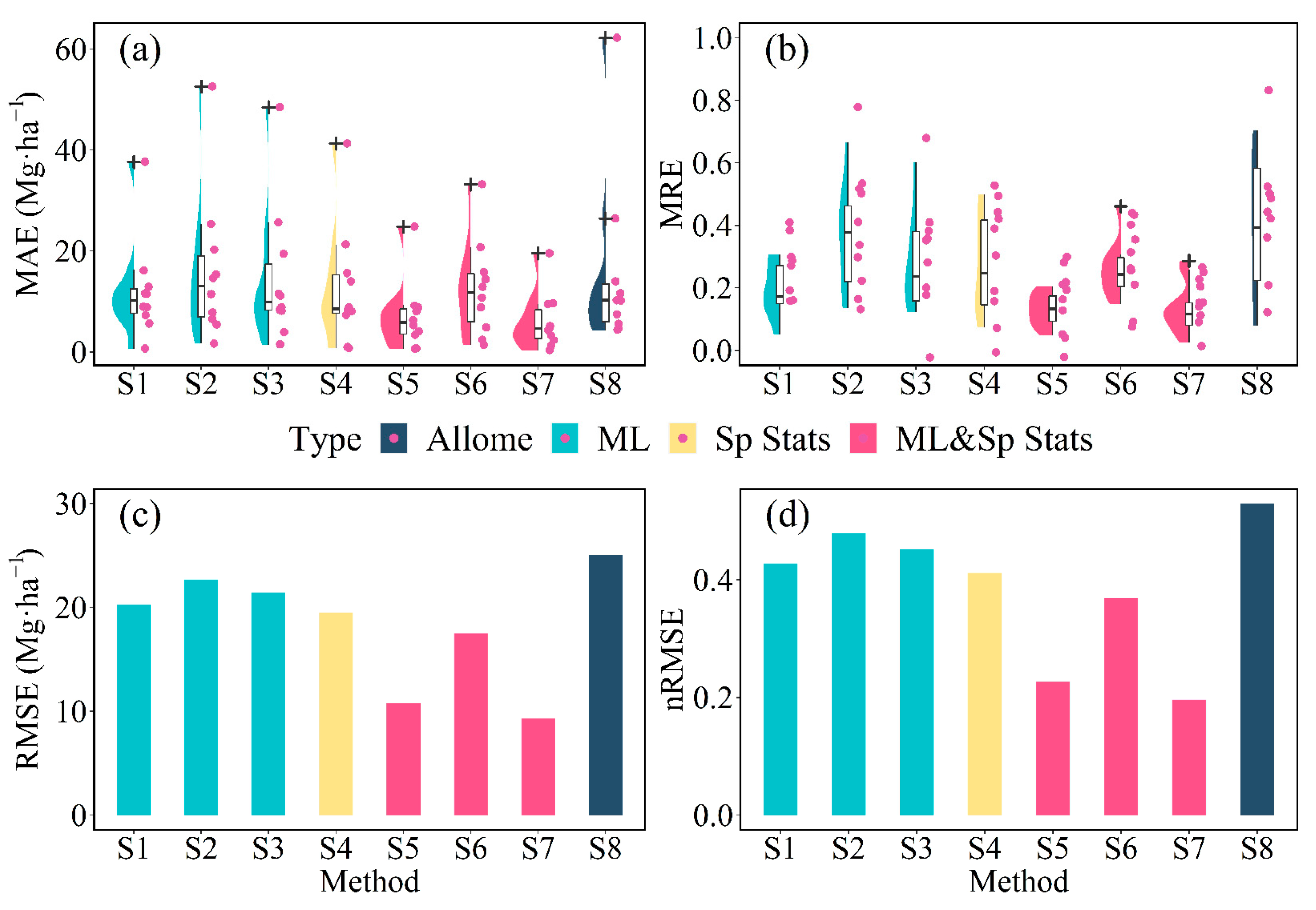
3.3. Performance of Plot-Level Models
4. Discussion
4.1. Significance and Challenges of Accurate Plot-Level AGB Estimates
4.2. Why a Combined Model Outperforms a Single Machine Learning or Spatial Statistical Model
4.3. Why the RF & P-BSHADE Method Outperforms Other Combined Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bustamante, M.M.C.; Roitman, I.; Aide, T.M.; Alencar, A.; Anderson, L.O.; Aragão, L.; Asner, G.P.; Barlow, J.; Berenguer, E.; Chambers, J.; et al. Toward an integrated monitoring framework to assess the effects of tropical forest degradation and recovery on carbon stocks and biodiversity. Glob. Chang. Biol. 2015, 22, 92–109. [Google Scholar] [CrossRef]
- Chen, Q.; Laurin, G.V.; Valentini, R. Uncertainty of remotely sensed aboveground biomass over an African tropical forest: Propagating errors from trees to plots to pixels. Remote Sens. Environ. 2015, 160, 134–143. [Google Scholar] [CrossRef]
- Sileshi, G.W. A critical review of forest biomass estimation models, common mistakes and corrective measures. For. Ecol. Manag. 2014, 329, 237–254. [Google Scholar] [CrossRef]
- Mauya, E.W.; Ene, L.T.; Bollandsås, O.M.; Gobakken, T.; Naesset, E.; Malimbwi, R.E.; Zahabu, E. Modelling aboveground forest biomass using airborne laser scanner data in the miombo woodlands of Tanzania. Carbon Balance Manag. 2015, 10, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gleason, C.J.; Im, J. Forest biomass estimation from airborne LiDAR data using machine learning approaches. Remote Sens. Environ. 2012, 125, 80–91. [Google Scholar] [CrossRef]
- Benítez, F.L.; Anderson, L.O.; Formaggio, A.R. Evaluation of geostatistical techniques to estimate the spatial distribution of aboveground biomass in the Amazon rainforest using high-resolution remote sensing data. Acta Amaz. 2016, 46, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Propastin, P. Modifying geographically weighted regression for estimating aboveground biomass in tropical rainforests by multispectral remote sensing data. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 82–90. [Google Scholar] [CrossRef]
- Van Der Laan, C.; Verweij, P.; Quiñones, M.J.; Faaij, A.P. Analysis of biophysical and anthropogenic variables and their relation to the regional spatial variation of aboveground biomass illustrated for North and East Kalimantan, Borneo. Carbon Balance Manag. 2014, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babcock, C.; Finley, A.O.; Bradford, J.B.; Kolka, R.; Birdsey, R.; Ryan, M.G. LiDAR based prediction of forest biomass using hierarchical models with spatially varying coefficients. Remote Sens. Environ. 2015, 169, 113–127. [Google Scholar] [CrossRef] [Green Version]
- Babcock, C.; Finley, A.O.; Cook, B.D.; Weiskittel, A.; Woodall, C.W. Modeling forest biomass and growth: Coupling long-term inventory and LiDAR data. Remote Sens. Environ. 2016, 182, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Gorgens, E.; Montaghi, A.; Rodriguez, L.C. A performance comparison of machine learning methods to estimate the fast-growing forest plantation yield based on laser scanning metrics. Comput. Electron. Agric. 2015, 116, 221–227. [Google Scholar] [CrossRef]
- Zhao, K.; Popescu, S.; Meng, X.; Pang, Y.; Agca, M. Characterizing forest canopy structure with lidar composite metrics and machine learning. Remote Sens. Environ. 2011, 115, 1978–1996. [Google Scholar] [CrossRef]
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach, 3rd ed.; Pearson Education Limited: London, UK, 2016. [Google Scholar]
- Frey, U.J.; Klein, M.; Deissenroth, M. Modelling complex investment decisions in Germany for renewables with different machine learning algorithms. Environ. Model. Softw. 2019, 118, 61–75. [Google Scholar] [CrossRef]
- Du, H.; Zhou, G.; Fan, W.; Ge, H.; Xu, X.; Shi, Y.; Fan, W. Spatial heterogeneity and carbon contribution of aboveground biomass of moso bamboo by using geostatistical theory. Plant Ecol. 2009, 207, 131–139. [Google Scholar] [CrossRef]
- Viana, H.; Aranha, J.; Lopes, D.; Cohen, W.B. Estimation of crown biomass of Pinus pinaster stands and shrubland above-ground biomass using forest inventory data, remotely sensed imagery and spatial prediction models. Ecol. Model. 2012, 226, 22–35. [Google Scholar] [CrossRef]
- Mitchard, E.T.A.; Feldpausch, T.R.; Brienen, R.J.W.; Lopez-Gonzalez, G.; Monteagudo, A.; Baker, T.R.; Lewis, S.L.; Lloyd, J.; Quesada, C.A.; Gloor, M.; et al. Markedly divergent estimates of A mazon forest carbon density from ground plots and satellites. Glob. Ecol. Biogeogr. 2014, 23, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Hengl, T.; Heuvelink, G.B.; Stein, A. A generic framework for spatial prediction of soil variables based on regression-kriging. Geoderma 2004, 120, 75–93. [Google Scholar] [CrossRef] [Green Version]
- Schabenberger, O.; Gotway, C.A. Statistical Methods for Spatial Data Analysis; Chapman & Hall0CRC: Boca Raton, FL, USA, 2005. [Google Scholar]
- Paul, K.I.; Roxburgh, S.H.; Chave, J.; England, J.; Zerihun, A.; Specht, A.; Lewis, T.; Bennett, L.; Baker, T.G.; Adams, M.; et al. Testing the generality of above-ground biomass allometry across plant functional types at the continent scale. Glob. Chang. Biol. 2016, 22, 2106–2124. [Google Scholar] [CrossRef]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.A.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S.; et al. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, D.; Rademacher, J.; Chen, J.; Crow, T.; Bresee, M.; Le Moine, J.; Ryu, S.-R. Estimating aboveground biomass using Landsat 7 ETM+ data across a managed landscape in northern Wisconsin, USA. Remote Sens. Environ. 2004, 93, 402–411. [Google Scholar] [CrossRef]
- Roman, K.; Barwicki, J.; Rzodkiewicz, W.; Dawidowski, M. Evaluation of Mechanical and Energetic Properties of the Forest Residues Shredded Chips during Briquetting Process. Energies 2021, 14, 3270. [Google Scholar] [CrossRef]
- Cliff, A.; Ord, V.J. Spatial Processes: Model and Applications; Pion Ltd.: London, UK, 1981. [Google Scholar]
- Wang, J.F.; Li, X.H.; Christakos, G.; Liao, Y.L.; Zhang, T.; Gu, X.; Zheng, X.Y. Geographical detectors-based health risk assessment and its application in the neural tube defects study of the Heshun region, China. Int. J. Geogr. Inf. Sci. 2010, 24, 107–127. [Google Scholar] [CrossRef]
- Wang, J.F.; Zhang, T.L.; Fu, B.J. A measure of spatial stratified heterogeneity. Ecol. Indic. 2016, 67, 250–256. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, C.; Zuo, S.; Li, Z. Scaling up of biomass simulation for Eucalyptus plantations based on landsenses ecology. Int. J. Sustain. Dev. World Ecol. 2017, 24, 135–148. [Google Scholar] [CrossRef]
- Cracknell, M.; Reading, A. Geological mapping using remote sensing data: A comparison of five machine learning algorithms, their response to variations in the spatial distribution of training data and the use of explicit spatial information. Comput. Geosci. 2014, 63, 22–33. [Google Scholar] [CrossRef] [Green Version]
- Fassnacht, F.; Hartig, F.; Latifi, H.; Berger, C.; Hernández, J.; Corvalán, P.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- Were, K.; Bui, D.T.; Dick, Ø.B.; Singh, B.R. A comparative assessment of support vector regression, artificial neural networks, and random forests for predicting and mapping soil organic carbon stocks across an Afromontane landscape. Ecol. Indic. 2015, 52, 394–403. [Google Scholar] [CrossRef]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Sayad, S. Support Vector Machine-Regression (SVR). Available online: https://www.saedsayad.com/support_vector_machine_reg.htm (accessed on 28 November 2021).
- Elanayar, V.T.S.; Shin, Y. Radial basis function neural network for approximation and estimation of nonlinear stochastic dynamic systems. IEEE Trans. Neural Netw. 1994, 5, 594–603. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.-G.; Wang, J.-F.; Zhao, Y.; Jia, L. A B-SHADE based best linear unbiased estimation tool for biased samples. Environ. Model. Softw. 2013, 48, 93–97. [Google Scholar] [CrossRef]
- Xu, C.-D.; Wang, J.; Hu, M.; Li, Q. Interpolation of Missing Temperature Data at Meteorological Stations Using P-BSHADE*. J. Clim. 2013, 26, 7452–7463. [Google Scholar] [CrossRef]
- Meyer, H.; Reudenbach, C.; Wöllauer, S.; Nauss, T. Importance of spatial predictor variable selection in machine learning applications –Moving from data reproduction to spatial prediction. Ecol. Model. 2019, 411, 108815. [Google Scholar] [CrossRef] [Green Version]
- Pohjankukka, J.; Pahikkala, T.; Nevalainen, P.; Heikkonen, J. Estimating the prediction performance of spatial models via spatial k-fold cross validation. Int. J. Geogr. Inf. Sci. 2017, 31, 2001–2019. [Google Scholar] [CrossRef]
- Roberts, D.R.; Bahn, V.; Ciuti, S.; Boyce, M.; Elith, J.; Guillera-Arroita, G.; Hauenstein, S.; Lahoz-Monfort, J.J.; Schroder, B.; Thuiller, W.; et al. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]
- Valavi, R.; Elith, J.; Lahoz-Monfort, J.J.; Guillera-Arroita, G. blockCV: An r package for generating spatially or environmentally separated folds for k -fold cross-validation of species distribution models. Methods Ecol. Evol. 2019, 10, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Marvin, D.C.; Asner, G.; Knapp, D.E.; Anderson, C.; Martin, R.E.; Sinca, F.; Tupayachi, R. Amazonian landscapes and the bias in field studies of forest structure and biomass. Proc. Natl. Acad. Sci. USA 2014, 111, E5224–E5232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Probst, P.; Wright, M.N.; Boulesteix, A. Hyperparameters and tuning strategies for random forest. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, 1301. [Google Scholar] [CrossRef] [Green Version]
| Item | Mean ± Standard Deviation | Median | Range (Minimum, Maximum) |
|---|---|---|---|
| AGB (t/ha) | 47.34 ± 34.46 | 46.64 | (1.02, 135.79) |
| Longitude | 117.48 ± 0.02 | 117.47 | (117.446, 117.503) |
| Latitude | 24.71 ± 0.01 | 24.71 | (24.694, 24.721) |
| Altitude (m) | 269.2 ± 82.5 | 313.5 | (135, 389) |
| Stand density (stems/ha) | 852.5 ± 251.8 | 800.0 | (450, 1375) |
| DBH (cm) | 12.29 ± 4.48 | 13.19 | (2.19, 17.99) |
| H (m) | 12.98 ± 4.72 | 14.42 | (2.83, 18.23) |
| Age (years) | 5.5 ± 2.92 | 5.5 | (1, 10) |
| Number | Age (Years) | a | b | R2 |
|---|---|---|---|---|
| 1 | 1–2 | 0.1538 | 0.6993 | 0.99 |
| 2 | 3–5 | 0.0377 | 0.9244 | 0.98 |
| 3 | 6–10 | 0.0689 | 0.8489 | 0.88 |
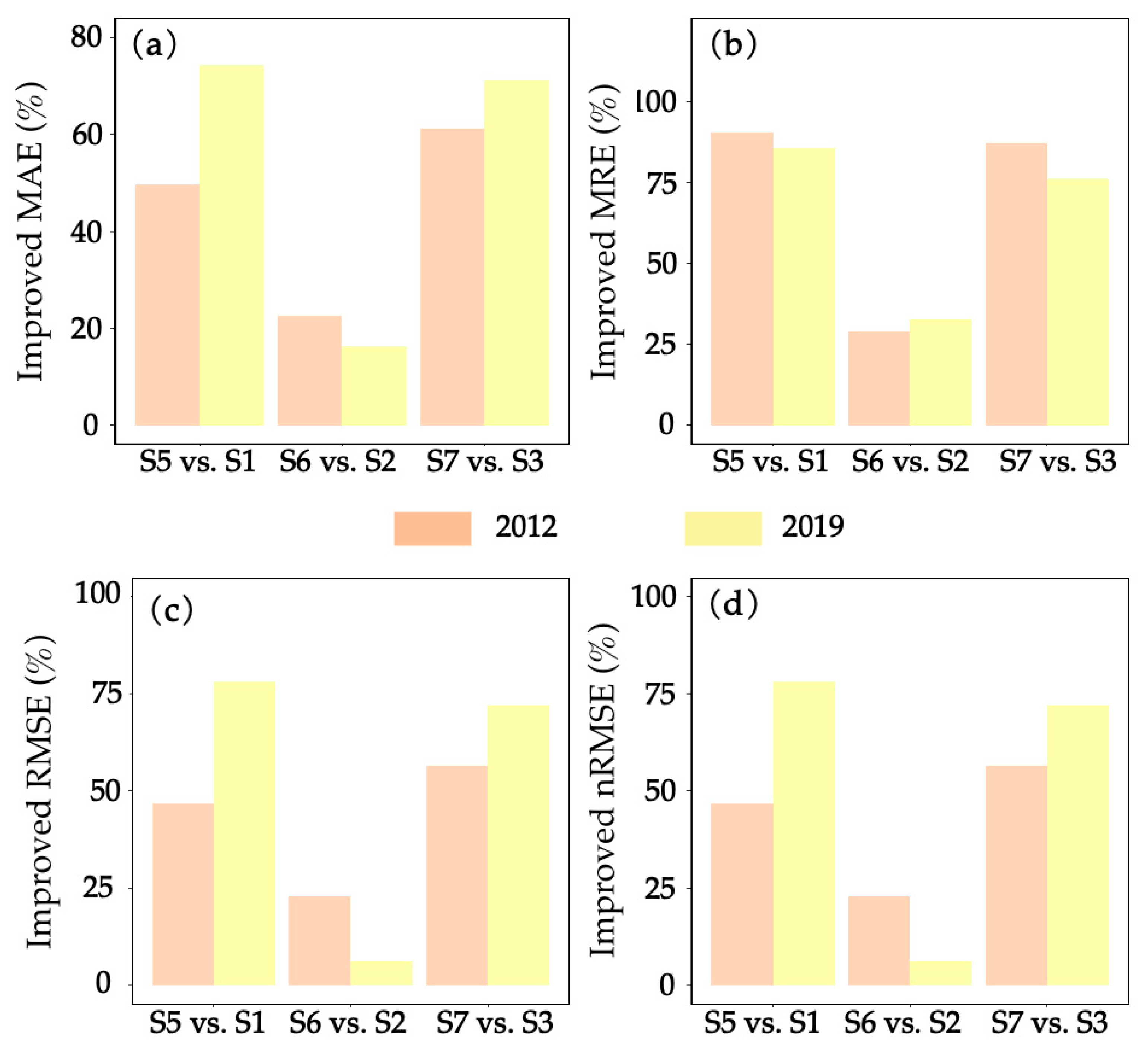
| Method | p-Value for MAE | p-Value for MRE | p-Value for RMSE | p-Value for nRMSE | ||||
|---|---|---|---|---|---|---|---|---|
| 2012 | 2019 | 2012 | 2019 | 2012 | 2019 | 2012 | 2019 | |
| S1 vs. S5 | 0.209 | 0.005 *** | 0.289 | 0.108 | 0.148 | 0.009 *** | 0.168 | 0.009 *** |
| S2 vs. S6 | 0.525 | 0.582 | 0.090 * | 0.269 | 0.588 | 0.809 | 0.134 | 0.817 |
| S3 vs. S7 | 0.081 * | 0.029 ** | 0.216 | 0.033 ** | 0.091 * | 0.040 ** | 0.089 * | 0.003 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, S.; Zheng, X.; Gao, L.; Xu, C.; Zuo, S.; Chen, Q.; Wei, X.; Ren, Y. Improving Plot-Level Model of Forest Biomass: A Combined Approach Using Machine Learning with Spatial Statistics. Forests 2021, 12, 1663. https://doi.org/10.3390/f12121663
Dai S, Zheng X, Gao L, Xu C, Zuo S, Chen Q, Wei X, Ren Y. Improving Plot-Level Model of Forest Biomass: A Combined Approach Using Machine Learning with Spatial Statistics. Forests. 2021; 12(12):1663. https://doi.org/10.3390/f12121663
Chicago/Turabian StyleDai, Shaoqing, Xiaoman Zheng, Lei Gao, Chengdong Xu, Shudi Zuo, Qi Chen, Xiaohua Wei, and Yin Ren. 2021. "Improving Plot-Level Model of Forest Biomass: A Combined Approach Using Machine Learning with Spatial Statistics" Forests 12, no. 12: 1663. https://doi.org/10.3390/f12121663
APA StyleDai, S., Zheng, X., Gao, L., Xu, C., Zuo, S., Chen, Q., Wei, X., & Ren, Y. (2021). Improving Plot-Level Model of Forest Biomass: A Combined Approach Using Machine Learning with Spatial Statistics. Forests, 12(12), 1663. https://doi.org/10.3390/f12121663









