Targeted CRISPR/Cas9-Based Knock-Out of the Rice Orthologs TILLER ANGLE CONTROL 1 (TAC1) in Poplar Induces Erect Leaf Habit and Shoot Growth
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Identification of Putative PpeTAC1 Homologs in Poplar
2.3. Guide RNA (gRNA) Design and Agrobacterium-Mediated Poplar Transformation
2.4. Molecular Analyses of Transgenic Plants and Sequencing of CRISPR/Cas9-Induced Mutations
2.5. Morphological Investigations of Glasshouse Grown Plants
2.6. Data Analysis and Statistics
3. Results
3.1. Regeneration and Molecular Analysis of Putative TAC1 Mutants
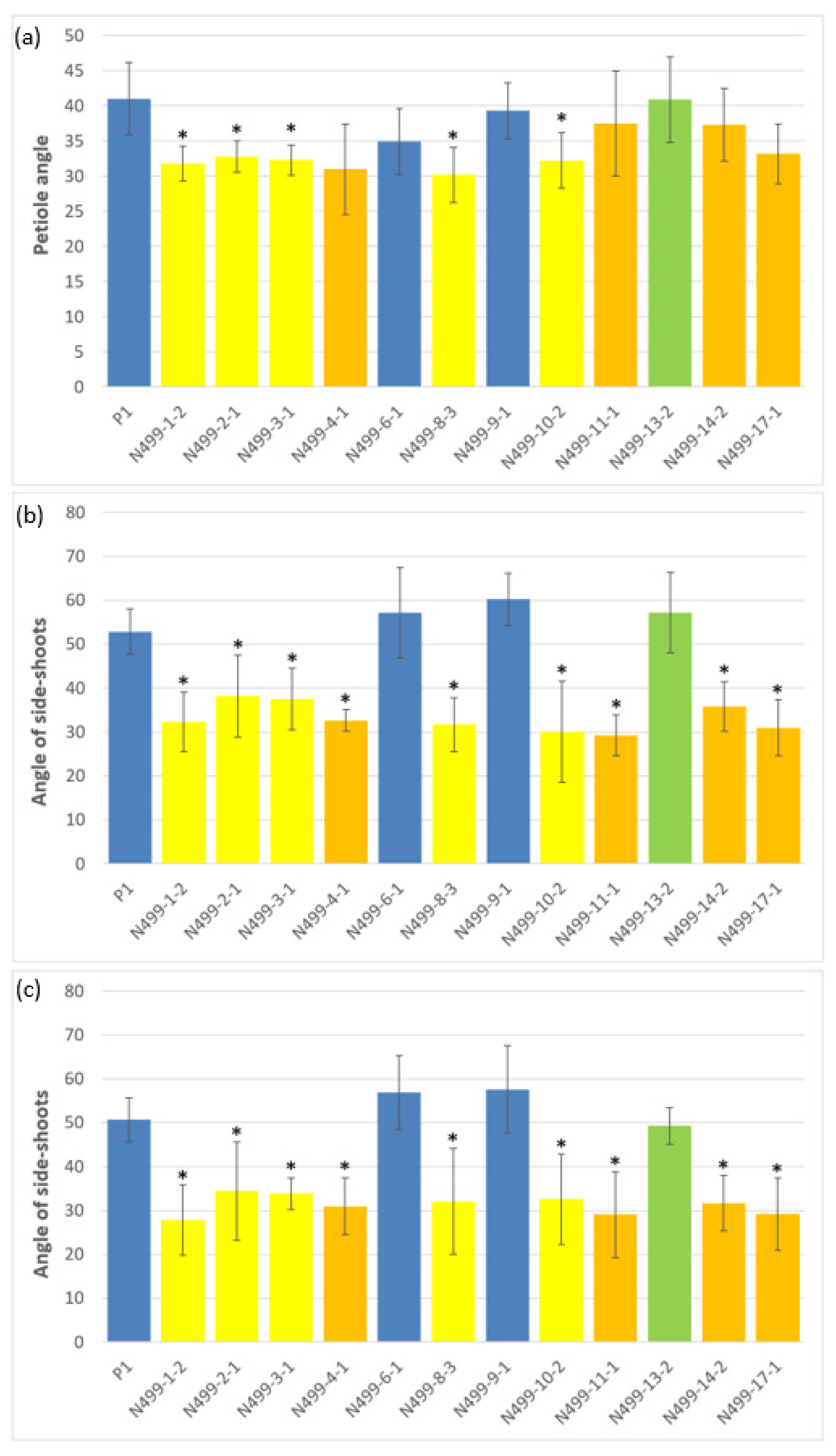
3.2. Morphological Investigations of In Vitro and Glasshouse Grown Plants
4. Discussion and Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Reinhardt, D.; Kuhlemeier, C. Plant architecture. EMBO Rep. 2002, 3, 846–851. [Google Scholar] [CrossRef]
- Fladung, M.; Bossinger, G.; Roeb, G.W.; Salamini, F. Correlated alterations in leaf and flower morphology and rate of photosynthesis in a midribless (mbl) mutant of Panicum maximum (Jacq.). Planta 1991, 184, 356–361. [Google Scholar] [CrossRef]
- Fladung, M. Genetic variants of Panicum maximum (Jacq.) in C4 photosynthetic traits. J. Plant Physiol. 1994, 143, 165–172. [Google Scholar] [CrossRef]
- Godin, C. Representing and encoding plant architecture: A review. Ann. For. Sci. 2000, 57, 413–438. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J. Molecular basis of plant architecture. Ann. Rev. Plant Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef]
- Costes, E. Physiology and genetics of plant architecture. Ann. Plant Rev. Online 2019, 2, 1031–1068. [Google Scholar]
- Barthélémy, D.; Caraglio, Y. Plant architecture: A dynamic, multilevel and comprehensive approach to plant form, structure and ontogeny. Ann. Bot. 2007, 99, 375–407. [Google Scholar] [CrossRef] [Green Version]
- Chelakkot, R.; Mahadevan, L. On the growth and form of shoots. J. R. Soc. Interface 2017, 14, 20170001. [Google Scholar] [CrossRef] [Green Version]
- Nybom, N. Mutation types in barley. Acta Agricult. Scand. 1954, 4, 430–456. [Google Scholar] [CrossRef]
- Kharkwal, M.C.; Pandey, R.N.; Pawar, S.E. Mutation breeding for crop improvement. In Plant Breeding; Jain, H.K., Kharkwal, M.C., Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 601–645. [Google Scholar]
- Pathirana, R. Plant mutation breeding in agriculture. Plant Sci. Rev. 2011, 6, 107–126. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnolog. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Holme, I.B.; Gregersen, P.L.; Brinch-Pedersen, H. Induced genetic variation in crop plants by random or targeted mutagenesis: Convergence and differences. Front. Plant Sci. 2019, 10, 1468. [Google Scholar] [CrossRef]
- Lamo, K.; Bhat, D.J.; Kour, K.; Solanki, S.P.S. Mutation studies in fruit crops: A review. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3620–3633. [Google Scholar] [CrossRef]
- Fladung, M.; Ziegenhagen, B. M13 DNA fingerprinting can be used in studies on phenotypical revisions of forest tree mutants. Trees 1998, 12, 310–314. [Google Scholar] [CrossRef]
- Fladung, M.; Tusch, A.; Markussen, T.; Ziegenhagen, B. Analysis of morphological mutants in Picea. In Proceedings of the Applications of Biotechnology to Forest Genetics (Biofor 99), Vitoria-Gasteiz, Spain, 22–25 September 1999; Espinel, S., Ritter, E., Eds.; pp. 167–170. [Google Scholar]
- Flachowsky, H.; Hanke, M.V.; Peil, A.; Strauss, S.H.; Fladung, M. A review on transgenic approaches to accelerate breeding of woody plants. Plant Breed. 2009, 128, 217–226. [Google Scholar] [CrossRef]
- Teichmann, T.; Muhr, M. Shaping plant architecture. Front. Plant Sci. 2015, 6, 233. [Google Scholar] [CrossRef] [Green Version]
- Zawaski, C.; Kadmiel, M.; Pickens, J.; Ma, C.; Strauss, S.H.; Busov, V. Repression of gibberellin biosynthesis or signaling produces striking alterations in poplar growth, morphology, and flowering. Planta 2011, 234, 1285–1298. [Google Scholar] [CrossRef]
- Buhl, C.; Strauss, S.H.; Lindroth, R.L. Genetic down-regulation of gibberellin results in semi-dwarf poplar but few non-target effects on chemical resistance and tolerance to defoliation. J. Plant Ecol. 2019, 12, 124–136. [Google Scholar] [CrossRef] [Green Version]
- Busov, V.B.; Meilan, R.; Pearce, D.W.; Ma, C.; Rood, S.B.; Strauss, S.H. Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 2003, 132, 1283–1291. [Google Scholar] [CrossRef] [Green Version]
- Weigel, D.; Nilsson, O. A developmental switch sufficient for flower initiation in diverse plants. Nature 1995, 377, 495–500. [Google Scholar] [CrossRef]
- Rottmann, W.H.; Meilan, R.; Sheppard, L.A.; Brunner, A.M.; Skinner, J.S.; Ma, C.; Cheng, S.; Jouanin, L.; Pilate, G.; Strauss, S.H. Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 2000, 22, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.M.; Chen, S.; Lin, L.; Wei, R.; Li, H.Y.; Liu, G.F.; Jiang, J. Genome-wide analysis of a TaLEA-introduced transgenic Populus simonii × Populus nigra dwarf mutant. Int. J. Mol. Sci. 2012, 13, 2744–2762. [Google Scholar] [CrossRef]
- Bruegmann, T.; Fladung, M. Overexpression of both flowering time genes AtSOC1 and SaFUL revealed huge influence onto plant habitus in poplar. Tree Genet. Genom. 2019, 15, 1–13. [Google Scholar] [CrossRef]
- Fladung, M. Transformation of diploid and tetraploid potato clones with the rolC gene of Agrobacterium rhizogenes and characterization of transgenic plants. Plant Breed. 1990, 104, 295–304. [Google Scholar] [CrossRef]
- Schmülling, T.; Fladung, M.; Großmann, K.; Schell, J. Hormonal content and sensitivity of transgenic tobacco and potato plants expressing single rol genes of Agrobacterium rhizogenes T-DNA. Plant J. 1993, 3, 371–382. [Google Scholar] [CrossRef]
- Fladung, M.; Muhs, H.J.; Ahuja, M.R. Morphological changes observed in transgenic Populus carrying the rolC gene from Agrobacterium rhizogenes. Silv. Genet. 1996, 45, 349–354. [Google Scholar]
- Fladung, M.; Kumar, S.; Ahuja, M.R. Genetic transformation of Populus genotypes with different chimaeric gene constructs: Transformation efficiency and molecular analysis. Transgen. Res. 1997, 6, 111–121. [Google Scholar] [CrossRef]
- Fladung, M.; Polak, O. Ac/Ds-transposon activation tagging in poplar: A powerful tool for gene discovery. BMC Genom. 2012, 13, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechol. 2000, 18, 784–788. [Google Scholar] [CrossRef]
- Dünisch, O.; Funada, R.; Nakaba, S.; Fladung, M. Influence of overexpression of a gibberellin 20-oxidase gene on the kinetics of xylem cell development in hybrid poplar (Populus tremula L × P. tremuloides Michx). Holzforschung 2006, 60, 608–617. [Google Scholar] [CrossRef]
- Jeon, H.W.; Cho, J.S.; Park, E.J.; Han, K.H.; Choi, Y.I.; Ko, J.H. Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 2016, 14, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Bruegmann, T.; Wetzel, H.; Hettrich, K.; Smeds, A.; Willför, S.; Kersten, B.; Fladung, M. Knockdown of PCBER1, a gene of neolignan biosynthesis, resulted in increased poplar growth. Planta 2019, 249, 515–525. [Google Scholar] [CrossRef]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S.; et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef]
- Richards, R.A.; Cavanagh, C.R.; Riffkin, P. Selection for erect canopy architecture can increase yield and biomass of spring wheat. Field Crops Res. 2019, 244, 107649. [Google Scholar] [CrossRef]
- Burgess, A.J.; Retkute, R.; Herman, T.; Murchie, E.H. Exploring relationships between canopy architecture, light distribution, and photosynthesis in contrasting rice genotypes using 3D canopy reconstruction. Front. Plant Sci. 2017, 8, 734. [Google Scholar] [CrossRef]
- Mantilla-Perez, M.B.; Salas Fernandez, M.G. Differential manipulation of leaf angle throughout the canopy: Current status and prospects. J. Exp. Bot. 2017, 68, 5699–5717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, T.R.; Sheehy, J.E. Erect leaves and photosynthesis in rice. Science 1999, 283, 1456. [Google Scholar] [CrossRef]
- Scorza, R.; Lightner, G.W.; Liverani, A. The pillar peach tree and growth habit analysis of compact x pillar progeny. J. Amer. Soc. Hortic. Sci. 1989, 114, 991–995. [Google Scholar]
- Kelsey, D.F.; Brown, S.K. ‘McIntosh Wijcik’: A columnar mutation of ‘McIntosh’ apple proving useful in physiology and breeding research. Fruit Var. J. 1992, 46, 83–87. [Google Scholar]
- Tworkoski, T.; Scorza, R. Root and shoot characteristics of peach trees with different growth habits. J. Amer. Soc. Hortic. Sci. 2001, 126, 785–790. [Google Scholar] [CrossRef] [Green Version]
- Dardick, C.; Callahan, A.; Horn, R.; Ruiz, K.B.; Zhebentyayeva, T.; Hollender, C.; Whitaker, M.; Abbott, A.; Scorza, R. PpeTAC1 promotes the horizontal growth of branches in peach trees and is a member of a functionally conserved gene family found in diverse plants species. Plant J. 2013, 75, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Hollender, C.A.; Waite, J.M.; Tabb, A.; Raines, D.; Chinnithambi, S.; Dardick, C. Alteration of TAC1 expression in Prunus species leads to pleiotropic shoot phenotypes. Horticult. Res. 2018, 5, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Papageorgiou, A.C.; Panetsos, K.P.; Hattemer, H.H. Genetic differentiation of natural Mediterranean cypress (Cupressus sempervirens L.) populations in Greece. For. Genet. 1994, 1, 1–12. [Google Scholar]
- Hoffman, M.H.A. Cultivar classification of Taxus L. (Taxaceae). Acta Horticult. 2004, 634, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Kwantlen Polytechnic University. School of Horticulture Plant Database—Populus tremula ‘Erecta’. Available online: https://plantdatabase.kpu.ca/plant/plantDetail/284 (accessed on 22 November 2021).
- Xu, X.; Tong, L.; Li, F.; Kang, S.; Qu, Y. Sap flow of irrigated Populus alba var. pyramidalis and its relationship with environmental factors and leaf area index in an arid region of Northwest China. J. For. Res. 2011, 16, 144–152. [Google Scholar] [CrossRef]
- Zsuffa, L. The genetics of Populus nigra L. Ann. For. 1974, 6, 29–53. [Google Scholar]
- Schroeder, W.; Soolanayakanahally, R.; Lindquist, C. AC Sundancer™ Poplar. Can. J. Plant Sci. 2013, 93, 1285–1287. [Google Scholar] [CrossRef]
- Wood, C.D. “A Most Dangerous Tree”: The Lombardy Poplar in landscape gardening. Arnoldia 1994, 54, 24–30. [Google Scholar]
- Vanden Broeck, A.; Cox, K.; Brys, R.; Castiglione, S.; Cicatelli, A.; Guarino, F.; Heinze, B.; Steenackers, M.; Vander Mijnsbrugge, K. Variability in DNA methylation and generational plasticity in the Lombardy Poplar, a single genotype worldwide distributed since the Eighteenth century. Front. Plant Sci. 2018, 9, 1635. [Google Scholar] [CrossRef] [Green Version]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Bruegmann, T.; Deecke, K.; Fladung, M. Evaluating the efficiency of gRNAs in CRISPR/Cas9 mediated genome editing in poplars. Int. J. Mol. Sci. 2019, 20, 3623. [Google Scholar] [CrossRef] [Green Version]
- Leple, J.C.; Brasileiro, A.C.M.; Michel, M.F.; Delmotte, F.; Jouanin, L. Transgenic poplars: Expression of chimeric genes using four different constructs. Plant Cell Rep. 1992, 11, 137–141. [Google Scholar] [CrossRef]
- Briones, M.V.; Hoenicka, H.; Cañas, L.A.; Beltrán, J.P.; Hanelt, D.; Sharry, S.; Fladung, M. Efficient evaluation of a gene containment system for poplar through early flowering induction. Plant Cell Rep. 2020, 39, 577–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucl. Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andronescu, M.; Condon, A.; Hoos, H.H.; Mathews, D.H.; Murphy, K.P. Efficient parameter estimation for RNA secondary structure prediction. Bioinformatics 2007, 23, i19–i28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuker, M.; Stiegler, P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucl. Acids Res. 1981, 9, 133–148. [Google Scholar] [CrossRef]
- Mader, M.; Le Paslier, M.C.; Bounon, R.; Bérard, A.; Faivre Rampant, P.; Fladung, M.; Leplé, J.C.; Kersten, B. Whole-genome draft assembly of Populus tremula × Populus alba clone INRA 717-1B4. Silv. Genet. 2016, 65, 74–79. [Google Scholar] [CrossRef] [Green Version]
- Bruegmann, T.; Polak, O.; Deecke, K.; Nietsch, J.; Fladung, M. Poplar Transformation. In Transgenic Plants; Kumar, S., Barone, P., Smith, M., Eds.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; pp. 165–177. [Google Scholar]
- Bennett, T.; Leyser, O. Something on the side: Axillary meristems and plant development. Plant Mol. Biol. 2006, 60, 843–854. [Google Scholar] [CrossRef]
- Henry, A. The Black poplars. Gard. Chron. 1914, 56, 46–47. [Google Scholar]
- Doebley, J.; Wang, R.L. Genetics and the evolution of plant form: An example from maize. Cold Spring Harbor Symp. Quant. Biol. 1997, 62, 361–367. [Google Scholar]
- Yu, B.; Lin, Z.; Li, H.; Li, X.; Li, J.; Wang, Y.; Zhang, X.; Zhu, Z.; Zhai, W.; Wang, X.; et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007, 52, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Hallé, F.; Oldeman, R.A.; Tomlinson, P.B. Opportunistic tree architecture. In Tropical Trees and Forests; Hallé, F., Oldeman, R.A., Tomlinson, P.B., Eds.; Springer: Berlin/Heidelberg, Germany, 1978; pp. 269–331. [Google Scholar]
- Carreño, I.; Dolle, T. The Court of Justice of the European Union’s judgment on mutagenesis and international trade: A case of GMO, mutagenesis and international trade. Glob. Trade Cust. J. 2019, 14, 91–101. [Google Scholar]


| Putative Transgenic Line | Ramet | NPTII | Cas9 |
|---|---|---|---|
| N499-1 | −2 | + | + |
| N499-2 | −1 | + | + |
| N499-3 | −1 | + | + |
| N499-4 | −1 | + | + |
| N499-5 | −1 | - | - |
| N499-6 | −1 | - | - |
| N499-7 | −1 | - | - |
| N499-8 | −3 | + | + |
| N499-9 | −1 | - | - |
| N499-10 | −2 | + | + |
| N499-11 | −1 | + | + |
| N499-12 | −1 | - | - |
| N499-13 | −2 | + | + |
| N499-14 | −2 | + | + |
| N499-15 | −1 | - | - |
| N499-16 | −1 | - | - |
| N499-17 | −1 | + | + |
| N499-18 | −1 | - | - |
| N499-19 | −1 | + | + |
| P1/Putative Transgenic Line | Potri.014G102600 (“TAC-14”) | Potri.002G175300 (“TAC-2”) | ||
|---|---|---|---|---|
| 2R | 4F | 2R | 4F | |
| P1 (Wildtype) | - | - | - | - |
| N499-1-2 | GT del hom | - | G ins hom | - |
| N499-2-1 | T ins hom | - | R ins hom | - |
| N499-3-1 | T ins hom | - | R ins hom | - |
| N499-4-1 | K ins hom | - | ? ins/del het | - |
| N499-5-1 | - | - | n.d. | - |
| N499-6-1 | - | - | - | - |
| N499-7-1 | - | - | n.d. | - |
| N499-8-3 | K ins hom | - | R ins hom | - |
| N499-9-1 | - | - | - | - |
| N499-10-2 | T ins hom | - | A ins hom | - |
| N499-11-1 | K ins hom | - | ? ins/del het | - |
| N499-12-1 | - | - | n.d. | - |
| N499-13-2 | ? ins/del het | - | ? ins/del het | - |
| N499-14-2 | T ins hom | - | ? ins/del het | - |
| N499-15-1 | - | - | n.d. | - |
| N499-16-1 | - | - | - | - |
| N499-17-1 | T ins hom | - | ? ins/del het | - |
| N499-18-1 | - | - | - | - |
| N499-19-1 | T ins hom | - | K ins hom | - |
| Lines | Potri.014G102600 (“TAC-14”): Genomic Target Region 2R | Type of Mutation |
|---|---|---|
| P1, not-transgenic regenerated lines * | .…..GAAACAAGTAGCCCTGGT-GGATGTGCTTGATGGTTG…… | Wildtype |
| N499-1-2 | ……GAAACAAGTAGCCCTG - - - GGATGTGCTTGATGGTTG…… | GT-deletion, hom |
| N499-2-1, -3-1, -10-2, -14-2, -17-1 | ……GAAACAAGTAGCCCTGGTTGGATGTGCTTGATGGTTG……. | T-insertion, hom |
| N499-3-3 | ……GAAACAAGTAGCCCTGGTGGGATGTGCTTGATGGTTG……. | G-insertion, hom |
| N499-10-3 | ……GAAACAAGTAGCCCTGGTAGGATGTGCTTGATGGTTG……. | A-insertion, hom |
| N499-4-1, -8-3, -11-1 | ……GAAACAAGTAGCCCTGGTKGGATGTGCTTGATGGTTG……. | G/T-insertion, hom |
| N499-13-2 | ……GAAACAAGTAGCCCTGGT-GGATGTGCTTGATGGTTG…….……??????????????????……. | ? bp insertion or deletion, het |
| Potri.002G175300 (“TAC-2”): Genomic Target Region 2R | ||
| P1, not-transgenic regenerated lines * | ……GAAACAAGTAGCCCTGGT-GGATGTGCTTGATGGTTG…… | Wildtype |
| N499-1-2 | ……GAAACAAGTAGCCCTGGTGGGATGTGCTTGATGGTTG……. | G-insertion, hom |
| N499-10-2 | ……GAAACAAGTAGCCCTGGTAGGATGTGCTTGATGGTTG……. | A-insertion, hom |
| N499-2-1, -3-1, -8-3, -10-3 | ……GAAACAAGTAGCCCTGGTRGGATGTGCTTGATGGTTG……. | A/G-insertion, hom |
| N499-4-1, -11-1, -13-2, -14-2, -17-1 | ……GAAACAAGTAGCCCTGGT-GGATGTGCTTGATGGTTG……. ……??????????????????……. | ? bp insertion or deletion, het |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fladung, M. Targeted CRISPR/Cas9-Based Knock-Out of the Rice Orthologs TILLER ANGLE CONTROL 1 (TAC1) in Poplar Induces Erect Leaf Habit and Shoot Growth. Forests 2021, 12, 1615. https://doi.org/10.3390/f12121615
Fladung M. Targeted CRISPR/Cas9-Based Knock-Out of the Rice Orthologs TILLER ANGLE CONTROL 1 (TAC1) in Poplar Induces Erect Leaf Habit and Shoot Growth. Forests. 2021; 12(12):1615. https://doi.org/10.3390/f12121615
Chicago/Turabian StyleFladung, Matthias. 2021. "Targeted CRISPR/Cas9-Based Knock-Out of the Rice Orthologs TILLER ANGLE CONTROL 1 (TAC1) in Poplar Induces Erect Leaf Habit and Shoot Growth" Forests 12, no. 12: 1615. https://doi.org/10.3390/f12121615
APA StyleFladung, M. (2021). Targeted CRISPR/Cas9-Based Knock-Out of the Rice Orthologs TILLER ANGLE CONTROL 1 (TAC1) in Poplar Induces Erect Leaf Habit and Shoot Growth. Forests, 12(12), 1615. https://doi.org/10.3390/f12121615






