Transcriptome Reveals the Specificity of Phyllostachys edulis ‘Pachyloen’ Shoots at Different Developmental Stages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Extraction, cDNA Library Construction, and Sequencing
2.3. Sequence Data Analysis and Assembly
2.4. Differential Expression Analysis
2.5. Quantitative Real-Time PCR (qRT-PCR) Validation of Differential Expression
3. Results
3.1. Read Mapping to the Reference Genome Dataset
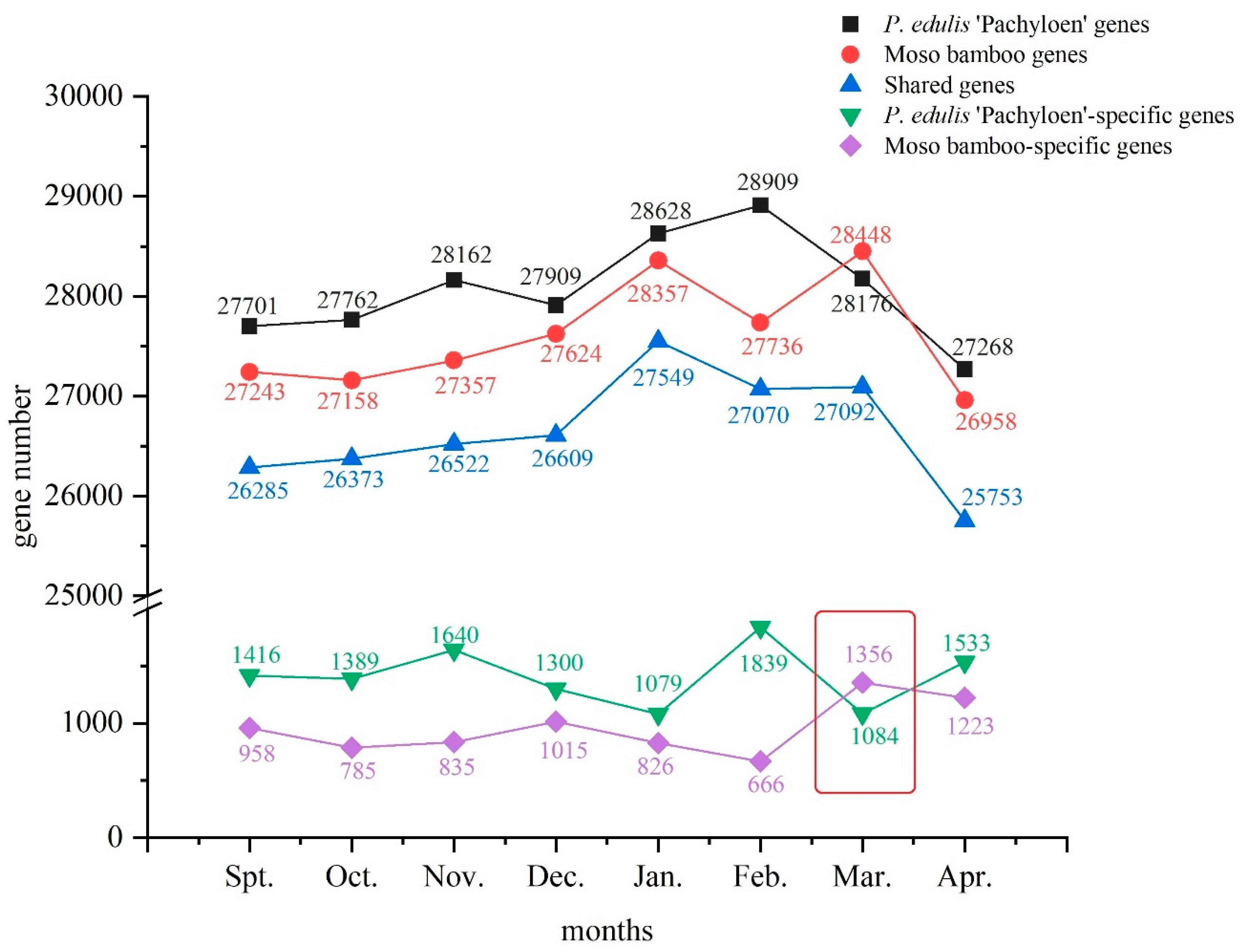
3.2. Trend of Alterations in Gene Expression Quantity
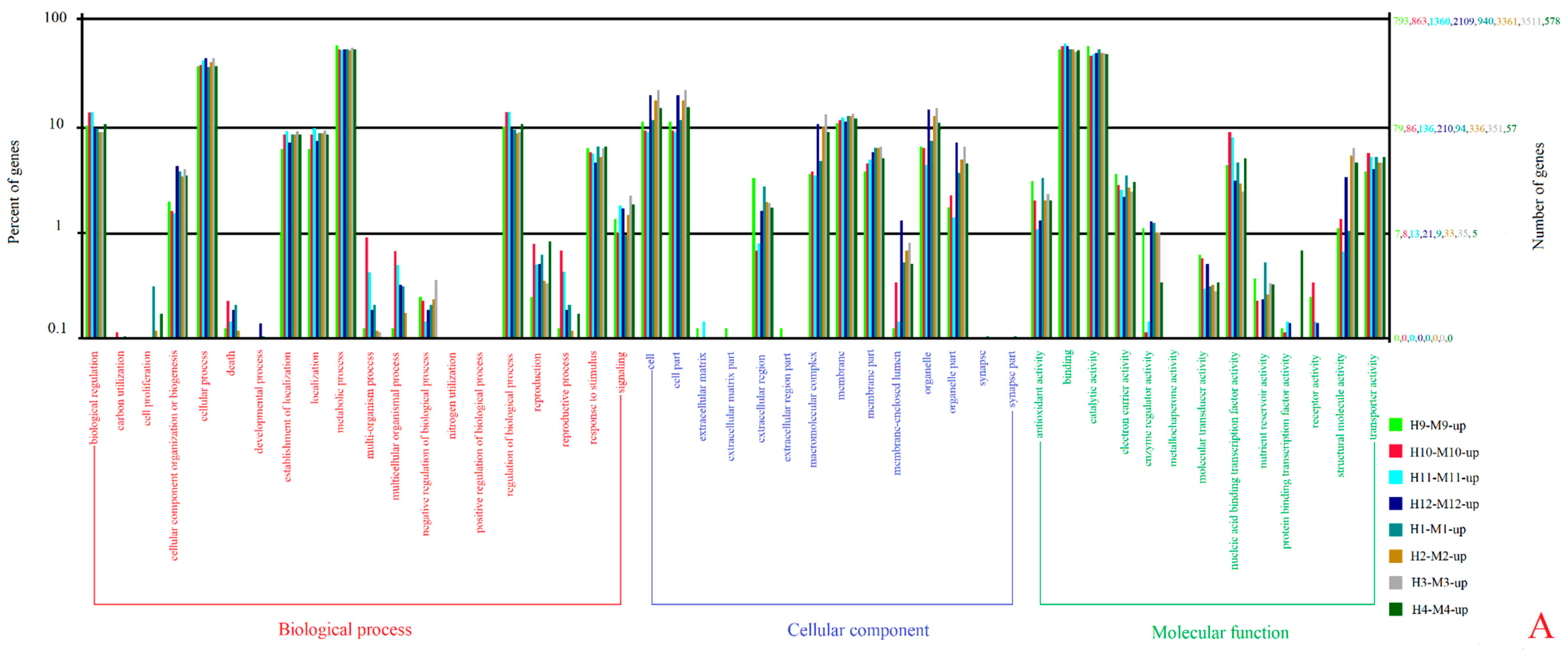
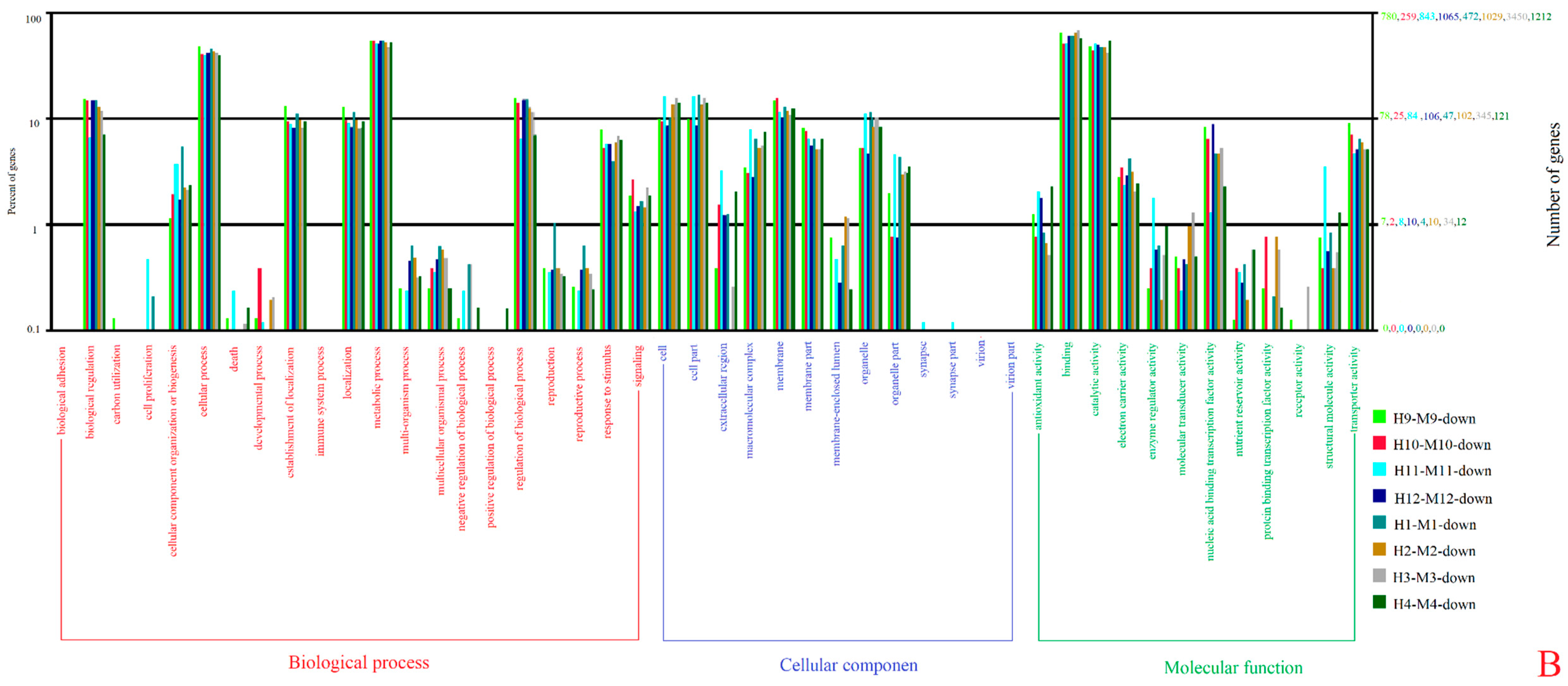
3.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analyses
3.4. Quantitative Real-Time PCR (qRT-PCR) Validation of Differential Expression
4. Discussion
4.1. Molecular Characteristics of Development in P. edulis ‘Pachyloen’
4.2. Gene Expression during the Development of P. edulis ‘Pachyloen’
4.3. Genetic Regulation of Important Moso Bamboo Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chang, S.T.; Wu, J.H. Green-color conservation of ma bamboo (Dendrocalamus latiflorus) treated with chromium-based reagents. J. Wood Sci. 2000, 46, 40–44. [Google Scholar] [CrossRef]
- Dileep, P.; Narayanankutty, S.K. A novel method for preparation of nanosilica from bamboo leaves and its green modification as a multi-functional additive in styrene butadiene rubber. Mater. Today Commun. 2020, 24, 100957. [Google Scholar] [CrossRef]
- Kassahun, T. Review of Bamboo Value Chain in Ethiopia. J. Biol. Agric. Healthc. 2014, 2, 52–67. [Google Scholar]
- Xie, J.; Hong, M. Effects of ecological factors on growth of Arundinaria spanostachya shoots in Liziping National Nature Reserve, China. Glob. Ecol. Conserv. 2020, 23, e01121. [Google Scholar] [CrossRef]
- Zakaria, S.A.; Latif, R.; Bahauddin, A. Malay Wisdom in the Motifs of Bamboo Shoots. SHS Web Conf. 2018, 45, 02004. [Google Scholar] [CrossRef] [Green Version]
- Piazza, M.; Lobovikov, M.; Paudel, S.; Ren, H.; Wu, J. World bamboo resources: A thematic study prepared in the framework of the Global Forest Resources Assessment 2005. Non Wood For. Prod. 2007, 18. [Google Scholar] [CrossRef]
- Fu, T.; Ke, J.H.; Zhou, S.; Xie, G.H. Estimation of the quantity and availability of forestry residue for bioenergy production in China. Resour. Conserv. Recycl. 2020, 162, 104993. [Google Scholar] [CrossRef]
- Lin, X.C.; Ruan, X.S.; Lou, Y.F.; Guo, X.Q.; Fang, W. Genetic similarity among cultivars of Phyllostachys pubescens. Plant Syst. Evol. 2009, 277, 67–73. [Google Scholar] [CrossRef]
- Liese, W.; Khl, M. Bamboo–The Plant and Its Uses; Springer: Berlin, Germany, 2015. [Google Scholar]
- Guo, Q.R.; Fang-Ming, H.U.; Tian-Zhen, D.U.; Yang, G.Y.; Chen, H.W.; Zhu, J.Y. Leaves Physiological and Biochemical Characters of Phyllostachys edulis cv. Pachyloen. J. Cent. South For. Univ. 2005, 25, 7–11. [Google Scholar]
- Qiang; Wei; Junjie; Cao; Weijie; Qian; Mengjian; Xu; Zhongru; Li. Establishment of an efficient micropropagation and callus regeneration system from the axillary buds of Bambusa ventricosa. Plant Cell Tissue Organ Cult. 2015, 122, 1–8. [Google Scholar] [CrossRef]
- Zhang, H.; Ying, Y.-Q.; Wang, J.; Zhao, X.-H.; Zeng, W.; Beahan, C.; He, J.-B.; Chen, X.-Y.; Bacic, A.; Song, L.-L.; et al. Transcriptome analysis provides insights into xylogenesis formation in Moso bamboo (Phyllostachys edulis) shoot. Sci. Rep. 2018, 8, 3951. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Zhang, C.; Zhang, Y.; Hu, T.; Mu, S.; Li, X.; Gao, J. Transcriptome Sequencing and Analysis of the Fast Growing Shoots of Moso Bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e78944. [Google Scholar] [CrossRef] [PubMed]
- Miyashima, S.; Sebastian, J.; Lee, J.Y.; Helariutta, Y. Stem cell function during plant vascular development. Semin. Cell Dev. Biol. 2012, 32, 178–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Li, N.; Ma, X.; Gupta, V.K.; Zhang, D.; Li, T.; Dai, X. The Ectopic Overexpression of the Cotton Ve1 and Ve2-Homolog Sequences Leads to Resistance Response to Verticillium Wilt in Arabidopsis. Front. Plant Sci. 2017, 8, 844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, F.S.; Green, E.D.; Guttmacher, A.E.; Guyer, M.S. A vision for the future of genomics research. Nature 2003, 422, 835–847. [Google Scholar] [CrossRef]
- Morozova, O.; Marra, M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics 2008, 92, 255–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiang, W.; Chen, J.; Lin, G.; Ding, Y.; Cao, J.; Feng, J.; Dong, X.; Mao, L.; Sun, H.; Yu, F. Exploring key cellular processes and candidate genes regulating the primary thickening growth of Moso underground shoots. New Phytol. 2017, 214, 81–96. [Google Scholar]
- Wang, Y.; Gao, Y.; Zhang, H.; Wang, H.; Liu, X.; Xu, X.; Zhang, Z.; Kohnen, M.V.; Hu, K.; Wang, H.; et al. Genome-Wide Profiling of Circular RNAs in the Rapidly Growing Shoots of Moso Bamboo (Phyllostachys edulis). Plant Cell Physiol. 2019, 60, 1354–1373. [Google Scholar] [CrossRef]
- Xu, X.; Lou, Y.; Yang, K.; Shan, X.; Zhu, C.; Gao, Z. Identification of Homeobox Genes Associated with Lignification and Their Expression Patterns in Bamboo Shoots. Biomolecules 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Wu, L.; Pan, Y.; Zhong, H. Extraction, Optical Properties and Bio-Imaging of Fluorescent Composition From Moso Bamboo Shoots. J. Renew. Mater. 2019, 7, 1209–1219. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Xie, L.; Zheng, H.; Cai, M.; Cheng, Z.; Bai, Y.; Li, J.; Gao, J. Transcriptome profiling of postharvest shoots identifies PheNAP2- and PheNAP3-promoted shoot senescence. Tree Physiol. 2019, 39, 2027–2044. [Google Scholar] [CrossRef]
- Zhang, X.M.; Zhao, L.; Larson-Rabin, Z.; Li, D.Z.; Guo, Z.H.; Nitabach, M.N. De Novo Sequencing and Characterization of the Floral Transcriptome of Dendrocalamus latiflorus (Poaceae: Bambusoideae). PLoS ONE 2012, 7, e42082. [Google Scholar] [CrossRef] [PubMed]
- Shou, Y.T.; Zhu, Y.H.; Ding, Y.L. Transcriptome analysis of lateral buds from Phyllostachys edulis rhizome during germination and early shoot stages. BMC Plant Biol. 2020, 20, 229. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.C.; Chou, M.L.; Yue, J.J.; Hsu, C.T.; Chang, W.J.; Ko, S.S.; Liao, D.C.; Huang, Y.T.; Chen, J.J.; Yuan, J.L. BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues- characterization of transcriptome and study of MADS genes in bamboo floral development. BMC Plant Biol. 2014, 14, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rougemont, J.; Amzallag, A.; Iseli, C.; Farinelli, L.; Xenarios, I.; Naef, F. Probabilistic base calling of Solexa sequencing data. BMC Bioinf. 2008, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M.; Blazquez, M.A. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.; Jung, K.H.; Lee, D.E.; Lee, D.Y.; Lee, J.; An, K.; Kang, H.G.; An, G. The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol. Cells 2006, 21, 147. [Google Scholar]
- Demura, T.; Fukuda, H. Transcriptional regulation in wood formation. Trends Plant Sci. 2007, 12, 64–70. [Google Scholar] [CrossRef]
- MARCO, F.; ELISA, B.; FRANCESCA, R.; GIULIANO, C.; VANIA, M.; DANIELE, B.; RITA, B.; CLAUDIO, P. stem fasciated, a Recessive Mutation in Sunflower (Helianthus annuus), Alters Plant Morphology and Auxin Level. Ann. Bot. 2006, 4, 715–730. [Google Scholar]
- Jia, N.; Liu, X.; Gao, H. A DNA2 homolog is required for DNA damage repair, cell cycle regulation, and meristem maintenance in plants. Plant Physiol. 2016, 171, 318–333. [Google Scholar] [CrossRef] [Green Version]
- Madoka, Y.; Kashiwagi, T.; Hirotsu, N.; Ishimaru, K. Indian rice “Kasalath” contains genes that improve traits of Japanese premium rice “Koshihikari”. Theor. Appl. Genet. 2008, 116, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Ookawa, T.; Aya, K.; Ochiai, Y.; Hirasawa, T.; Ebitani, T.; Takarada, T.; Yano, M.; Yamamoto, T.; Fukuoka, S.; et al. Isolation of a Novel Lodging Resistance QTL Gene Involved in Strigolactone Signaling and Its Pyramiding with a QTL Gene Involved in Another Mechanism. Mol. Plant 2015, 8, 303–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiryu, T.; Miyoshi, Y.; Furuta, Y. The Mechanism of Improvement of Physical Properties of Moso Bamboo (Phyllostachys pubescens) with Increasing Age I. Relationship between modulus of elasticity in bending and cell wall thickness and/or percentage of cell walls. Mokuzai Gakkaishi 2016, 62, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Chiu, W.B.; Lin, C.H.; Chang, C.J.; Hsieh, M.H.; Wang, A.Y. Molecular characterization and expression of four cDNAs encoding sucrose synthase from green bamboo Bambusa oldhamii. New Phytol. 2006, 170, 53–63. [Google Scholar] [CrossRef]
- Hsieh, L.S.; Ma, G.J.; Yang, C.C.; Lee, P.D. Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry 2010, 71, 1999–2009. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, X.; Ding, Y.; Fei, Z.; Jiao, C.; Fan, M.; Yao, B.; Xin, P.; Chu, J.; Wei, Q. Cellular and molecular characterization of a thick-walled variant reveal a pivotal role of shoot apical meristem in transverse development of bamboo culm. J. Exp. Bot. 2019, 70, 3911–3926. [Google Scholar] [CrossRef]
- Li, L.; Cheng, Z.; Ma, Y.; Bai, Q.; Li, X.; Cao, Z.; Wu, Z.; Gao, J. The association of hormone signalling genes, transcription and changes in shoot anatomy during moso bamboo growth. Plant Biotechnol. J. 2018, 16, 72–85. [Google Scholar] [CrossRef]
- Wang, H.Y.; Cui, K.; He, C.Y.; Zeng, Y.F.; Liao, S.X.; Zhang, J.G. Endogenous hormonal equilibrium linked to bamboo culm development. Genet. Mol. Res. 2015, 14, 11312–11323. [Google Scholar] [CrossRef]
- Wang, K.; Peng, H.; Lin, E.; Jin, Q.; Hua, X.; Yao, S.; Bian, H.; Han, N.; Pan, J.; Wang, J.; et al. Identification of genes related to the development of bamboo rhizome bud. J. Exp. Bot. 2010, 61, 551–561. [Google Scholar] [CrossRef] [Green Version]
- Jian, G.; Ying, Z.; Zhang, C.; Qi, F.; Li, X.; Mu, S.; Peng, Z.; Emmanuel, G. Characterization of the Floral Transcriptome of Moso Bamboo (Phyllostachys edulis) at Different Flowering Developmental Stages by Transcriptome Sequencing and RNA-Seq Analysis. PLoS ONE 2014, 9, e98910. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Jiang, Z. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Samples | Time and Date | Samples | Time and Date |
|---|---|---|---|
| H9 | 9 A.M. on 21 September 2019 | H1 | 9 A.M. on 22 January 2020 |
| M9 | M1 | ||
| H10 | 9 A.M. on 22 October 2019 | H2 | 9 A.M. on 21 February 2020 |
| M10 | M2 | ||
| H11 | 9 A.M. on 21 November 2019 | H3 | 9 A.M. on 21 March 2020 |
| M11 | M3 | ||
| H12 | 9 A.M. on 22 December 2019 | H4 | 9 A.M. 22 on April 2020 |
| M12 | M4 |
| Sample | Clean Reads | Mapping Genome Reads | Mapping Rates (%) | Mapping Genes Reads | Unique Mapping Reads |
|---|---|---|---|---|---|
| H9 | 21,449,638 | 10,555,822 | 79.21 | 9,172,353 | 8,919,360 |
| H10 | 26,155,564 | 19,043,559 | 72.81 | 9,225,730 | 9,025,293 |
| H11 | 19,442,088 | 9,421,445 | 78.46 | 9,809,217 | 8,571,281 |
| H12 | 27,091,394 | 20,156,532 | 74.40 | 9,039,050 | 8,709,974 |
| H1 | 43,677,560 | 33,908,109 | 77.63 | 12,139,871 | 11,885,126 |
| H2 | 33,790,364 | 26,486,050 | 78.38 | 9,039,050 | 8,826,300 |
| H3 | 24,424,416 | 18,079,544 | 74.02 | 7,771,250 | 7,545,415 |
| H4 | 25,466,886 | 20,125,400 | 79.03 | 7,657,497 | 7,487,547 |
| M9 | 18,729,148 | 12,364,644 | 66.02 | 6,827,775 | 6,618,518 |
| M10 | 32,557,142 | 24,291,385 | 74.61 | 10,545,047 | 10,318,143 |
| M11 | 15,304,348 | 12,304,338 | 80.04 | 5,686,732 | 5,498,670 |
| M12 | 31,482,700 | 23,532,358 | 74.75 | 10,078,339 | 9,877,400 |
| M1 | 44,347,518 | 31,581,328 | 71.21 | 17,076,767 | 16,717,652 |
| M2 | 33,122,658 | 24,422,403 | 73.73 | 11,468,718 | 11,242,241 |
| M3 | 41,049,276 | 3,026,449 | 73.73 | 13,936,705 | 13,639,031 |
| M4 | 13,722,470 | 11,111,002 | 80.97 | 4,326,952 | 4,096,310 |
| Terms | H9 | H10 | H11 | H12 | H1 | H2 | H3 | H4 | |
|---|---|---|---|---|---|---|---|---|---|
| BP | Metabolic process | 370 | 407 | 465 | 318 | 270 | 504 | 296 | 412 |
| 26% * | 29% | 28% | 24% | 25% | 27% | 27% | 26% | ||
| Cellular process | 255 | 285 | 335 | 212 | 169 | 317 | 164 | 316 | |
| 18% | 20% | 20% | 16% | 15% | 17% | 15% | 20% | ||
| CC | Cell | 84 | 63 | 75 | 79 | 50 | 97 | 47 | 120 |
| 5% | 4% | 4% | 6% | 4% | 5% | 4% | 7% | ||
| Cell part | 84 | 63 | 75 | 79 | 50 | 97 | 47 | 120 | |
| 5% | 4% | 4% | 6% | 4% | 5% | 4% | 7% | ||
| Membrane | 78 | 85 | 87 | 77 | 58 | 104 | 61 | 82 | |
| 5% | 6% | 5% | 5% | 5% | 5% | 5% | 5% | ||
| MF | Binding | 408 | 477 | 560 | 355 | 283 | 552 | 312 | 509 |
| 28% | 34% | 34% | 27% | 26% | 30% | 28% | 33% | ||
| Catalytic activity | 363 | 388 | 472 | 303 | 278 | 502 | 320 | 366 | |
| 25% | 27% | 28% | 23% | 25% | 27% | 29% | 23% |
| No. | Pathway | ID | Total Genes in Each Pathway | Total DEGs in Each Pathway | Percentage (%) |
|---|---|---|---|---|---|
| 1 | Biosynthesis of siderophore group non-ribosomal peptides | ko01053 | 1 | 1 | 100.00 |
| 2 | Photosynthesis-antenna proteins | ko00196 | 30 | 26 | 86.67 |
| 3 | Glycerolipid metabolism | ko00561 | 81 | 6 | 74.07 |
| 4 | Phenylpropanoid biosynthesis | ko00564 | 128 | 71 | 55.47 |
| 5 | 1- and 2-Methylnaphthalene degradation | ko00624 | 8 | 4 | 50.00 |
| 6 | Flavone and flavonol biosynthesis | ko00944 | 4 | 2 | 50.00 |
| 7 | Flavonoid biosynthesis | ko00941 | 35 | 15 | 42.86 |
| 8 | Linoleic acid metabolism | ko00591 | 13 | 5 | 38.46 |
| 9 | Phenylalanine metabolism | ko00360 | 143 | 51 | 35.67 |
| 10 | Glycosphingolipid biosynthesis-globo series | ko00603 | 15 | 5 | 33.33 |
| No. | Pathway | ID | No. | Pathway | ID |
|---|---|---|---|---|---|
| 1 | C5-Branched dibasic acid metabolism | ko00660 | 15 | Selenoamino acid metabolism | ko00450 |
| 2 | Butanoate metabolism | ko00650 | 16 | Glycosaminoglycan degradation | ko00531 |
| 3 | Glyoxylate and dicarboxylate metabolism | ko00630 | 17 | alpha-Linolenic acid metabolism | ko00592 |
| 4 | Sulfur metabolism | ko00920 | 18 | Ubiquinone and other terpenoid-quinone biosynthesis | ko00130 |
| 5 | Biosynthesis of unsaturated fatty acids | ko01040 | 19 | Carotenoid biosynthesis | ko00906 |
| 6 | Steroid biosynthesis | ko00100 | 20 | Cysteine and methionine metabolism | ko00270 |
| 7 | Tyrosine metabolism | ko00350 | 21 | Flavone and flavonol biosynthesis | ko00944 |
| 8 | Glycosphingolipid biosynthesis—ganglio series | ko00604 | 22 | Limonene and pinene degradation | ko00903 |
| 9 | Glycosphingolipid biosynthesis—globo series | ko00603 | 23 | Naphthalene and anthracene degradation | ko00626 |
| 10 | Pantothenate and CoA biosynthesis | ko00770 | 24 | Cyanoamino acid metabolism | ko00460 |
| 11 | Biosynthesis of siderophore group non-ribosomal peptides | ko01053 | 25 | Tropane, piperidine and pyridine alkaloid biosynthesis | ko00960 |
| 12 | Novobiocin biosynthesis | ko00401 | 26 | ErbB signaling pathway | ko04012 |
| 13 | 1- and 2-Methylnaphthalene degradation | ko00624 | 27 | Cell cycle | ko04110 |
| 14 | Homologous recombination | ko03440 |
| Pathway | Gene ID |
|---|---|
| Tyrosine metabolism | PH01000462G0820, PH01001716G0420, PH01000603G0990, PH01001395G0320, PH01003894G0070, PH01001395G0350 |
| Sulfur metabolism | PH01002362G0220, PH01174972G0010, PH01000136G0860, PH01001450G0180 |
| Biosynthesis of unsaturated fatty acids | PH01002036G0150, PH01000826G0750, PH01001269G0550, PH01006014G0050,PH01001016G0360 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Zhang, Y.; Zhou, J.; Wang, G.; Guo, Q. Transcriptome Reveals the Specificity of Phyllostachys edulis ‘Pachyloen’ Shoots at Different Developmental Stages. Forests 2020, 11, 861. https://doi.org/10.3390/f11080861
Hu Y, Zhang Y, Zhou J, Wang G, Guo Q. Transcriptome Reveals the Specificity of Phyllostachys edulis ‘Pachyloen’ Shoots at Different Developmental Stages. Forests. 2020; 11(8):861. https://doi.org/10.3390/f11080861
Chicago/Turabian StyleHu, Yaping, Ying Zhang, Jie Zhou, Guibing Wang, and Qirong Guo. 2020. "Transcriptome Reveals the Specificity of Phyllostachys edulis ‘Pachyloen’ Shoots at Different Developmental Stages" Forests 11, no. 8: 861. https://doi.org/10.3390/f11080861
APA StyleHu, Y., Zhang, Y., Zhou, J., Wang, G., & Guo, Q. (2020). Transcriptome Reveals the Specificity of Phyllostachys edulis ‘Pachyloen’ Shoots at Different Developmental Stages. Forests, 11(8), 861. https://doi.org/10.3390/f11080861





