Abstract
Phyllostachys edulis ‘Pachyloen’ can have a stalk wall thickness of up to 2.5 cm at a height of 1.3 m, which is 1.8 times that of normal Moso bamboo (Phyllostachys edulis); this serves as an excellent cultivar, comprising both wood and bamboo shoots. We collected bamboo shoot samples of Phyllostachys edulis ‘Pachyloen’ and Moso bamboo on a monthly basis from September to April and used transcriptome sequencing to explore the differences in their development. The results showed that there were 666–1839 Phyllostachys edulis ‘Pachyloen’-specific genes at different developmental stages enriched in 20 biological processes, 15 cellular components, 12 molecular functions, and 137 metabolic pathways, 52 of which were significant. Among these, 27 metabolic pathways such as tyrosine metabolism and their uniquely expressed genes were found to play important roles in the thickening of Phyllostachys edulis ‘Pachyloen’. This study provides insights into the mechanisms underlying the thickening of the culm wall of Phyllostachys edulis ‘Pachyloen’.
1. Introduction
Bamboo plants are non-timber forest products of important economic, ecological, and cultural value [1,2,3,4,5]. Approximately 2.5 billion people worldwide rely economically on bamboo, and the annual trade generated by bamboo amounts to 2.5 billion dollars [6,7]. Bamboo plants are different from trees in that bamboo stems are hollow and there is no periodic radial growth process. Phyllostachys edulis ‘Pachyloen’ is a novel variety of Phyllostachys edulis (normal Moso bamboo) that exhibits several advantageous features of timber and bamboo shoot including bamboo wall thickening, subsolid culm, large biomass, good tasting shoot, and stable heredity [8]. It was first discovered in Jiangxi Province in southeastern China in 1995. The thickness of its culm wall is almost twice as that of the normal phenotype of the species, and its yields are comparatively higher [9]. The major biological characteristics of P. edulis ‘Pachyloen’ including the growth pattern of its bamboo shoots, photosynthetic physiology, transpiration dynamics, cold resistance, anatomical characteristics, and physical and mechanical properties have been studied systematically since its discovery [10,11,12].
The thickness of bamboo stalks is closely related to the development of bamboo shoots [13]. The walls of bamboo stalks are primarily composed of thin-walled basic tissue cells and vascular bundles, with thick-walled cells forming the main body. Vascular bundle-related genes regulate the growth and development of vascular bundles and improve plant resistance [14]. The expression of GbaVd1 and GbaVd2 genes has been associated with the increase in the degree of lignification of transgenic Arabidopsis lines [15]. Bamboo research has entered the era of functional genomics with the sequencing of the Moso bamboo genome [16,17]. In addition, the cellular mechanisms underlying the growth of the underground shoots of Moso bamboo have been systematically studied [18,19,20,21]. The molecular mechanisms underlying the regulation of bamboo flowering have also been elucidated using RNA-Seq [22,23,24,25]. However, the mechanism underlying the apparent thickening of the culm walls of P. edulis ‘Pachyloen’ remains unclear.
In the present study, the shoots of P. edulis ‘Pachyloen’ and Moso bamboo (control) in different growth stages were analyzed by transcriptome sequencing using the whole genome sequence of Moso bamboo as a reference. As a result, several uniquely expressed genes and their related pathways were discovered in P. edulis ‘Pachyloen’. The results presented in this study have important scientific and theoretical implications for the characterization of the mechanism underlying the thickening of bamboo culm walls.
2. Materials and Methods
2.1. Plant Materials
Six to fifteen pieces of P. edulis ‘Pachyloen’ shoots were collected between September 2019 and April 2020 on a monthly basis from the experimental bamboo forest (113.1124063° E, 28.2698183° N, 44.9 m above sea level) in Lukou Town, Changsha County, Hunan Province, China, based on the speed of bamboo shoot growth. Moso bamboo shoots were also collected for use as a control at the same time and under the same environmental conditions. The samples were collected at 1 cm from the apex of the shoot. The volume and degree of development of the experimental shoots were consistent across the samples. The collected shoots were immediately washed with distilled water, wrapped in foil, placed in a liquid nitrogen tank, and stored at −80 °C until RNA extraction. The samples of P. edulis ‘Pachyloen’ were marked as H9, H10, H11, H12, H1, H2, H3, and H4; similarly, the Moso bamboo samples were marked as M9, M10, M11, M12, M1, M2, M3, and M4. The sample collection time and dates are outlined in Table 1. Three biological repeats, each with three technical repeats, were performed for all samples.

Table 1.
Sample collection time and date.
2.2. RNA Extraction, cDNA Library Construction, and Sequencing
Tender shoots lacking sheaths (50–100 mg) were ground in liquid nitrogen. Total RNA was extracted using the TRIzol reagent (Invitrogen Scientific, Inc., Carlsbad, CA, USA) in accordance with the manufacturer’s protocol. The quality of the total RNA of the sample was detected using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) with an RNA integrity number (RIN) value of ≥7 required for all samples. The libraries were constructed using a VAHTS mRNA-Seq V2 Library Prep Kit for Illumina® (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China) in accordance with the manufacturer’s instructions. The libraries were then sequenced on an Illumina sequencing platform (HiSeq™ 2500). The Illumina raw sequencing data were submitted to the National Center for Biotechnology Information (NCBI) Short Reads Archive (SRA) database under accession number SRP169499.
2.3. Sequence Data Analysis and Assembly
The original image data were transformed into sequence data by base calling, which is defined as raw reads. Reads shorter than 50 bp, with a predicted error rate of >20%, or with full passes lower than 0 were removed [26]. The clean reads were aligned with the Moso bamboo reference genome. The data sources of the Moso bamboo reference genome are listed in Supplementary File S1. Coding sequences (CDS) of Moso bamboo with a mismatch of up to five bases were allowed to obtain the gene expression profile data for ‘Pachyloen’ at the different developmental periods.
2.4. Differential Expression Analysis
The differentially expressed genes (DEGs) (old change ≥ 2, FDR ≤ 0.001) detected at different developmental stages of ‘Pachyloen’ were subjected to Gene Ontology (GO) functional annotation and classification as well as to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway significance enrichment analysis. Hierarchical cluster analysis was used to annotate and analyze the GO results and explore patterns of gene expression in WEGO software [27]. R software (v3.6.1) was used to analyze the gene expression patterns of specific metabolic pathways.
2.5. Quantitative Real-Time PCR (qRT-PCR) Validation of Differential Expression
Ten putative genes (PH01000081G111, PH01000104G133, PH01000152G124, PH01000159G013, PH01000931G0650, PH01001141G0100, PH01001292G0400, PH01001299G0140, PH01001317G0160, and PH01001859G0010) were randomly selected for validation by quantitative real-time PCR (qRT-PCR) to validate the accuracy of the RNA-Seq results. The transcription shoots were analyzed by qRT-PCR in an ABI 7300 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) using SYBR Green Realtime PCR Master Mix. The tonoplast intrinsic protein (TIP41) gene was amplified using the forward primer 5′-AAAATCATTGTAGGCCATTGTCG-3′ and the reverse primer 5′-ACTAAATTAAGCCAGCGGGAGTG-3′ as an internal control [28]. One microgram of extracted RNA was reverse-transcribed using a Superscript II First Strand Synthesis Kit (Invitrogen) and random hexamer primers according to the manufacturer’s instructions. For each sample, three replicate experiments were performed at a final volume of 10 µL (3 µL of ddH2O, 1 µL of cDNA, 5 µL of SYBR Mix, 0.5 µL of primers, and 0.5 µL of ROX Reference Dye). The cycling conditions were as follows: 95 °C for 10 min, 95 °C for 10 s, and 62 °C for 10 s for 50 cycles. The qRT-PCR primers used are listed in Supplementary File S2. The amplification results were analyzed using the comparative cycle threshold (Ct) method with the 2−ΔΔCT formula. The qRT-PCR results were calculated based on the mean values from three replicate experiments. Significant differences between treatments were evaluated based on their standard deviation. The error bars represent the standard error of the three biological replicates.
3. Results
3.1. Read Mapping to the Reference Genome Dataset
The clean reads of H9–H4 and M9–M4 were mapped to the genes expressed in the Moso bamboo reference genome (Table 2). Approximately 27,000 to 28,000 genes were annotated to the reference gene in each sample, accounting for 88% of the genes in the Moso bamboo reference genome, which proved that the sequencing library constructed by cDNA fragments was comprised of high quality and reliable sequencing data. During the eight months of growth and development of P. edulis ‘Pachyloen’, the number of expressed genes was the highest in February at 28,911, and the lowest in April at 27,265. Similarly, for the Moso bamboo samples, the number of expressed genes in March was the highest at 28,448, and the lowest in April at 26,958.

Table 2.
The results of sample mapping to the reference genome.
The gene expression patterns of P. edulis ‘Pachyloen’ and Moso bamboo based on the gene coverage and randomness of reads mapped to the reference genome and genes are outlined in Supplementary File S3.
3.2. Trend of Alterations in Gene Expression Quantity
January and February are typically the coldest months of the year. Information on the climate of the collection sites during sampling is provided in Supplementary File S4. In winter, bamboo shoots enter a dormant stage, known as the overwintering period, to resist the harsh conditions in the outside world. Bamboo shoots then enter the germination stage in March, during which the number of genes being expressed increases, reflecting an increase in fertility. The gene expression number of P. edulis ‘Pachyloen’ and Moso bamboo (control) increased at first, and then decreased with changes in phenology (Figure 1). The number of genes expressed in P. edulis ‘Pachyloen’ each month was higher than Moso bamboo, except for March, which may be related to the physiological and metabolic activities of P. edulis ‘Pachyloen’.
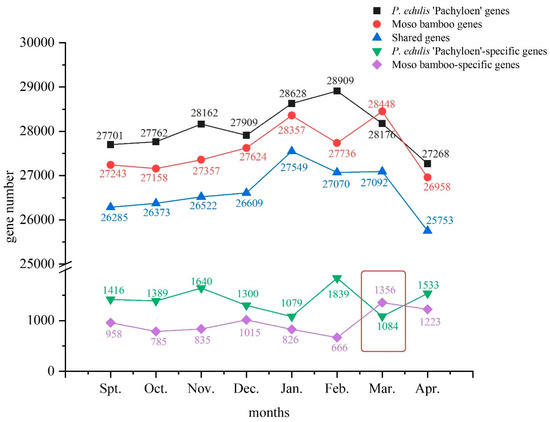
Figure 1.
The changes in the number of genes expressed each month.
The highest number of shared genes was 27,549, in the samples collected in January, and the lowest was 25,735 in the samples collected in April. Several P. edulis ‘Pachyloen’-specific genes were detected upon comparison with the genes from Moso bamboo as control genes. Information on the P. edulis ‘Pachyloen’-specific genes and Moso-specific genes is provided in Supplementary File S5. P. edulis ‘Pachyloen’ had more unique genes than Moso bamboo throughout its growth and development, except in March. The identification of P. edulis ‘Pachyloen’-specific genes and its functional annotation and metabolic pathway enrichment analysis play an important role in explaining the thickening mechanism of P. edulis ‘Pachyloen’.
3.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analyses
DEGs (FDR ≤ 0.001 and |log2Ratio| ≥ 1) were screened between P. edulis ‘Pachyloen’ and Moso bamboo for expression level (estimated using the reads per kilobase per million mapped reads (RPKM) value) (Figure 2). We obtained 2393 (H9/M9), 1719 (H10/M10), 3551 (H11/M11), 4976 (H12/M12), 2451 (H1/M1), 7076 (H2/M2), 11,033 (H3/M3), and 2832 (H4/M4) DEGs by comparing the RPKM values of the co-expressed genes between P. edulis ‘Pachyloen’ and Moso bamboo (control). The number of upregulated DEGs (21,647) in P. edulis ‘Pachyloen’ was significantly higher than the number of downregulated DEGs (14,384) compared to those in Moso bamboo. The number of DEGs was the highest including 5604 upregulated genes and 5429 downregulated genes in the H3/M3 comparison group, which indicates high levels of developmental activity in the bamboo shoots in March.
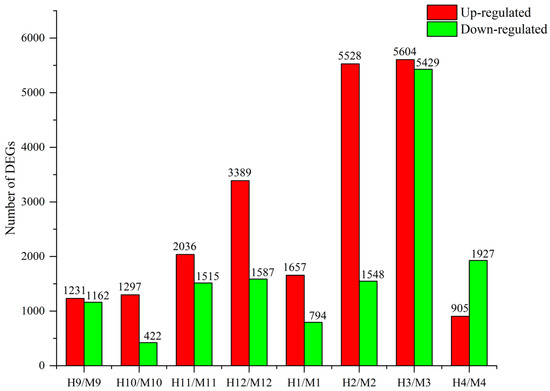
Figure 2.
Comparison of differentially expressed genes (DEGs) in different months.
The unigenes were mainly enriched in 22 terms of biological processes (BP), 16 cellular components (CC), and 13 molecular functions (MF), according to the GO enrichment analysis of the DEGs (Figure 3). “Cellular process”, “metabolic process”, “cell”, “cell part”, “membrane”, “binding”, and “catalytic activity” were rich in unigenes. The seven GO terms with the most enrichment of P. edulis ‘Pachyloen’-specific genes at the different developmental stages were counted for the comparison of the developmental differences between P. edulis ‘Pachyloen’ and Moso bamboo (Table 3).
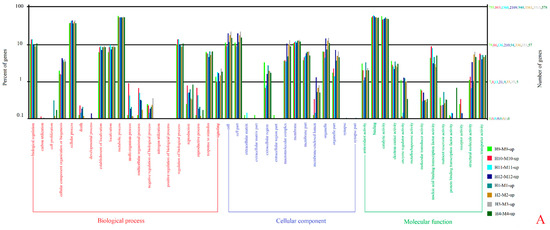
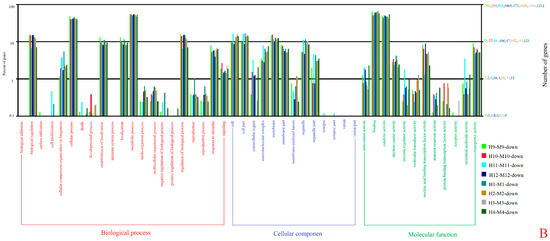
Figure 3.
GO classification of unigenes: (A) upregulated genes and (B) downregulated genes.

Table 3.
Numbers and percentages in gene ontology (GO) terms of P. edulis ‘Pachyloen’-specific genes.
The 2393 (H9/M9), 1719 (H10/M10), 3551 (H11/M11), 4976 (H12/M12), 2451 (H1/M1), 7076 (H2/M2), 11,033 (H3/M3), and 2832 (H4/M4) DEGs were subjected to pathway enrichment analysis using the KEGG pathway database. The co-expressed genes and Moso bamboo-specific genes were enriched in 68 and 35 significant metabolic pathways (p ≤ 0.05). Flavone and flavonol biosynthesis, phenylpropanoid biosynthesis, and phenylalanine metabolism were the top three of the number of DEGs annotated to each metabolic pathway as a percentage of the total number of genes in the metabolic pathway. The P. edulis ‘Pachyloen’-specific genes were enriched into 52 significantly enriched metabolic pathways throughout the development process (H9–H4; p ≤ 0.05) (Table 4). The metabolic pathways of the co-expressed genes, the P. edulis ‘Pachyloen’-specific genes, and the Moso bamboo-specific genes were analyzed to determine the key metabolic pathways involved in the development of bamboo thick-walled germplasm. As a result, 27 significant metabolic pathways were considered to be related to the thickening of P. edulis ‘Pachyloen’(Table 5), 15 metabolic pathways were only involved in the expression of P. edulis ‘Pachyloen’-specific genes, and 12 metabolic pathways were involved in DEGs and P. edulis ‘Pachyloen’-specific genes.

Table 4.
Significant enrichment of the top 10 P. edulis ‘Pachyloen’-specific gene metabolic pathways.

Table 5.
Significant enrichment pathways of P. edulis ‘Pachyloen’.
The free amino acid content and biomass of P. edulis ‘Pachyloen’ were significantly greater than that of the control (Moso bamboo). Therefore, we analyzed the pathways of the tyrosine metabolism, sulfur metabolism, and biosynthesis of unsaturated fatty acids from 27 significant metabolic pathways. Fifteen P. edulis ‘Pachyloen’-specific genes were found to be involved in the regulation of wall thickening (Table 6). Our findings on the other 24 metabolic pathways are provided in the Supplementary Materials.

Table 6.
P. edulis ‘Pachyloen’-specific genes in three significant pathways.
3.4. Quantitative Real-Time PCR (qRT-PCR) Validation of Differential Expression
The expression profiles of 10 randomly chosen genes are provided in Supplementary File S6. The relative gene expression was calculated using 2−△△Ct and amplified using a LightCycler®96 fluorescence quantitative PCR instrument for three biological replicates. The qRT-PCR verification of the 10 selected genes was consistent with the sequencing results.
4. Discussion
4.1. Molecular Characteristics of Development in P. edulis ‘Pachyloen’
In terms of morphology, the meristem at the apex of the underground stem of P. edulis ‘Pachyloen’ does not differentiate into medullary tissue; rather, it differentiates into vascular bundle tissues. The apical meristems of P. edulis ‘Pachyloen’ shoots are not similar to the low and wide ridge-shaped meristems of Moso bamboos; instead, they are tall and narrow finger-shaped meristems. Moreover, the meristems of thick-walled bamboo shoots are larger, with a greater number of cells. Larger apical meristems can produce more auxin, which acts as a promoter of the differentiation and proliferation of vascular tissue [29,30]. The same phenomenon occurs in sunflower and Arabidopsis mutants [31,32]. Certain studies have reported that wheat stalk thickness is controlled by a single gene, complementary genes, or multiple genes. The rice cultivar “Kasalath” has chromosomal fragments that increase stem resistance to enhance lodging resistance [33]. Yano et al. detected a quantitative trait locus (QTL) named SCM3, which regulates the thickness of rice stem and stem wall [34]. However, the molecular mechanism underlying bamboo culm wall thickening is yet to be completely elucidated. In this study, we detected 27 key metabolic pathways associated with P. edulis ‘Pachyloen’ culm wall thickening, including tyrosine metabolism, sulfur metabolism, and unsaturated fatty acid biosynthesis. Plant endogenous hormones such as cyclin A, cyclin B, auxin, and gibberellin appeared to strongly influence cell division, cell elongation, and cell cycle. The P. edulis ‘Pachyloen’ genes identified play important regulatory roles in different periods.
4.2. Gene Expression during the Development of P. edulis ‘Pachyloen’
The increase in the cell number of the culm wall plays an important role in promoting wall thickening in P. edulis ‘Pachyloen’ [35,36,37]. In this study, we investigated the changes in the morphology of thick-walled P. edulis ‘Pachyloen’ from shoots to bamboo using transcriptomic analysis to identify the key metabolic pathways involved in the formation of its thick culm walls. In general, the different growth and development states of plants are known to be directly reflected in their DEGs. In this study, the shoot of P. edulis ‘Pachyloen’ and Moso bamboo showed evident phenological characteristics and their underground growth process could be divided into differentiation periods: the overwintering period and germination period [38,39]. At the end of summer and early autumn, the side buds on the bamboo whip began to germinate and differentiate into shoots. In winter, the bamboo shoots entered the overwintering period and lay dormant. By the second year, the bamboo shoots continued to grow, and increased in size with rising temperatures. During this process, extremely complex physiological and biochemical reactions occurred in bamboo shoots [40,41]. We found that there was a lack of regular changes in the number of upregulated and downregulated DEGs related to the growth and development of P. edulis ‘Pachyloen’ between developmental stages.
From the winter (November to January) of the following year, the bamboo shoot buds were dormant, with a small number of genes being actively expressed due to low temperatures. The highest number of DEGs were expressed from March to April. Once the shoots had completed the underground development stage, cell proliferation increased and shoots emerged from the ground, marking the beginning of a rapid increase in cell volume. As bamboo shoots undergo the greatest number of changes when undergoing growth above ground, the number of downregulated genes was much higher during March and April than in other months.
4.3. Genetic Regulation of Important Moso Bamboo Traits
At present, there are no studies on the omics of bamboo culm wall thickening; however, studies have been conducted on the omics of rapid growth and flowering. Bamboo plants have the unique reproductive biological characteristics of a long flowering cycle and plant death after blooming. It has been reported in certain studies that different key genes play a central role in the regulation of different growth periods of flowers; 290 genes related to the transition from vegetative to reproductive growth and 47 genes related to the growth and development of flowers have been identified. MADS, GID1, GID2, LHY, and MYB were upregulated during flower development. The expression of Dof genes was observed to be significantly inhibited during anthesis and embryo formation, which suggests that Dof genes play a more significant role in the early stages of flower development [42]. Additionally, 6076 upregulated genes and 4613 downregulated genes were identified during the rapid growth stage of Moso bamboo. Concurrently, candidate genes related to transcription factors, plant hormones, cell cycle regulation, cell wall metabolism, and cell morphogenesis have also been identified [13]. There are 31,987 genes in the Moso bamboo reference genome [43]. It is obvious that a large number of single genes regulate multiple traits in Moso bamboo. The gene regulation network of traits should be continuously evaluated until it is completely characterized.
5. Conclusions
Transcriptome sequencing analysis was performed on the shoots of P. edulis ‘Pachyloen’ and Moso bamboo from September to April of the following year to determine the developmental differences at the omics level. In all, 666–1839 genes were uniquely expressed in P. edulis ‘Pachyloen’, while 1079–1356 genes were uniquely expressed in Moso bamboo. The P. edulis ‘Pachyloen’-specific genes were enriched in 20 BPs, 15 CCs, 12 MFs, and 137 metabolic pathways including 52 significant metabolic pathways. In total, 27 metabolic pathways such as tyrosine metabolism were considered important in the thickening of P. edulis ‘Pachyloen’.
This study provides a solid foundation for the further study of the thickening characteristics of P. edulis ‘Pachyloen’ as well as research on bamboo plants more generally.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/8/861/s1, Supplementary File S1: The website of the data source; Supplementary File S2: Primer; Supplementary File S3: Distribution of gene’s coverage identification for DEGs; Supplementary File S4: Daily maximum temperature; Supplementary File S5: Specific genes; Supplementary File S6: The expression profiles of 10 genes randomly chosen.
Author Contributions
Data curation, Y.H.; Formal analysis, Y.H. and Y.Z.; Funding acquisition, Q.G.; Investigation, J.Z. and G.W.; Writing—original draft, Y.H.; Writing—review & editing, Q.G. All authors read and approved the final version of the manuscript.
Funding
This work was kindly supported by research projects of the National Natural Science Foundation of China (31971648) and the Talent Introduction Project Study of Nanjing Forestry University (GXL2018001) on Ginkgo biloba and other important tree germplasm resources.
Acknowledgments
We thank the Co-Innovation Center for Sustainable Forestry in Southern China and International Center of Bamboo and Rattan for allowing us to use their research facilities.
Conflicts of Interest
We declare that there are no conflicts of interest.
References
- Chang, S.T.; Wu, J.H. Green-color conservation of ma bamboo (Dendrocalamus latiflorus) treated with chromium-based reagents. J. Wood Sci. 2000, 46, 40–44. [Google Scholar] [CrossRef]
- Dileep, P.; Narayanankutty, S.K. A novel method for preparation of nanosilica from bamboo leaves and its green modification as a multi-functional additive in styrene butadiene rubber. Mater. Today Commun. 2020, 24, 100957. [Google Scholar] [CrossRef]
- Kassahun, T. Review of Bamboo Value Chain in Ethiopia. J. Biol. Agric. Healthc. 2014, 2, 52–67. [Google Scholar]
- Xie, J.; Hong, M. Effects of ecological factors on growth of Arundinaria spanostachya shoots in Liziping National Nature Reserve, China. Glob. Ecol. Conserv. 2020, 23, e01121. [Google Scholar] [CrossRef]
- Zakaria, S.A.; Latif, R.; Bahauddin, A. Malay Wisdom in the Motifs of Bamboo Shoots. SHS Web Conf. 2018, 45, 02004. [Google Scholar] [CrossRef][Green Version]
- Piazza, M.; Lobovikov, M.; Paudel, S.; Ren, H.; Wu, J. World bamboo resources: A thematic study prepared in the framework of the Global Forest Resources Assessment 2005. Non Wood For. Prod. 2007, 18. [Google Scholar] [CrossRef]
- Fu, T.; Ke, J.H.; Zhou, S.; Xie, G.H. Estimation of the quantity and availability of forestry residue for bioenergy production in China. Resour. Conserv. Recycl. 2020, 162, 104993. [Google Scholar] [CrossRef]
- Lin, X.C.; Ruan, X.S.; Lou, Y.F.; Guo, X.Q.; Fang, W. Genetic similarity among cultivars of Phyllostachys pubescens. Plant Syst. Evol. 2009, 277, 67–73. [Google Scholar] [CrossRef]
- Liese, W.; Khl, M. Bamboo–The Plant and Its Uses; Springer: Berlin, Germany, 2015. [Google Scholar]
- Guo, Q.R.; Fang-Ming, H.U.; Tian-Zhen, D.U.; Yang, G.Y.; Chen, H.W.; Zhu, J.Y. Leaves Physiological and Biochemical Characters of Phyllostachys edulis cv. Pachyloen. J. Cent. South For. Univ. 2005, 25, 7–11. [Google Scholar]
- Qiang; Wei; Junjie; Cao; Weijie; Qian; Mengjian; Xu; Zhongru; Li. Establishment of an efficient micropropagation and callus regeneration system from the axillary buds of Bambusa ventricosa. Plant Cell Tissue Organ Cult. 2015, 122, 1–8. [Google Scholar] [CrossRef]
- Zhang, H.; Ying, Y.-Q.; Wang, J.; Zhao, X.-H.; Zeng, W.; Beahan, C.; He, J.-B.; Chen, X.-Y.; Bacic, A.; Song, L.-L.; et al. Transcriptome analysis provides insights into xylogenesis formation in Moso bamboo (Phyllostachys edulis) shoot. Sci. Rep. 2018, 8, 3951. [Google Scholar] [CrossRef]
- Peng, Z.; Zhang, C.; Zhang, Y.; Hu, T.; Mu, S.; Li, X.; Gao, J. Transcriptome Sequencing and Analysis of the Fast Growing Shoots of Moso Bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e78944. [Google Scholar] [CrossRef] [PubMed]
- Miyashima, S.; Sebastian, J.; Lee, J.Y.; Helariutta, Y. Stem cell function during plant vascular development. Semin. Cell Dev. Biol. 2012, 32, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, N.; Ma, X.; Gupta, V.K.; Zhang, D.; Li, T.; Dai, X. The Ectopic Overexpression of the Cotton Ve1 and Ve2-Homolog Sequences Leads to Resistance Response to Verticillium Wilt in Arabidopsis. Front. Plant Sci. 2017, 8, 844. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Green, E.D.; Guttmacher, A.E.; Guyer, M.S. A vision for the future of genomics research. Nature 2003, 422, 835–847. [Google Scholar] [CrossRef]
- Morozova, O.; Marra, M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics 2008, 92, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Qiang, W.; Chen, J.; Lin, G.; Ding, Y.; Cao, J.; Feng, J.; Dong, X.; Mao, L.; Sun, H.; Yu, F. Exploring key cellular processes and candidate genes regulating the primary thickening growth of Moso underground shoots. New Phytol. 2017, 214, 81–96. [Google Scholar]
- Wang, Y.; Gao, Y.; Zhang, H.; Wang, H.; Liu, X.; Xu, X.; Zhang, Z.; Kohnen, M.V.; Hu, K.; Wang, H.; et al. Genome-Wide Profiling of Circular RNAs in the Rapidly Growing Shoots of Moso Bamboo (Phyllostachys edulis). Plant Cell Physiol. 2019, 60, 1354–1373. [Google Scholar] [CrossRef]
- Xu, X.; Lou, Y.; Yang, K.; Shan, X.; Zhu, C.; Gao, Z. Identification of Homeobox Genes Associated with Lignification and Their Expression Patterns in Bamboo Shoots. Biomolecules 2019, 9. [Google Scholar] [CrossRef]
- Yang, J.; Wu, L.; Pan, Y.; Zhong, H. Extraction, Optical Properties and Bio-Imaging of Fluorescent Composition From Moso Bamboo Shoots. J. Renew. Mater. 2019, 7, 1209–1219. [Google Scholar] [CrossRef]
- Li, X.; Xie, L.; Zheng, H.; Cai, M.; Cheng, Z.; Bai, Y.; Li, J.; Gao, J. Transcriptome profiling of postharvest shoots identifies PheNAP2- and PheNAP3-promoted shoot senescence. Tree Physiol. 2019, 39, 2027–2044. [Google Scholar] [CrossRef]
- Zhang, X.M.; Zhao, L.; Larson-Rabin, Z.; Li, D.Z.; Guo, Z.H.; Nitabach, M.N. De Novo Sequencing and Characterization of the Floral Transcriptome of Dendrocalamus latiflorus (Poaceae: Bambusoideae). PLoS ONE 2012, 7, e42082. [Google Scholar] [CrossRef] [PubMed]
- Shou, Y.T.; Zhu, Y.H.; Ding, Y.L. Transcriptome analysis of lateral buds from Phyllostachys edulis rhizome during germination and early shoot stages. BMC Plant Biol. 2020, 20, 229. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.C.; Chou, M.L.; Yue, J.J.; Hsu, C.T.; Chang, W.J.; Ko, S.S.; Liao, D.C.; Huang, Y.T.; Chen, J.J.; Yuan, J.L. BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues- characterization of transcriptome and study of MADS genes in bamboo floral development. BMC Plant Biol. 2014, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rougemont, J.; Amzallag, A.; Iseli, C.; Farinelli, L.; Xenarios, I.; Naef, F. Probabilistic base calling of Solexa sequencing data. BMC Bioinf. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M.; Blazquez, M.A. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef]
- Moon, S.; Jung, K.H.; Lee, D.E.; Lee, D.Y.; Lee, J.; An, K.; Kang, H.G.; An, G. The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol. Cells 2006, 21, 147. [Google Scholar]
- Demura, T.; Fukuda, H. Transcriptional regulation in wood formation. Trends Plant Sci. 2007, 12, 64–70. [Google Scholar] [CrossRef]
- MARCO, F.; ELISA, B.; FRANCESCA, R.; GIULIANO, C.; VANIA, M.; DANIELE, B.; RITA, B.; CLAUDIO, P. stem fasciated, a Recessive Mutation in Sunflower (Helianthus annuus), Alters Plant Morphology and Auxin Level. Ann. Bot. 2006, 4, 715–730. [Google Scholar]
- Jia, N.; Liu, X.; Gao, H. A DNA2 homolog is required for DNA damage repair, cell cycle regulation, and meristem maintenance in plants. Plant Physiol. 2016, 171, 318–333. [Google Scholar] [CrossRef]
- Madoka, Y.; Kashiwagi, T.; Hirotsu, N.; Ishimaru, K. Indian rice “Kasalath” contains genes that improve traits of Japanese premium rice “Koshihikari”. Theor. Appl. Genet. 2008, 116, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Ookawa, T.; Aya, K.; Ochiai, Y.; Hirasawa, T.; Ebitani, T.; Takarada, T.; Yano, M.; Yamamoto, T.; Fukuoka, S.; et al. Isolation of a Novel Lodging Resistance QTL Gene Involved in Strigolactone Signaling and Its Pyramiding with a QTL Gene Involved in Another Mechanism. Mol. Plant 2015, 8, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Kiryu, T.; Miyoshi, Y.; Furuta, Y. The Mechanism of Improvement of Physical Properties of Moso Bamboo (Phyllostachys pubescens) with Increasing Age I. Relationship between modulus of elasticity in bending and cell wall thickness and/or percentage of cell walls. Mokuzai Gakkaishi 2016, 62, 61–66. [Google Scholar] [CrossRef][Green Version]
- Chiu, W.B.; Lin, C.H.; Chang, C.J.; Hsieh, M.H.; Wang, A.Y. Molecular characterization and expression of four cDNAs encoding sucrose synthase from green bamboo Bambusa oldhamii. New Phytol. 2006, 170, 53–63. [Google Scholar] [CrossRef]
- Hsieh, L.S.; Ma, G.J.; Yang, C.C.; Lee, P.D. Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry 2010, 71, 1999–2009. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, X.; Ding, Y.; Fei, Z.; Jiao, C.; Fan, M.; Yao, B.; Xin, P.; Chu, J.; Wei, Q. Cellular and molecular characterization of a thick-walled variant reveal a pivotal role of shoot apical meristem in transverse development of bamboo culm. J. Exp. Bot. 2019, 70, 3911–3926. [Google Scholar] [CrossRef]
- Li, L.; Cheng, Z.; Ma, Y.; Bai, Q.; Li, X.; Cao, Z.; Wu, Z.; Gao, J. The association of hormone signalling genes, transcription and changes in shoot anatomy during moso bamboo growth. Plant Biotechnol. J. 2018, 16, 72–85. [Google Scholar] [CrossRef]
- Wang, H.Y.; Cui, K.; He, C.Y.; Zeng, Y.F.; Liao, S.X.; Zhang, J.G. Endogenous hormonal equilibrium linked to bamboo culm development. Genet. Mol. Res. 2015, 14, 11312–11323. [Google Scholar] [CrossRef]
- Wang, K.; Peng, H.; Lin, E.; Jin, Q.; Hua, X.; Yao, S.; Bian, H.; Han, N.; Pan, J.; Wang, J.; et al. Identification of genes related to the development of bamboo rhizome bud. J. Exp. Bot. 2010, 61, 551–561. [Google Scholar] [CrossRef]
- Jian, G.; Ying, Z.; Zhang, C.; Qi, F.; Li, X.; Mu, S.; Peng, Z.; Emmanuel, G. Characterization of the Floral Transcriptome of Moso Bamboo (Phyllostachys edulis) at Different Flowering Developmental Stages by Transcriptome Sequencing and RNA-Seq Analysis. PLoS ONE 2014, 9, e98910. [Google Scholar] [CrossRef]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Jiang, Z. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).