Identification of Spruce Budworm Natural Enemies Using a qPCR-Based Molecular Sorting Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Development of a Barcode Sequence Database for Spruce Budworm Parasitoids
2.2. Development of a Barcode Sequence Database for Spruce Budworm Microsporidia
2.3. Development of the Molecular Sorting Tool
2.4. SYBR Green-Based Real-Time PCR Quantification of DNA
2.5. TaqMan Assay Validation Using Purified DNA Samples and gBlocks Gene Fragments
2.6. Testing of the Molecular Sorting Tool Using Field-Collected Budworm Larvae
2.7. Assessing the Limit of Detection
3. Results
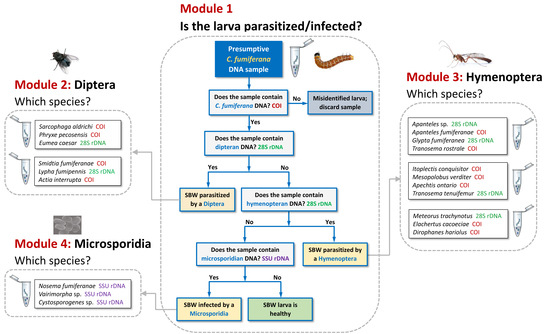
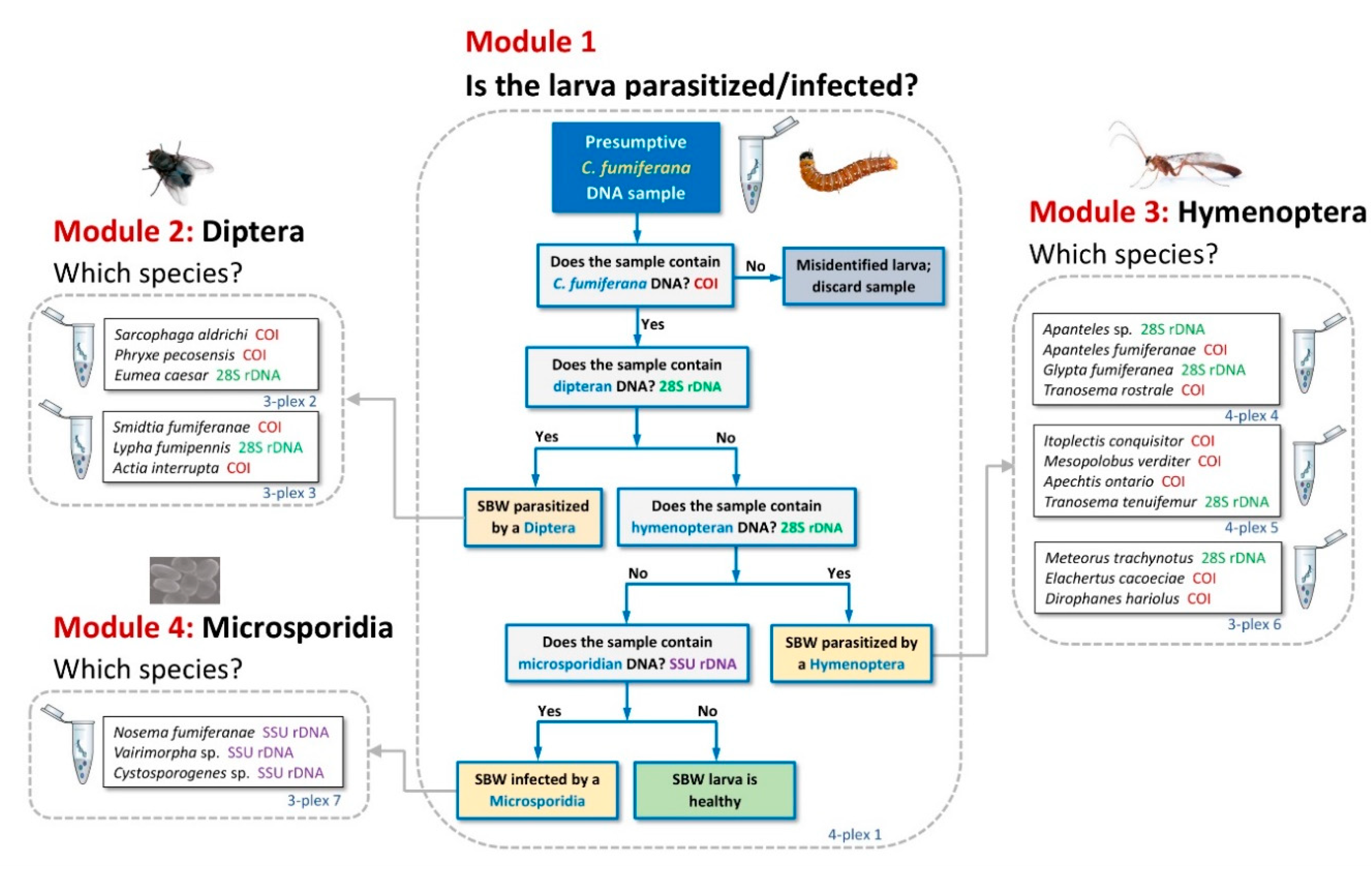
3.1. Tool Development and Architecture
3.2. Tool Validation Using Synthetic gBlocks® Fragments and DNA Extracts
3.2.1. Detection
3.2.2. Discrimination
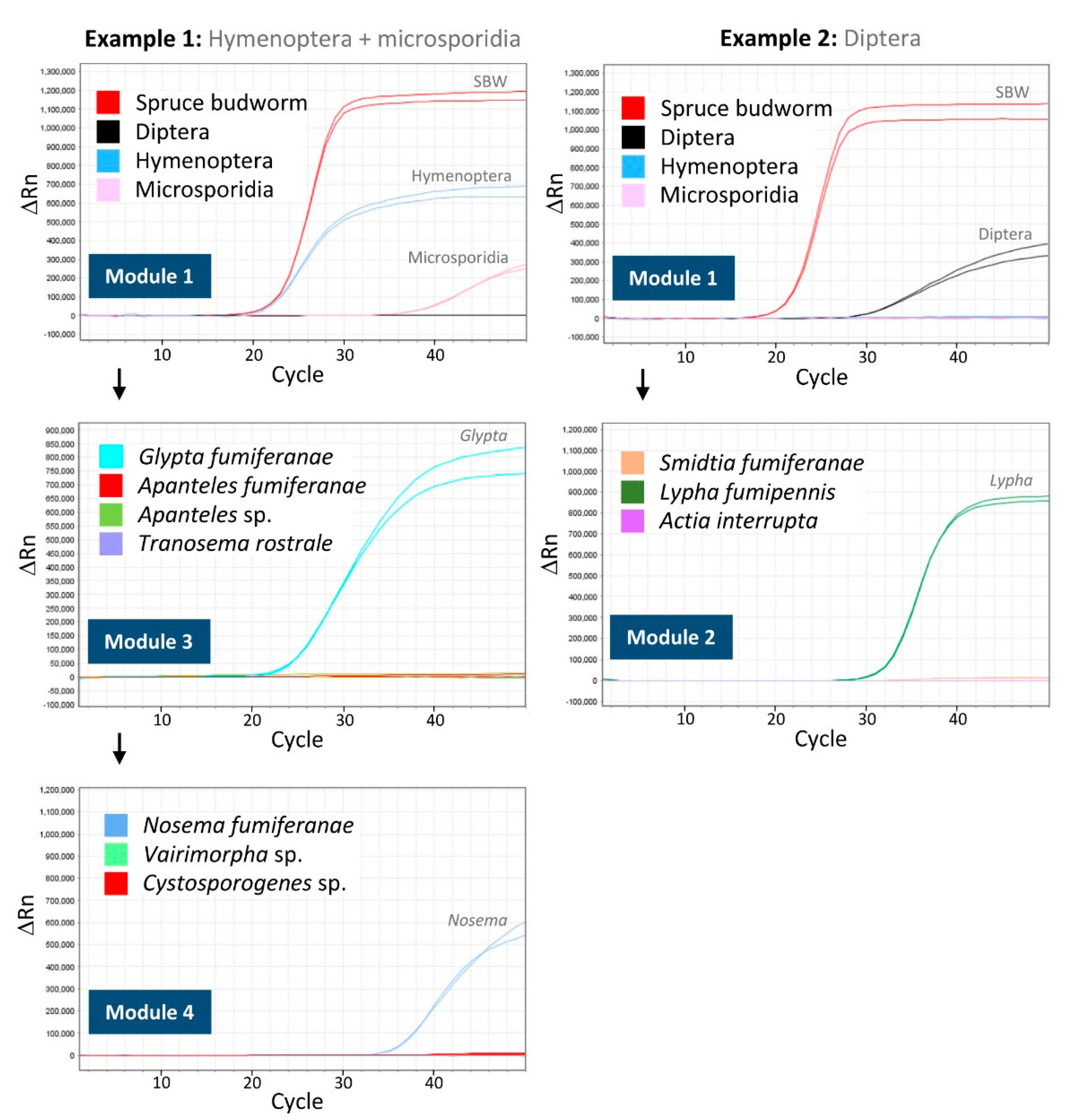
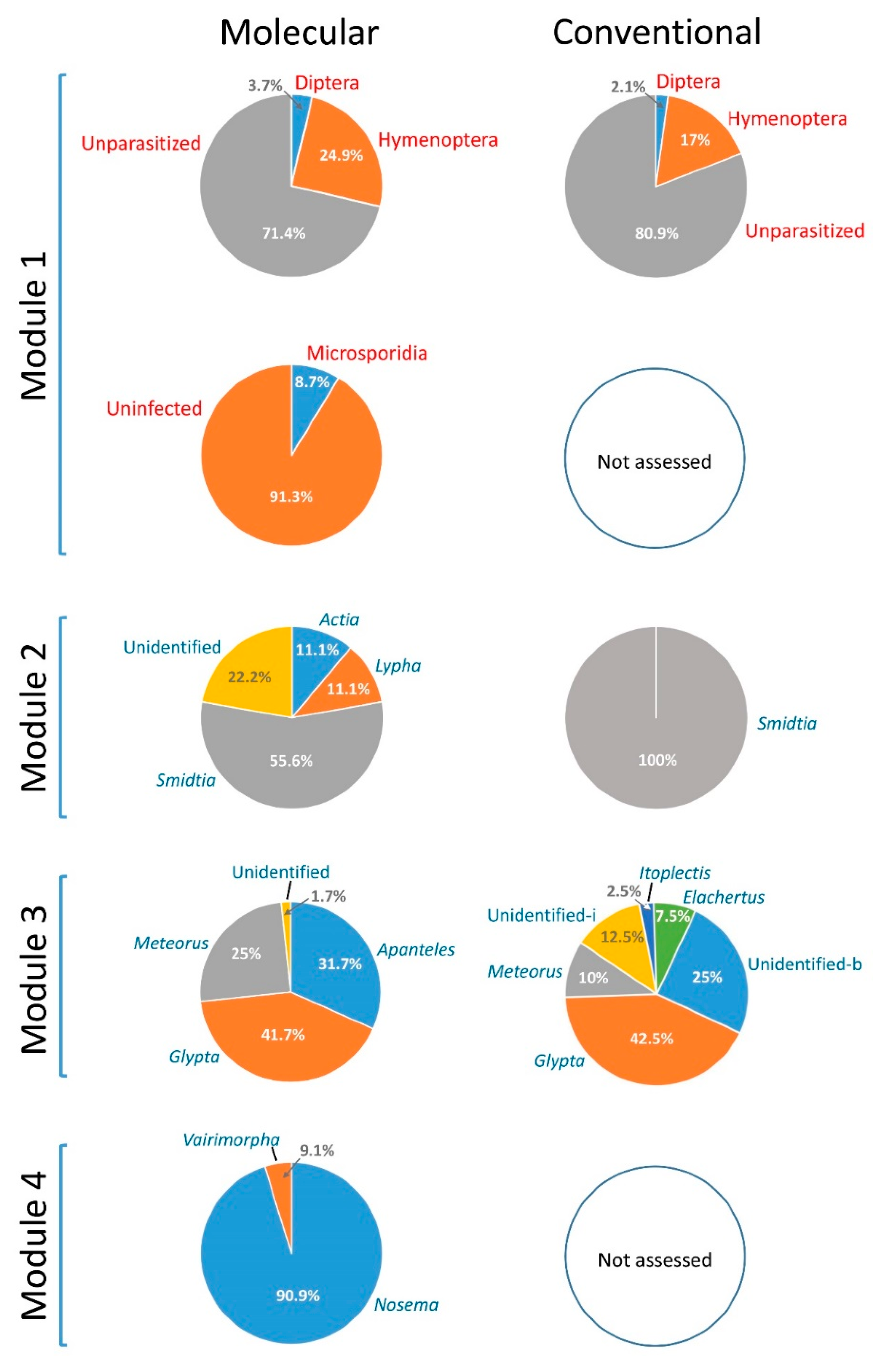
3.3. Comparative Performance of the Molecular Sorting Tool Using Field-Collected Larvae
3.4. Assessment of the Limit of Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johns, R.C.; Bowden, J.J.; Carleton, D.R.; Cooke, B.J.; Edwards, S.; Emilson, E.J.S.; James, P.M.A.; Kneeshaw, D.; MacLean, D.A.; Martel, V.; et al. A conceptual framework for the spruce budworm early intervention strategy: Can outbreaks be stopped? Forests 2019, 10, 910. [Google Scholar] [CrossRef]
- Régnière, J.; Cooke, B.J.; Béchard, A.; Dupont, A.; Therrien, P. Dynamics and management of rising outbreak spruce budworm populations. Forests 2019, 10, 748. [Google Scholar] [CrossRef]
- Royama, T. Population dynamics of the spruce budworm, Choristoneura fumiferana. Ecol. Monogr. 1984, 54, 429–462. [Google Scholar] [CrossRef]
- Royama, T.; Eveleigh, E.S.; Morin, J.R.B.; Pollock, S.J.; McCarthy, P.C.; McDougall, G.A.; Lucarotti, C.J. Mechanisms underlying spruce budworm outbreak processes as elucidated by a 14-year study in New Brunswick, Canada. Ecol. Monogr. 2017, 87, 600–631. [Google Scholar] [CrossRef]
- Régnière, J.; Nealis, V.G. Ecological mechanisms of population change during outbreaks of the spruce budworm. Ecol. Entomol. 2007, 32, 461–477. [Google Scholar] [CrossRef]
- Eveleigh, E.S.; McCann, K.S.; McCarthy, P.C.; Pollock, S.J.; Lucarotti, C.J.; Morin, B.; McDougall, G.A.; Strongman, D.B.; Huber, J.T.; Umbanhowar, J.; et al. Fluctuations in density of an outbreak species drive diversity cascades in food webs. Proc. Natl. Acad. Sci. USA. 2007, 104, 16976–16981. [Google Scholar] [CrossRef]
- Bouchard, M.; Martel, V.; Régnière, J.; Therrien, P.; Pinto Correia, D.L. Do natural enemies explain fluctuations in low-density spruce budworm populations? Ecology 2018, 99, 2047–2057. [Google Scholar] [CrossRef]
- Fernández-Triana, J.L.; Huber, J. Braconid parasitoids (Hymenoptera: Braconidae) of nearctic Choristoneura species (Lepidoptera: Tortricidae), with a summary of other parasitoid families attacking Choristoneura. Can. Entomol. 2010, 142, 295–343. [Google Scholar] [CrossRef]
- Sullivan, D.J.; Völkl, W. Hyperparasitism: Multitrophic ecology and behavior. Annu. Rev. Entomol. 1999, 44, 291–315. [Google Scholar] [CrossRef]
- Lucarotti, C.J.; Eveleigh, E.S.; Royama, T.; Morin, B.; McCarthy, P.; Ebling, P.M.; Kaupp, W.J.; Guertin, C.; Arella, M. Prevalence of baculoviruses in spruce budworm (Lepidoptera: Tortricidae) populations in New Brunswick. Can. Entomol. 2004, 136, 255–264. [Google Scholar] [CrossRef]
- Seehausen, M.L.; Labrecque, M.; Martel, V.; Régnière, J.; Mansour, A.; Smith, S.M. Reproductive biology and behavior of Tranosema rostrale (Hymenoptera: Ichneumonidae), a parasitoid of low density spruce budworm (Lepidoptera: Tortricidae) populations. J. Insect Behav. 2016, 29, 500–514. [Google Scholar] [CrossRef]
- Pook, V.G.; Athey, K.J.; Chapman, E.G.; Clutts-Stoelb, S.A.; Sharkey, M.J. New PCR primers enhance investigation of host-parasitoid food webs. Ent. Exp. Appl. 2017, 162, 309–314. [Google Scholar] [CrossRef]
- Smith, M.A.; Eveleigh, E.S.; McCann, K.S.; Merilo, M.T.; McCarthy, P.C.; Van Rooyen, K.I. Barcoding a quantified food web: Crypsis, concepts, ecology and hypotheses. PLoS ONE 2011, 6, e14424. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Fernández-Triana, J.L.; Eveleigh, E.; Gómez, J.; Guclu, C.; Hallwachs, W.; Hebert, P.D.; Hrcek, J.; Huber, J.T.; Janzen, D.; et al. DNA barcoding and the taxonomy of Microgastrinae wasps (Hymenoptera, Braconidae): Impacts after 8 years and nearly 20,000 sequences. Mol. Ecol. Res. 2013, 13, 168–176. [Google Scholar] [CrossRef]
- Kyei-Poku, G.; Gauthier, D.; van Frankenhuyzen, K. Molecular data and phylogeny of Nosema infecting lepidopteran forest defoliators in the genera Choristoneura and Malacosoma. J. Eukaryot. Microbiol. 2008, 55, 51–58. [Google Scholar] [CrossRef]
- Van Frankenhuyzen, K.; Ebling, P.; McCron, B.; Ladd, T.; Gauthier, D.; Vossbrinck, C. Occurrence of Cystosporogenes sp. (Protozoa, Microsporidia) in a multi-species insect production facility and its elimination from a colony of the eastern spruce budworm, Choristoneura fumiferana (Clem.) (Lepidoptera: Tortricidae). J. Invert. Pathol. 2004, 87, 16–28. [Google Scholar] [CrossRef]
- Stewart, D.; Zahiri, R.; Djoumad, A.; Freschi, L.; Lamarche, J.; Holden, D.; Cervantes, S.; Ojeda, D.I.; Potvin, A.; Nisole, A.; et al. A multi-species TaqMan PCR assay for the identification of Asian gypsy moths (Lymantria spp.) and other invasive lymantriines of biosecurity concern to North America. PLoS ONE 2016, 11, e0160878. [Google Scholar] [CrossRef]
- Stewart, D.; Nisole, A.; Djoumad, A.; Zahiri, R.; Lamarche, J.; Levesque, R.C.; Hamelin, R.C.; Cusson, M. A needle in a haystack: A multigene TaqMan assay for the detection of Asian gypsy moths in bulk pheromone trap samples. Biol. Invasions 2018, 21, 1843–1856. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data system (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Huber, J.T.; Eveleigh, E.; Pollock, S.; McCarthy, P. The chalcidoid parasitoids and hyperparasitoids (Hymenoptera: Chalcidoidea) of Choristoneura species (Lepidoptera: Tortricidae) in America north of Mexico. Can. Entomol. 1996, 128, 1167–1220. [Google Scholar] [CrossRef]
- Wilson, G.G. Transmission of Nosema fumiferanae (Microsporida) to its host Choristoneura fumiferana (Clem.). Z. Parasitenkd 1982, 68, 47–51. [Google Scholar] [CrossRef]
- Wilson, G.G. The transmission and effects of Nosema fumiferanae and Pleistophora schubergi (Microsporida) on Choristoneura fumiferana (Lepidoptera: Tortricidae). Proc. Ent. Soc. Ont. 1984, 115, 71–75. [Google Scholar]
- Van Frankenhuyzen, K.; Nystrom, C.; Liu, Y. Vertical transmission of Nosema fumiferanae (Microsporidia: Nosematidae) and consequences for distribution, post-diapause emergence and dispersal of second-instar larvae of the spruce budworm, Choristoneura fumiferana (Clem.) (Lepidoptera: Tortricidae). J. Invert. Pathol. 2007, 96, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Eveleigh, E.S.; Lucarotti, C.J.; McCarty, P.C.; Morin, B. Prevalence, transmission, and mortality associated with Nosema fumiferanae infections in field populations of spruce budworm Choristoneura fumiferana. Agr. For. Entomol. 2012, 14, 389–398. [Google Scholar] [CrossRef]
- Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5′—3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 1991, 88, 7276–7280. [Google Scholar] [CrossRef] [PubMed]
- Cusson, M.; Laforge, M.; Miller, D.; Cloutier, C.; Stoltz, D. Functional significance of parasitism-induced suppression of juvenile hormone esterase activity in developmentally delayed Choristoneura fumiferana larvae. Gen. Comp. Endocrinol. 2000, 117, 343–354. [Google Scholar] [CrossRef]
- Doucet, D.; Cusson, M. Role of calyx fluid in alterations of immunity in Choristoneura fumiferana larvae parasitized by Tranosema rostrale. Comp. Biochem. Physiol. 1996, 114, 311–317. [Google Scholar] [CrossRef]
- Doucet, D.; Cusson, M. Alteration of developmental rate and growth of Choristoneura fumiferana parasitized by Tranosema rostrale: Role of the calyx fluid. Entomol. Exp. Appl. 1996, 81, 21–30. [Google Scholar] [CrossRef]
- Djoumad, A.; Dallaire, F.; Lucarotti, C.J.; Cusson, M. Characterization of the polydnaviral TrV gene family: TrV1 expression inhibits in vitro cell proliferation. J. Gen. Virol. 2013, 94, 1134–1144. [Google Scholar] [CrossRef]
- Rasoolizadeh, A.; Béliveau, C.; Stewart, D.; Cloutier, C.; Cusson, M. Tranosema rostrale ichnovirus repeat element genes display distinct transcriptional patterns in caterpillar and wasp hosts. J. Gen. Virol. 2009, 90, 1505–1514. [Google Scholar] [CrossRef]
- Seehausen, M.L.; Cusson, M.; Régnière, J.; Bory, M.; Stewart, D.; Djoumad, A.; Smith, S.M.; Martel, V. High temperature induces downregulation of polydnavirus gene transcription in lepidopteran host and enhances accumulation of host immunity gene transcripts. J. Insect Physiol. 2017, 98, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Roe, A.D.; Demidovich, M.; Dedes, J. Origins and history of laboratory insect stocks in a multispecies insect production facility, with the proposal of standardized nomenclature and designation of formal standard names. J. Insect Sci. 2017, 18, 1. [Google Scholar] [CrossRef]
- McMorran, A. A synthetic diet for the spruce budworm Choristoneura fumiferana. Can. Entomol. 1965, 97, 58–62. [Google Scholar] [CrossRef]
- Miller, C.A.; Renault, T.R. Notes on the biology of Synetaeris tenuifemur Walley (Hymenoptera: Ichneumonidae). Can. Entomol. 1963, 95, 24–28. [Google Scholar] [CrossRef]
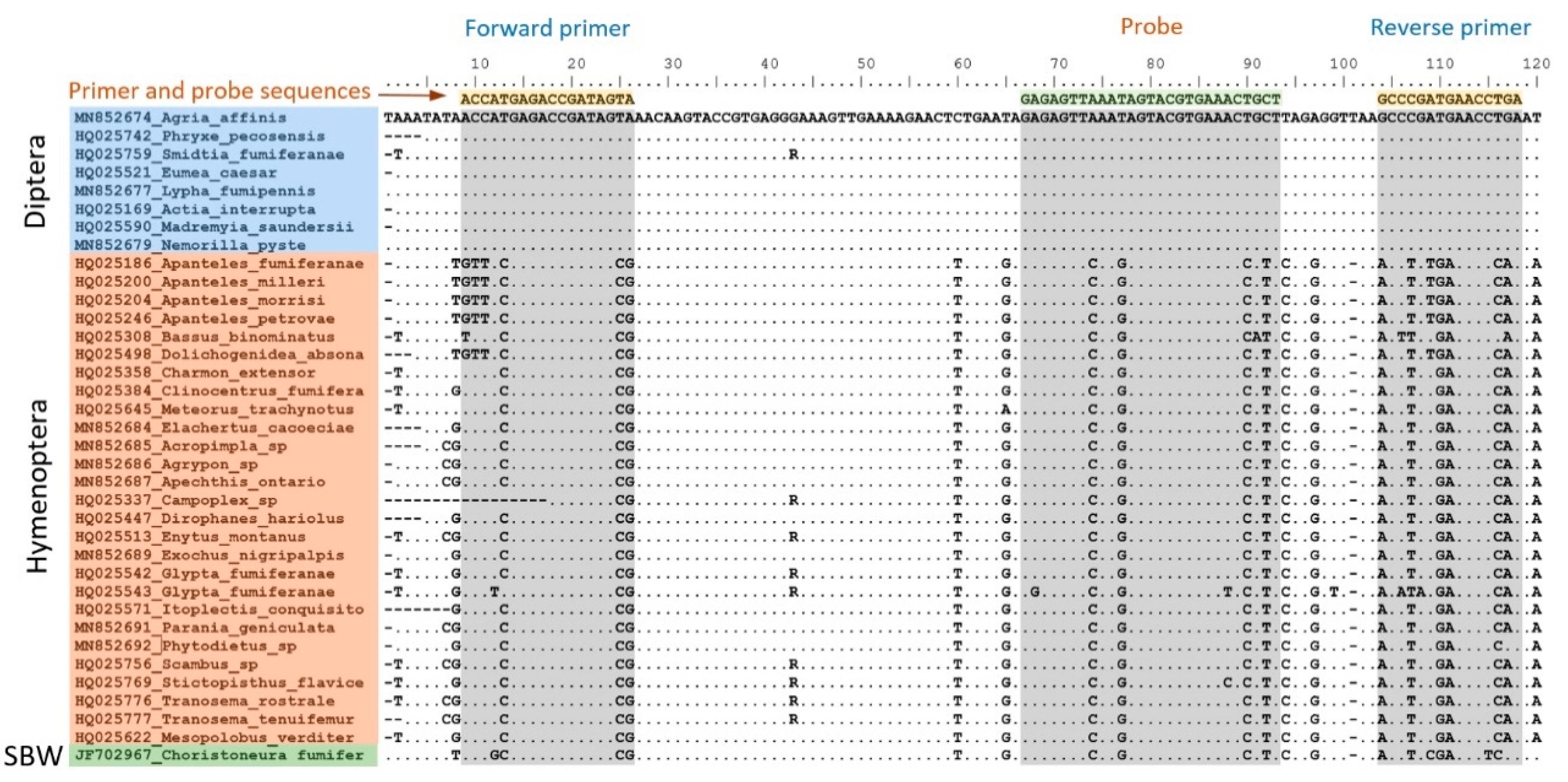
| Parasitoid Taxon | Source and Number of Marker Sequences Used 1 | ||||
|---|---|---|---|---|---|
| 28S rDNA | COI | ||||
| GenBank | New | GenBank | New | ||
| Frequent | Diptera (Sarcophagidae) | ||||
| Sarcophaga aldrichi | − | − | 6 | − | |
| Diptera (Tachinidae) | |||||
| Eumea caesar | 8 | − | 8 | − | |
| Lypha fumipennis | − | 3 | 1 | 3 | |
| Phryxe pecosensis | 8 | − | 7 | − | |
| Smidtia (= Winthemia) fumiferanae | − | 7 | − | 7 | |
| Actia interrupta | 10 | 2 | 10 | 2 | |
| Hymenoptera (Braconidae) | |||||
| Apanteles fumiferanae | 22 | − | 22 | − | |
| Meteorus trachynotus | 6 | − | 12 | − | |
| Hymenoptera (Eulophidae) | |||||
| Elachertus cacoeciae | − | 1 | 3 | 1 | |
| Hymenoptera (Ichneumonidae) | |||||
| Apechthis ontario | − | 2 | 11 | 2 | |
| Glypta fumiferanae | 4 | 15 | 25 | 15 | |
| Itoplectis conquisitor | 2 | 2 | 15 | 2 | |
| Dirophanes maculicornis | 15 | 1 | 15 | 1 | |
| Tranosema rostrale | 2 | 7 | 25 | 10 | |
| Tranosema tenuifemur | 7 | − | 8 | − | |
| Mesopolobus verditer | 1 | 1 | 13 | 1 | |
| Occasional | Diptera (Sarcophagidae) | ||||
| Pseudosarcophaga affinis | − | 1 | 6 | 1 | |
| Diptera (Tachinidae) | |||||
| Madremya saundersii | 1 | 2 | 1 | 2 | |
| Nemorilla pystes | − | 1 | − | ||
| Hymenoptera (Braconidae) | |||||
| Apanteles morrisi | 1 | − | 5 | − | |
| Apanteles petrovae | 1 | − | 3 | − | |
| Bassus binominata | 1 | − | 1 | − | |
| Charmon extensor | 4 | − | 1 | − | |
| Dolichogenidea absona | 2 | 2 | 2 | 2 | |
| Macrocentrus fumiferanae | 6 | − | 4 | − | |
| Hymenoptera (Ichneumonidae) | |||||
| Enytus montanus | 5 | − | 2 | − | |
| Exochus nigripalpis | − | 2 | 1 | 1 | |
| Less common | Hymenoptera (Braconidae) | ||||
| Apanteles milleri | 1 | − | 1 | − | |
| Hymenoptera (Ichneumonidae) | |||||
| Acropimpla sp. | − | 1 | − | 1 | |
| Agrypon prismaticum | − | 2 | − | 2 | |
| Campoplex spp. | 1 | − | 1 | − | |
| Gelis spp. | 2 | 2 | 20 | 2 | |
| Parania geniculata | − | 2 | − | 2 | |
| Phytodietus spp. | − | 2 | 5 | 2 | |
| Scambus sp. | 2 | − | 2 | − | |
| Stictopisthus flaviceps | 1 | − | 1 | − | |
| Target Group | Family | Assay Code 1 | Assay Name | Fluorophore | Position 2 |
|---|---|---|---|---|---|
| Lepidoptera | Tortricidae | 4-plex 1A | SBW COI | Texas Red | 273-400 |
| Diptera | Not applicable | 4-plex 1B | Diptera GEN 28S | Cy5 Tao | 8-119 |
| Hymenoptera | Not applicable | 4-plex 1C | Hymenoptera GEN 28S | FAM LNA | 96-273 |
| Microsporidia | Not applicable | 4-plex 1D | Microsporidia SSU rDNA | HEX Zen | 706-94 |
| Diptera | Sarcophagidae | 3-plex 2A | Sarcophaga aldrichi COI | Cy5 Tao | 432-560 |
| Diptera | Tachinidae | 3-plex 2B | Phryxe pecosensis COI | FAM LNA | 295-380 |
| Diptera | Tachinidae | 3-plex 2C | Eumea caesar 28S | HEX LNA | 195-362 |
| Diptera | Tachinidae | 3-plex 3A | Smidtia fumiferanae COI | HEX ZEN | 526-672 |
| Diptera | Tachinidae | 3-plex 3B | Lypha fumipennis 28S | FAM LNA | 107-285 |
| Diptera | Tachnidae | 3-plex 3C | Actia interrupta COI | Cy5 Tao | 344-489 |
| Hymenoptera | Braconidae | 4-plex 4A | Apanteles sp. 28S | HEX LNA | 143-303 |
| Hymenoptera | Braconidae | 4-plex 4B | Apanteles fumiferanae COI | Cy5 LNA | 242-364 |
| Hymenoptera | Ichneumonidae | 4-plex 4C | Glypta fumiferanae 28S | Texas Red | 141-313 |
| Hymenoptera | Ichneumonidae | 4-plex 4D | Tranosema rostrale COI | FAM Zen | 365-511 |
| Hymenoptera | Ichneumonidae | 4-plex 5A | Itoplectis conquisitor COI | Texas Red | 31-218 |
| Hymenoptera | Pteromalidae | 4-plex 5B | Mesopolobus verditer COI | Cy5 Tao | 245-373 |
| Hymenoptera | Ichneumonidae | 4-plex 5C | Apechthis ontario COI | HEX Zen | 381-561 |
| Hymenoptera | Ichneumonidae | 4-plex 5D | Tranosema tenuifemur 28S | FAM LNA | 99-215 |
| Hymenoptera | Braconidae | 3-plex 6A | Meteorus trachynotus 28S | Cy5 Tao | 162-243 |
| Hymenoptera | Eulophidae | 3-plex 6B | Elachertus cacoeciae COI | HEX Zen | 287-432 |
| Hymenoptera | Ichneumonidae | 3-plex 6C | Dirophanes hariolus COI | FAM LNA | 463-552 |
| Microsporidia | Nosematidae | 3-plex 7A | Nosema SSU rDNA | FAM Zen | 805-940 |
| Microsporidia | Nosematidae | 3-plex 7B | Vairimorpha SSU rDNA | Cy5 Tao | 247-353 |
| Microsporidia | Glugeidae | 3-plex 7C | Cystosporogenes SSU rDNA | HEX Zen | 78-201 |
| Assay | Amplicon Length | Ct Value @ 10 fg | Test Species Used 1 |
|---|---|---|---|
| SBW COI | 128bp | 20.10 | |
| Hymenoptera GEN 28S | 178bp | 20.86 | Apanteles fumiferanae |
| Diptera GEN 28S | 112bp | 22.66 | Lypha fumipennis |
| Microsporidia SSU rDNA | 89bp | 21.82 | Nosema fumiferanae |
| Sarcophaga aldrichi COI | 128bp | 21.48 | |
| Phryxe pecosensis COI | 110bp | 20.58 | |
| Eumea caesar 28S | 168bp | 20.23 | |
| Smidtia fumiferanae COI | 121bp | 20.42 | |
| Lypha fumipennis 28S | 177bp | 20.91 | |
| Actia interrupta COI | 146bp | 22.96 | |
| Apanteles sp 28S | 161bp | 20.56 | |
| Apanteles fumiferanae COI | 123bp | 23.41 | |
| Glypta fumiferanae 28S | 173bp | 20.58 | |
| Tranosema rostrale COI | 157bp | 22.35 | |
| Itoplectis conquisitor COI | 188bp | 21.66 | |
| Mesopolobus verditer COI | 129bp | 20.18 | |
| Apechthis ontario COI | 181bp | 21.03 | |
| Tranosema tenuifemur 28S | 106bp | 20.80 | |
| Meteorus trachynotus 28S | 78bp | 20.36 | |
| Elachertus cacoeciae COI | 146bp | 24.38 | |
| Dirophanes hariolus COI | 90bp | 20.12 | |
| Nosema SSU rDNA | 136bp | 20.58 | |
| Vairimorpha SSU rDNA | 107bp | 21.14 | |
| Cystosporogenes SSU rDNA | 124bp | 20.51 |
| Target Group | Sample Species | Hym 28S 1-plex | Hym 28S 4-plex | Hym 28S 4-plex + SBW | Dipt 28S 1-plex | Dipt 28S 4-plex | Dipt 28S 4-plex + SBW | SBW COI 4-plex + SBW | Microspo 1-plex | Microspo 4-plex + SBW |
|---|---|---|---|---|---|---|---|---|---|---|
| Diptera | Sarcophaga aldrichi 1 | - | - | - | - | - | - | - | - | No Ct |
| Diptera | Phryxe pecosensis | - | No Ct | No Ct | 27.12 | 26.76 | 26.76 | 20.52 | - | No Ct |
| Diptera | Eumea caesar 2 | - | No Ct | No Ct | 28.33 | 27.60 | 27.79 | 20.05 | - | No Ct |
| Diptera | Smidtia fumiferanae | - | No Ct | No Ct | 27.56 | 27.28 | 27.20 | 20.59 | - | No Ct |
| Diptera | Lypha fumipennis | - | No Ct | No Ct | 28.64 | 28.24 | 28.35 | 20.58 | - | No Ct |
| Diptera | Actia interrupta | - | No Ct | No Ct | 27.16 | 26.68 | 26.65 | 20.46 | - | No Ct |
| Diptera | Agria affinis 3 | - | - | No Ct | - | - | 27.86 | 20.47 | - | No Ct |
| Diptera | Madremya saundersii | - | - | No Ct | - | - | 28.96 | 20.46 | - | No Ct |
| Hymenoptera | Apanteles fumiferanae2 | 28.48 | 27.91 | 28.24 | - | No Ct | No Ct | 20.51 | - | No Ct |
| Hymenoptera | Glypta fumiferanae | 27.09 | 26.48 | 27.02 | - | No Ct | No Ct | 20.63 | - | No Ct |
| Hymenoptera | Tranosema rostrales | 28.56 | 27.87 | 28.41 | - | No Ct | No Ct | 20.43 | - | No Ct |
| Hymenoptera | Itoplectis conquisitor | 28.34 | 27.65 | 28.20 | - | No Ct | No Ct | 20.55 | - | No Ct |
| Hymenoptera | Mesopolobus verditer | 28.87 | 28.16 | 28.80 | - | No Ct | No Ct | 20.44 | - | No Ct |
| Hymenoptera | Apechthis ontario | 27.63 | 26.93 | 27.77 | - | No Ct | No Ct | 20.46 | - | No Ct |
| Hymenoptera | Tranosema tenuifemur 2 | 27.76 | 27.49 | 27.47 | - | No Ct | No Ct | 20.10 | - | No Ct |
| Hymenoptera | Meteorus trachynotus | 26.85 | 26.54 | 26.80 | - | No Ct | No Ct | 20.54 | - | No Ct |
| Hymenoptera | Elachertus cacoeciae | 29.95 | 28.85 | 29.28 | - | No Ct | No Ct | 20.63 | - | No Ct |
| Hymenoptera | Dirophanes hariolus | 28.69 | 27.95 | 28.23 | - | No Ct | No Ct | 20.38 | - | No Ct |
| Hymenoptera | Acropimpla sp. | - | - | 27.56 | - | - | No Ct | 20.30 | - | No Ct |
| Hymenoptera | Agrypon sp. | - | - | 30.12 | - | - | No Ct | 20.54 | - | No Ct |
| Hymenoptera | Campoplex sp. | - | - | 29.06 | - | - | No Ct | 20.53 | - | No Ct |
| Hymenoptera | Dolochogenidae absona | - | - | 28.24 | - | - | No Ct | 20.30 | - | No Ct |
| Hymenoptera | Enytus montanus | - | - | 28.07 | - | - | No Ct | 20.38 | - | No Ct |
| Hymenoptera | Exochus nigripalpis | - | - | 28.17 | - | - | No Ct | 20.56 | - | No Ct |
| Hymenoptera | Parania geniculata | - | - | 27.12 | - | - | No Ct | 20.34 | - | No Ct |
| Hymenoptera | Phytodietus sp. | - | - | 29.40 | - | - | No Ct | 20.45 | - | No Ct |
| Microsporidia | Nosema fumiferanae4 | - | - | No Ct | - | - | No Ct | 20.23 | 27.55 | 27.13 |
| Microsporidia | Vairimorpha sp. 4 | - | - | No Ct | - | - | No Ct | 20.06 | 28.73 | 28.49 |
| Microsporidia | Cystosporogenes sp. 4 | - | - | No Ct | - | - | No Ct | 20.39 | 28.19 | 27.99 |
| Lepidoptera | Choristoneura fumiferana 2 | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | 20.64 | No Ct | No Ct |
| Module | Assay | Ct Value Simplex | Ct Value Multiplex | Ct Value Multiplex + SBW |
|---|---|---|---|---|
| 2 | Sarcophaga aldrichi COI 1 | 27.68 | 27.98 | 28.51 |
| Phryxe pecosensis COI | 28.56 | 28.50 | 28.45 | |
| Eumea caesar 28S 2 | 26.18 | 26.56 | 26.44 | |
| Smidtia fumiferanae COI | 33.25 | 33.60 | 33.33 | |
| Lypha fumipennis 28S | 30.47 | 30.10 | 30.30 | |
| Actia interrupta COI | 29.09 | 29.21 | 29.15 | |
| 3 | Apanteles sp. 28S 2 | 29.02 | 28.65 | 29.14 |
| Apanteles fumiferanae COI 2 | 30.45 | 31.30 | 30.53 | |
| Glypta fumiferanae 28S | 29.04 | 28.56 | 28.01 | |
| Tranosema rostrale COI | 31.05 | 31.33 | 31.05 | |
| Itoplectis conquisitor COI | 32.65 | 32.52 | 31.92 | |
| Mesopolobus verditer COI | 31.67 | 31.15 | 32.10 | |
| Apechthis ontario COI | 32.32 | 31.38 | 31.13 | |
| Tranosema tenuifemur 28S 2 | 28.33 | 28.20 | 28.23 | |
| Meteorus trachynotus 28S | 28.60 | 28.46 | 28.53 | |
| Elachertus cacoeciae COI | 31.65 | 31.73 | 31.84 | |
| Dirophanes hariolus COI | 29.90 | 30.14 | 30.64 | |
| 4 | Nosema SSU rDNA 3 | 26.48 | 26.43 | 26.81 |
| Vairimorpha SSU rDNA 3 | 28.78 | 28.62 | 28.84 | |
| Cystosporogenes SSU rDNA 3 | 27.66 | 27.71 | 27.62 |
| Days After Oviposition | Proportion Detected |
|---|---|
| 0 | 0/8 |
| 1 | 7/8 |
| 2 | 6/8 |
| 5 | 6/8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nisole, A.; Stewart, D.; Kyei-Poku, G.; Nadeau, M.; Trudeau, S.; Huron, P.; Djoumad, A.; Kamenova, S.; Smith, M.A.; Eveleigh, E.; et al. Identification of Spruce Budworm Natural Enemies Using a qPCR-Based Molecular Sorting Approach. Forests 2020, 11, 621. https://doi.org/10.3390/f11060621
Nisole A, Stewart D, Kyei-Poku G, Nadeau M, Trudeau S, Huron P, Djoumad A, Kamenova S, Smith MA, Eveleigh E, et al. Identification of Spruce Budworm Natural Enemies Using a qPCR-Based Molecular Sorting Approach. Forests. 2020; 11(6):621. https://doi.org/10.3390/f11060621
Chicago/Turabian StyleNisole, Audrey, Don Stewart, George Kyei-Poku, Marianne Nadeau, Simon Trudeau, Paule Huron, Abdelmadjid Djoumad, Stefaniya Kamenova, M. Alex Smith, Eldon Eveleigh, and et al. 2020. "Identification of Spruce Budworm Natural Enemies Using a qPCR-Based Molecular Sorting Approach" Forests 11, no. 6: 621. https://doi.org/10.3390/f11060621
APA StyleNisole, A., Stewart, D., Kyei-Poku, G., Nadeau, M., Trudeau, S., Huron, P., Djoumad, A., Kamenova, S., Smith, M. A., Eveleigh, E., Johns, R. C., Martel, V., & Cusson, M. (2020). Identification of Spruce Budworm Natural Enemies Using a qPCR-Based Molecular Sorting Approach. Forests, 11(6), 621. https://doi.org/10.3390/f11060621