Soil Microbial Community Assembly and Interactions Are Constrained by Nitrogen and Phosphorus in Broadleaf Forests of Southern China
Abstract
1. Background
2. Materials and Methods
2.1. Site Description and Soil Sample Collection
2.2. Plant and Environmental Variables Measurements
2.3. Soil Microbial DNA Extraction, Purification and Quantitation
2.4. Illumina Sequencing and GeoChip Experiments and Raw Data Processing
2.5. Statistical Analyses
3. Results
3.1. Diversity and Similarity of Soil Microbial Communities
3.2. Soil Microbial Taxonomic Distribution
3.3. Functional Genes Relevant to Nitrogen and Carbon Cycling
3.4. Molecular Ecological Networks Aanalysis of Functional Genes
3.5. Soil Microbial Community Assembly Processes and Quantitative Spatial Turnover
4. Discussion
Availability of data and materials
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SOM | soil organic matter; |
| TN | total nitrogen; |
| TK | total potassium, |
| TP | total phosphorus, |
| AN | available nitrogen, |
| AP | available phosphorus; |
| OUT | operational taxonomic units; |
| MRPP | multiple response permutation procedure; |
| NMDS | non-metric multidimensional scaling; |
| Βnti | β-nearest taxon index; |
| MEN | molecular ecological network; |
| RMT | random matrix theory; |
| avgCC | average clustering coefficients; |
| GD | average path; |
| avgK | average degree. |
References
- Pan, Y.; Birdsey, R.A.; Fang, J.; Houghton, R.; Kauppi, P.E.; Kurz, W.A.; Phillips, O.L.; Shvidenko, A.; Lewis, S.L.; Canadell, J.G.; et al. A large and persistent carbon sink in the world’s forests. Science 2011, 333, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Schluter, D.; Pennell, M.W. Speciation gradients and the distribution of biodiversity. Nature 2017, 546, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Sadam, A.; Zambrano, M.; Valencia, R.; Bahram, M. Low diversity and high host preference of ectomycorrhizal fungi in western Amazonia, a neotropical biodiversity hotspot. ISME J. 2010, 4, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Buée, M.; Deveau, A.; Mieszkin, S.; Martin, F. Ecology of the forest microbiome: Highlights of temperate and boreal ecosystems. Soil Biol. Biochem. 2016, 103, 471–488. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Krashevska, V.; Klarner, B.; Widyastuti, R.; Maraun, M.; Scheu, S. Impact of tropical lowland rainforest conversion into rubber and oil palm plantations on soil microbial communities. Biol. Fert. Soils 2015, 51, 697–705. [Google Scholar] [CrossRef]
- Sall, S.N.; Masse, D.; Ndour, N.Y.B.; Chotte, J.-L. Does cropping modify the decomposition function and the diversity of the soil microbial community of tropical fallow soil? Appl. Soil Ecol. 2006, 31, 211–219. [Google Scholar] [CrossRef]
- Hamaoui, G.S.; Rodrigues, J.L.M.; Bohannan, B.J.M.; Tiedje, J.M.; Nüsslein, K. Land-use change drives abundance and community structure alterations of thaumarchaeal ammonia oxidizers in tropical rainforest soils in Rondônia, Brazil. Appl. Soil Ecol. 2016, 107, 48–56. [Google Scholar] [CrossRef]
- Paula, F.S.; Rodrigues, J.L.; Zhou, J.; Wu, L.; Mueller, R.C.; Mirza, B.S.; Bohannan, B.J.; Nusslein, K.; Deng, Y.; Tiedje, J.M.; et al. Land use change alters functional gene diversity, composition and abundance in Amazon forest soil microbial communities. Mol. Ecol. 2014, 23, 2988–2999. [Google Scholar] [CrossRef]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B.H. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef]
- Vellend, M. Conceptual Synthesis in Community Ecology. Q. Rev. of Biol. 2010, 85, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Santoferrara, L.F.; Grattepanche, J.D.; Katz, L.A.; McManus, G.B. Patterns and processes in microbial biogeography: do molecules and morphologies give the same answers? ISME J. 2016, 10, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.M.; Myers, J.A. Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. B: Biol. Sci. 2011, 366, 2351–2363. [Google Scholar] [CrossRef]
- Long, W.; Yang, X.; Li, D. Patterns of species diversity and soil nutrients along a chronosequence of vegetation recovery in Hainan Island, South China. Ecol. Res. 2012, 27, 561–568. [Google Scholar] [CrossRef]
- Lu, R. Soil Agricultural Chemical Analysis Methods; Agricultural Sci-Tech. Press: Beijing, China, 1999; pp. 106–135. [Google Scholar]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef]
- Zhao, M.; Xue, K.; Wang, F.; Liu, S.; Bai, S.; Sun, B.; Zhou, J.; Yang, Y. Microbial mediation of biogeochemical cycles revealed by simulation of global changes with soil transplant and cropping. ISME J. 2014, 8, 2045–2055. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Zhao, M.; Sun, B.; Wu, L.; Gao, Q.; Wang, F.; Wen, C.; Wang, M.; Liang, Y.; Hale, L.; Zhou, J. Zonal Soil Type Determines Soil Microbial Responses to Maize Cropping and Fertilization. mSystems 2016, 1, e00075-16. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wen, C.; Qin, Y.; Yin, H.; Tu, Q.; Van Nostrand, J.D.; Yuan, T.; Yuan, M.; Deng, Y.; Zhou, J. Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol. 2015, 15, 125. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Van Nostrand, J.D.; Yin, H.; Wu, L.; Yuan, T.; Zhou, J. Hybridization of Environmental Microbial Community Nucleic Acids by GeoChip. Methods Mol. Biol. 2016, 1399, 183–196. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Liu, D.; Wu, H.; Lu, X.; Fang, Y.; Cheng, W.; Luo, W.; Jiang, P.; Shi, J.; et al. Aridity threshold in controlling ecosystem nitrogen cycling in arid and semi-arid grasslands. Nat. Commun. 2014, 5, 4799. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, Y.; Wang, S.; Xu, D.; Yu, H.; Wu, L.; Lin, Q.; Hu, Y.; Li, X.; He, Z.; et al. The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 2014, 8, 430–440. [Google Scholar] [CrossRef]
- Kruskal, J.B. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 1964, 29, 1–27. [Google Scholar] [CrossRef]
- Smouse, P.E.; Long, J.C.; Sokal, R.R. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 1986, 35, 627–632. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2016. [Google Scholar]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Tu, Q.; Zhi, X. Functional molecular ecological networks. mBio 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Houlton, B.Z.; Marklein, A.R.; Bai, E. Representation of nitrogen in climate change forecasts. Nat. Clim. Chang. 2015, 5, 398–401. [Google Scholar] [CrossRef]
- Hall, S.J.; Matson, P.A. Nitrogen oxide emissions after nitrogen additions in tropical forests. Nature 1999, 400, 152–155. [Google Scholar] [CrossRef]
- Schimel, J. Ecosystem consequences of microbial diversity and community structure. In Arctic and Alpine Biodiversity: Patterns, Causes and Ecosystem Consequences; Chapin, F.S., Korner, C., Eds.; Springer: Berlin, Germany, 1995; pp. 239–254. [Google Scholar]
- Zemunik, G.; Davies, S.J.; Turner, B.L. Soil drivers of local-scale tree growth in a lowland tropical forest. Ecology 2018, 99, 2844–2852. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xia, B.; Treves, D.S.; Wu, L.Y.; Marsh, T.L.; O’Neill, R.V.; Palumbo, A.V.; Tiedje, J.M. Spatial and Resource Factors Influencing High Microbial Diversity in Soil. Appl. Environ. Microbiol. 2002, 68, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Lewis, S.L.; Lloyd, J.; Sitch, S.; Mitchard, E.T.; Laurance, W.F. Changing ecology of tropical forests: evidence and drivers. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 529–549. [Google Scholar] [CrossRef]
- Kim, M.; Kim, W.S.; Tripathi, B.M.; Adams, J. Distinct bacterial communities dominate tropical and temperate zone leaf litter. Microbiol. Ecol. 2014, 67, 837–848. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Bohannan, B.J.; Brown, J.H.; Colwell, R.K.; Fuhrman, J.A.; Green, J.L.; Horner-Devine, M.C.; Kane, M.; Krumins, J.A.; Kuske, C.R. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 4, 102–112. [Google Scholar] [CrossRef]
- Andersson, A.F.; Riemann, L.; Bertilsson, S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010, 4, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Martiny, J.B.; Eisen, J.A.; Penn, K.; Allison, S.D.; Horner-Devine, M.C. Drivers of bacterial beta-diversity depend on spatial scale. Proc. Natl. Acad. Sci. USA 2011, 108, 7850–7854. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, Y.; Wang, M.; Sun, X.; Cong, J.; Deng, Y.; Lu, H.; Yuan, T.; Van Nostrand, J.D.; Li, D.; et al. Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests. Mol. Ecol. 2015, 24, 5175–5185. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, S.; Stouffer, D.B.; Uzzi, B.; Bascompte, J. Strong contributors to network persistence are the most vulnerable to extinction. Nature 2011, 478, 233–235. [Google Scholar] [CrossRef]
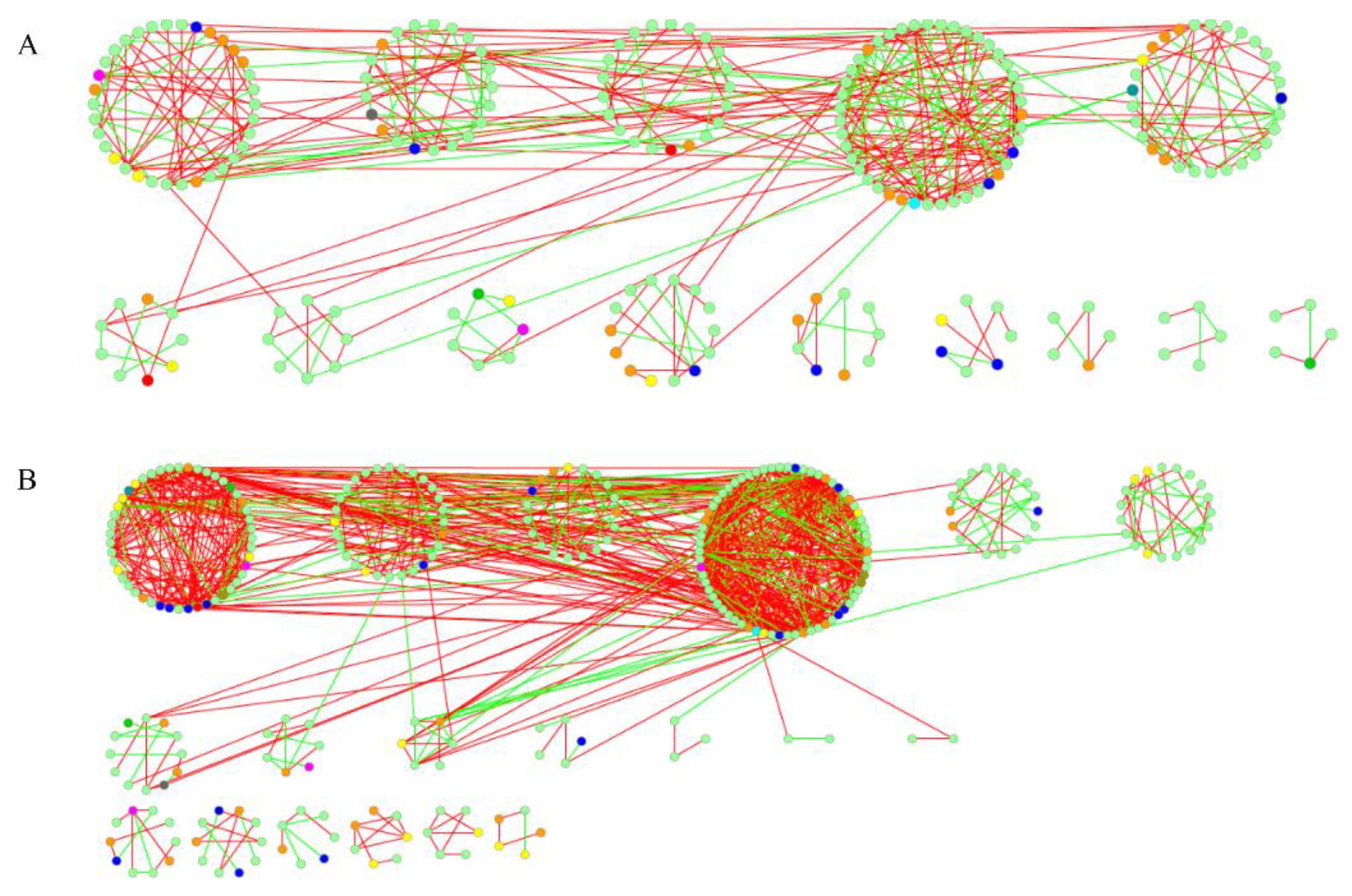
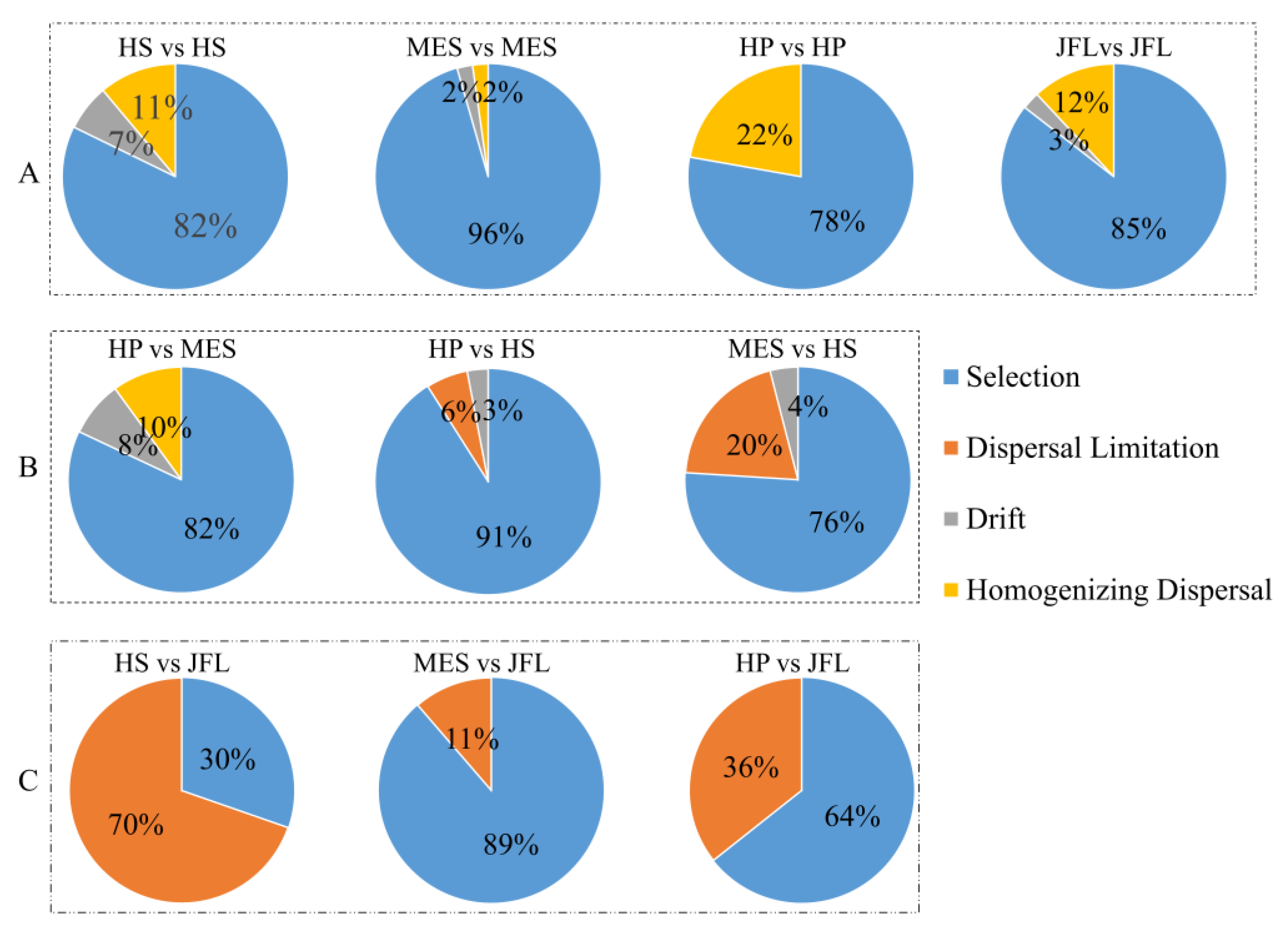
| Site | Taxa | Functional Gene |
|---|---|---|
| HS | 6.35 ± 0.12a | 10.21 ± 0.02a |
| MES | 6.22 ± 0.12b | 10.15 ± 0.11ab |
| HP | 6.39 ± 0.08a | 10.14 ± 0.03ab |
| JFL | 5.92 ± 0.12c | 10.04 ± 0.19b |
| HS | MES | HP | JFL | |
|---|---|---|---|---|
| HS | - | 0.295 *** | 0.266 *** | 0.294 *** |
| MES | 0.067 * | - | 0.253 ** | 0.293 *** |
| HP | 0.043 *** | 0.064 ** | - | 0.280 *** |
| JFL | 0.093 *** | 0.104 * | 0.091 ** | - |
| Network Properties | nifH | nirK | nosZ | |||||||||
| HS | MES | HP | JFL | HS | MES | HP | JFL | HS | MES | HP | JFL | |
| R2 | 0.80 | 0.90 | 0.95 | 0.87 | 0.88 | 0.78 | 0.95 | 0.70 | 0.86 | 0.91 | 0.91 | 0.90 |
| Network Size (n) | 330 | 244 | 344 | 214 | 207 | 191 | 208 | 137 | 273 | 224 | 280 | 192 |
| Modularity | 0.74 | 0.78 | 0.81 | 0.52 | 0.60 | 0.68 | 0.75 | 0.40 | 0.73 | 0.64 | 0.83 | 0.52 |
| avgK | 2.67 | 2.75 | 2.64 | 5.25 | 3.99 | 3.71 | 2.74 | 7.21 | 3.17 | 3.64 | 2.60 | 5.73 |
| avgCC | 0.16 | 0.20 | 0.18 | 0.29 | 0.21 | 0.25 | 0.17 | 0.34 | 0.18 | 0.19 | 0.22 | 0.34 |
| GD | 5.56 | 5.79 | 6.84 | 4.89 | 4.48 | 5.37 | 6.43 | 3.30 | 6.11 | 5.05 | 7.64 | 4.08 |
| Connectedness | 0.35 | 0.50 | 0.41 | 0.55 | 0.46 | 0.64 | 0.58 | 0.67 | 0.52 | 0.53 | 0.58 | 0.68 |
| Network Properties | amyA | cellulose | xylanase | |||||||||
| HS | MES | HP | JFL | HS | MES | HP | JFL | HS | MES | HP | JFL | |
| R2 | 0.96 | 0.97 | 0.92 | 0.92 | 0.91 | 0.89 | 0.89 | 0.91 | 0.90 | 0.97 | 0.93 | 0.90 |
| Network Size (n) | 967 | 532 | 939 | 495 | 338 | 290 | 367 | 268 | 357 | 260 | 366 | 228 |
| Modularity | 0.93 | 0.90 | 0.91 | 0.89 | 0.79 | 0.61 | 0.83 | 0.58 | 0.72 | 0.77 | 0.85 | 0.62 |
| avgK | 1.97 | 2.03 | 2.04 | 2.24 | 2.50 | 3.70 | 2.50 | 5.32 | 3.35 | 2.92 | 2.41 | 5.01 |
| avgCC | 0.14 | 0.14 | 0.13 | 0.17 | 0.18 | 0.26 | 0.18 | 0.27 | 0.22 | 0.20 | 0.15 | 0.30 |
| GD | 8.48 | 6.55 | 8.69 | 8.79 | 6.39 | 5.19 | 7.76 | 4.59 | 6.87 | 5.91 | 10.06 | 4.54 |
| Connectedness | 0.17 | 0.13 | 0.20 | 0.23 | 0.31 | 0.48 | 0.41 | 0.64 | 0.43 | 0.44 | 0.50 | 0.62 |
| Network Properties | chitinase | phenol_oxidase | ||||||||||
| HS | MES | HP | JFL | HS | MES | HP | JFL | |||||
| R2 | 0.90 | 0.82 | 0.87 | 0.86 | 0.77 | 0.88 | 0.86 | 0.83 | ||||
| Network Size (n) | 311 | 309 | 376 | 241 | 188 | 187 | 188 | 140 | ||||
| Modularity | 0.82 | 0.63 | 0.87 | 0.81 | 0.73 | 0.46 | 0.79 | 0.62 | ||||
| avgK | 2.45 | 4.42 | 2.37 | 4.12 | 3.15 | 5.56 | 2.59 | 3.97 | ||||
| avgCC | 0.17 | 0.23 | 0.15 | 0.27 | 0.24 | 0.29 | 0.20 | 0.26 | ||||
| GD | 5.40 | 5.18 | 7.87 | 4.98 | 5.28 | 4.07 | 5.62 | 4.21 | ||||
| Connectedness | 0.23 | 0.57 | 0.46 | 0.57 | 0.48 | 0.61 | 0.45 | 0.68 | ||||
| Environmental Variable | Microbial Taxonomic Composition | Network Properties | ||||
|---|---|---|---|---|---|---|
| Modularity | Average Degree (avgK) | Average Clustering Coefficient (avgCC) | Average Path Distance (GD) | Connectedness | ||
| Plant Richness | 0.18*** | 0.25 | 0.41 | 0.42 | −0.04 | 0.13 |
| Elevation | 0.02 | 0.05 | −0.11 | −0.56 | −0.04 | −0.20 |
| Soil Temperature | −0.01 | 0.03 | −0.15 | −0.52 | −0.07 | −0.13 |
| Annual Mean Temperature | 0.34*** | 0.62* | 0.63** | 0.58* | 0.09 | 0.29 |
| Annual Mean Precipitation | 0.64*** | −0.34 | −0.20 | 0.33 | −0.32 | 0.18 |
| Soil pH | 0.02 | 0.00 | −0.09 | −0.51 | 0.11 | −0.19 |
| Soil Moisture | 0.37*** | 0.36 | 0.56* | 0.56* | 0.14 | 0.09 |
| Total Nitrogen | −0.45 | −0.07 | −0.21 | −0.36 | 0.31 | −0.27 |
| Total Potassium | 0.43*** | 0.35 | 0.55* | 0.65* | 0.1 | 0.20 |
| Total Phosphorus | 0.11** | 0.30 | 0.41 | 0.5* | 0.33 | 0.03 |
| Soil Organic Carbon | −0.36 | −0.15 | −0.32 | −0.45 | 0.21 | −0.25 |
| Available Nitrogen | −0.26 | −0.01 | −0.12 | −0.25 | 0.3 | −0.25 |
| Available Phosphorus | −0.12 | 0.14 | 0.15 | 0.09 | 0.29 | −0.13 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Cong, J.; Cheng, J.; Qi, Q.; Sheng, Y.; Ning, D.; Lu, H.; Wyckoff, K.N.; Deng, Y.; Li, D.; et al. Soil Microbial Community Assembly and Interactions Are Constrained by Nitrogen and Phosphorus in Broadleaf Forests of Southern China. Forests 2020, 11, 285. https://doi.org/10.3390/f11030285
Zhao M, Cong J, Cheng J, Qi Q, Sheng Y, Ning D, Lu H, Wyckoff KN, Deng Y, Li D, et al. Soil Microbial Community Assembly and Interactions Are Constrained by Nitrogen and Phosphorus in Broadleaf Forests of Southern China. Forests. 2020; 11(3):285. https://doi.org/10.3390/f11030285
Chicago/Turabian StyleZhao, Mengxin, Jing Cong, Jingmin Cheng, Qi Qi, Yuyu Sheng, Daliang Ning, Hui Lu, Kristen N. Wyckoff, Ye Deng, Diqiang Li, and et al. 2020. "Soil Microbial Community Assembly and Interactions Are Constrained by Nitrogen and Phosphorus in Broadleaf Forests of Southern China" Forests 11, no. 3: 285. https://doi.org/10.3390/f11030285
APA StyleZhao, M., Cong, J., Cheng, J., Qi, Q., Sheng, Y., Ning, D., Lu, H., Wyckoff, K. N., Deng, Y., Li, D., Zhou, J., & Zhang, Y. (2020). Soil Microbial Community Assembly and Interactions Are Constrained by Nitrogen and Phosphorus in Broadleaf Forests of Southern China. Forests, 11(3), 285. https://doi.org/10.3390/f11030285






