Abstract
Subtropical and tropical broadleaf forests play important roles in conserving biodiversity and regulating global carbon cycle. Nonetheless, knowledge about soil microbial diversity, community composition, turnover and microbial functional structure in sub- and tropical broadleaf forests is scarce. In this study, high-throughput sequencing was used to profile soil microbial community composition, and a micro-array GeoChip 5.0 was used to profile microbial functional gene distribution in four sub- and tropical broadleaf forests (HS, MES, HP and JFL) in southern China. The results showed that soil microbial community compositions differed dramatically among all of four forests. Soil microbial diversities in JFL were the lowest (5.81–5.99) and significantly different from those in the other three forests (6.22–6.39). Furthermore, microbial functional gene interactions were the most complex and closest, likely in reflection to stress associated with the lowest nitrogen and phosphorus contents in JFL. In support of the importance of environmental selection, we found selection (78–96%) dominated microbial community assembly, which was verified by partial Mantel tests showing significant correlations between soil phosphorus and nitrogen content and microbial community composition. Taken together, these results indicate that nitrogen and phosphorus are pivotal in shaping soil microbial communities in sub- and tropical broadleaf forests in southern China. Changes in soil nitrogen and phosphorus, in response to plant growth and decomposition, will therefore have significant changes in both microbial community assembly and interaction.
1. Background
Forests play important roles in biodiversity conservation, providing a variety of critical resources and ecosystem services to humans, and generally serve as carbon sinks [1]. Subtropical and tropical forests, which contain up to 55% of global carbon stocks, dominate southern China and have favorable climatic conditions with regard to temperature, light and water [1]. Due to these advantageous climatic conditions, the region has high biodiversity in plants and animals [2]. Soil microbial information in subtropical and tropical forests of southern China, however, is relatively scarce, despite their important ecological functions in driving biogeochemical elemental cycles [3].
Soil microbes in forests are significant contributors to global carbon and nitrogen cycles due to their high metabolic activity and large populations of microorganisms [4]. Therefore, a deep analysis of soil microbial communities and their roles in ecological processes would improve our understanding of biodiversity as well as elemental biogeochemical cycles [5]. Soil microbial community composition and diversity in sub- and tropical forests are subjected to influence from land-use changes [6,7]. Microbial functional traits and groups—such as functional gene diversity, composition and abundance, and aerobic ammonia-oxidizing communities—in the Amazon Rainforest were also altered by land-use changes [8,9]. Therefore, it is a crucial to conduct a baseline study to document soil microbial biogeography in sub- and tropical forests, which is a prerequisite for understanding microbial responses to environmental changes and their ecological consequences.
Microbial biogeographic patterns are maintained using community assembly processes [10]. Vellend [11] has proposed four processes—selection, drift, dispersal, and mutation/diversification in shaping microbial communities. The selection process, also referred to as deterministic process or niche process, has a considerable influence in shaping microbial community distribution [12,13]. Selection determines the microbial community via abiotic factors such as various environmental impacts, including soil pH, carbon, and nitrogen [14,15], and biotic factors such as commensalism, mutualism, and parasitism [12]. In contrast, drift refers to stochastic changes of species abundance, and it is the most important process in shaping of microbial community distribution when the selection process is weak [16]. Species diversity among communities is influenced by dispersal, which determines species movement and colonization of a new location [10]. Microorganisms can evolve through diversification or mutation, particularly in inhospitable environments [12]. However, it remains unclear how deterministic and stochastic processes collectively shape microbial community distributions in broadleaf forests of southern China.
This study aimed to identify the soil microbial taxonomic composition through 16S rRNA sequencing and functional gene structure through GeoChip array in a four broadleaf forest of China. Given that soil nitrogen and phosphorus contents are important factors that affect vegetation diversity/recovery in southern China [17], we hypothesize that soil microbial diversities in forests with low nitrogen and phosphorus levels are low. As a consequence, the corresponding soil microbial networks are more complex, with the microbes in these networks more connected than those in the other networks, and microbes are closer to each other due to stronger pressure from soil nutrient limitations. We performed microbial community assembly analysis to examine whether environmental selection was important for microbial community assembly, and partial Mantel tests to identify the major environmental factors associated with differences in microbial communities.
2. Materials and Methods
2.1. Site Description and Soil Sample Collection
This study utilized four natural broadleaf forests in national natural reserves in China. The four forest reserves are located in Huangsang (HS), Hunan Province (110°04′ E and 26°23′ N) with a subtropical climate, Maoershan (MES), Guangxi Province (110°28′ E and 25°54′ N) with a subtropical climate, Huaping (HP), Guangxi Province (109°56′ E and 25°33′ N) with a subtropical climate, and Jianfengling (JFL), Hainan Province (108°53′ E and 18°43′ N) with a tropical climate. In 2012, Soil samples of HS, MES, and HP were collected from ten replicate plots per forest, and JFL soil samples were collected in three locations with 10 plots per location. Soil plots were 20 × 20 m with about 20 m between two adjacent plots. At each plot, ten to fifteen surface soil cores (0–10 cm) with over 1 m distance from each other were taken and mixed to form one sample after the removal of the litter layer. Soil was sieved through 2-mm mesh and then stored at −80 °C for DNA extraction or at 4 °C for soil chemistry characteristic analyses.
2.2. Plant and Environmental Variables Measurements
Recordings of species and richness of tree and shrub communities were taken in each plot. Arbor was chosen if plant breast diameter was over 5 cm and height was over 1.3 m, and shrub was chosen if breast diameter was less than 5 cm.
We collected annual average temperatures and annual mean precipitations through the WorldClim global climate dataset and then analyzed the data with ArcGIS version 9.3 (ESRI, Redlands, USA). Soil slurry pH measurements required a pH meter (Mettler Toledo Instruments, Shanghai, China). Soil environmental variables were measured by traditional methods [18]. In brief, soil organic carbon (SOC) was determined by potassium dichromate-outside heating method with ferrous sulfate titrating extra potassium dichromate, total nitrogen (TN) by Kjeldahl digestion, and available nitrogen (AN) by the Illinois Soil Nitrogen Test (ISNT) diffusion method. Total potassium (TK) and total phosphorus (TP) were digested with microwave, and available phosphorus (AP) was extracted with sodium bicarbonate. Then TK, TP and AP were determined by an inductively coupled plasma optical emission spectrometer (ICP-OES, Optima 5300DV, PerkinElmer, Manhattan, USA).
2.3. Soil Microbial DNA Extraction, Purification and Quantitation
Soil microbial DNA was extracted using a freeze-grinding method from 5 g soil as previously described [19]. Crude DNA was purified by agarose gel electrophoresis and quantified by a FLUOstar Optima microplate reader (BMG Labtech, Jena, Germany) [20].
2.4. Illumina Sequencing and GeoChip Experiments and Raw Data Processing
We targeted the V4 region of microbial 16S rRNA genes using 515F (5’-GTGCCAGCMGCCGCGGTAA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) primers [21]. The PCR processes and conditions previously described by Zhao et al. [22] were followed. The purified DNA library was sequenced on an Illumina MiSeq platform (Illumina, San Diego, USA) [23]. The Galaxy pipeline (http://zhoulab5.rccc.ou.edu:8080/root/login?redirect=%2Froot) was used for data processing as previously described [22]. Low-quality sequences were discarded, and remaining sequences were trimmed to ~250 base pairs. The UCHIME method was used to remove chimeric sequences [24]. Sequences were then classified into operational taxonomic units (OTUs) with 97% similarity, and taxonomic information was assigned using the RDP 16S rRNA gene classifier [25]. A total of 23,854 sequences were rarefied per sample for downstream analyses.
GeoChip 5.0 (https://www.glomics.com/gch-tech.html), which contained more than 57,000 oligonucleotide probes from 393 genes at the time of use, was used to determine soil microbial functional gene structure [26]. The purified soil microbial DNA was labelled with Cy 3 and hybridized with GeoChip 5.0 in an Agilent hybridization oven (Agilent Technologies Inc., Santa Clara, USA) at 67 °C for 24 hours [27]. Arrays were scanned with a NimbleGen MS200 Microarray Scanner (Roche NimbleGen, Inc., Madison, WI, USA), further using an Agilent Feature Extraction program for image extraction. The signal intensity was normalized by dividing the total signal intensity of a sample, and then multiplying a constant and logarithmic transformation [28].
2.5. Statistical Analyses
Normalized data from Illumina sequencing and GeoChip technologies were used for statistical analyses. Microbial α-diversity utilized the Shannon-Weaver index, followed by calculations of differences in microbial community structure using the multiple response permutation procedure (MRPP). Non-metric multidimensional scaling (NMDS) analyses were performed using the Bray-Curtis dissimilarity to examine microbial distribution patterns [29]. Partial Mantel tests assisted the examination of correlations between environmental variables and microbial community compositions [30]. All aforementioned analyses were completed using the “vegan” package in R software [31].
A method developed by Stegen et al. [32] was used to quantify the relative importance of selection, drift and dispersal on microbial community assembly. The abundance-weighted β-mean-nearest taxon distance (βMNTD) was used to quantify the phylogenetic distance between microbial communities. This test also generated the observed βMNTD (βMNTDobs) and a null distribution βMNTD (βMNTDnull). The β-nearest taxon index (βNTI) gives the magnitude of deviation between βMNTDobs and βMNTDnull. A value of | βNTI | > 2 indicates that selection governs the community assembly between pairwise microbial communities. The Bray-Curtis-based Raup-Crick (RCbray) index, which is the magnitude of deviation between observed Bray-Curtis and expected Bray-Curtis, is introduced when | βNTI | < 2. In the context of | βNTI | < 2, a value of RCbray > +0.95 indicates that observed turnover is governed by dispersal limitation acting alongside Drift. RCbray < -0.95 indicates the influence of homogenizing dispersal, and | RCbray | < 0.95 indicates the influence of drift acting alone. Analyses were completed using the “picante” package in R.
Functional molecular ecological networks (MEN) were analyzed with a subset of GeoChip data using the Molecular Ecological Network Analyses Pipeline (http://129.15.40.240/MENA/main.cgi) [33]. A random matrix theory (RMT)-based conceptual framework was used to make sure the network was automatically defined and robust to noise.
3. Results
3.1. Diversity and Similarity of Soil Microbial Communities
High-throughput sequencing and microarray approaches were employed to assess soil microbial taxonomic composition and functional gene structure, resulting in a total of 1,438,800 sequences and 31,937 microbial functional genes in all samples, respectively. Soil taxonomic diversities in JFL were the lowest (5.81–5.99), and significantly (P < 0.05) different from those in any other forests (6.22–6.39) (Table 1). Similarly, soil microbial functional gene diversities were the lowest in JFL (9.90–10.16).

Table 1.
Soil microbial Shannon diversities at different sites.
MRPP analyses indicated that soil taxonomic compositions and functional structures were significantly (P < 0.05) different among all of four forests (Table 2). Results of Non-metric Multidimensional Scaling (NMDS) showed that soil bacterial communities were clustered by individual forests (Figure S1).

Table 2.
Analysis of soil microbial similarity (multiple response permutation procedure, MRPP) based on the Bray-Curtis index between two study sites. Pairwise comparisons between soil taxonomic compositions and microbial functional structures.
3.2. Soil Microbial Taxonomic Distribution
The relative abundance of Proteobacteria, Acidobacteria, Verrucomicrobia, and Actinobacteria accounted for 83.5–88.4% of the total abundance in the four forests (Figure S2). The relative abundance of Acidobacteria was the most abundant in the HS (35.1%) and MES (39.4%) soils. Proteobacteria in JFL was the most abundant (44.2%), which was significantly (P < 0.01) higher than those in the other forests (33.4–38.7%). On the contrary, the relative abundance of Firmicutes in JFL (0.7%) was about half of that in the other forests (1.1–1.9%).
A total of 372 genera were detected. The relative abundance of 49 genera were significantly (P < 0.05) higher in JFL than those in the other forests, of which 62.5% were from Proteobacteria. For example, Rhodomicrobium, Afipia and Nevskia from Proteobacteria were 1.5–4.3 times greater in JFL than those in the other forests. In contrast, 66 genera were significantly lower in relative abundance in JFL than those in the other forests. About 19% of the 66 genera were from the phyla Acidobacteria, Chloroflexi, Gemmatimonadetes or Verrucomicrobia. For example, subgroups 6, 7, 10, 14, 25 in Acidobacteria were 2.7–9.0 times smaller in JFL those in the other forests.
3.3. Functional Genes Relevant to Nitrogen and Carbon Cycling
The relative abundance of nitrogen cycling genes was lowest in JFL (24241) and significantly (P < 0.05) different from those in the other forests (26132–27855). Nearly all genes—including the nifH gene-encoding nitrogenase, the amoA gene-encoding ammonia monooxygenase, and the gdh gene-encoding glutamate dehydrogenase—displayed lower intensities in JFL (Figure S3A). The only exception was the functional gene encoding nitrite reductase derived from protist, which was of a similar intensity across the four forests.
The total signal intensities of carbon cycling genes were also significantly (P = 0.001) lower in JFL (80010) than those in the other forests (85944–91500). Most carbon cycling genes (~85%) in JFL were more than 0.1 times lower in intensity than those in the other forests (Figure S3B). Such examples include the amyA gene-encoding alpha amylase (0.10 times lower in JFL), the chitinase encoding gene (0.11 times lower in JFL), the rubisco gene encoding carboxylase (0.10 times lower in JFL), and the mcrA gene encoding methyl coenzyme M reductase (0.13 times lower in JFL). These results showed the signal intensities of many functional genes involved in carbon and nitrogen cycling were significantly lower in JFL than in the three forests, and different microbial metabolic activities might be happening.
3.4. Molecular Ecological Networks Aanalysis of Functional Genes
Functional molecular ecological networks (MENs) construction used eight typical nitrogen or carbon cycling genes. All of the networks showed topological features similar to those of complex systems, including modular (higher modularity of empirical networks than corresponding random networks, Table S1), and scale free (R2 of power law = 0.70–0.97, indicting degree distributions fitting the power law model well, Table 3). Average Clustering Coefficients (avgCC) was the highest in JFL (0.17–0.34), and especially prominent for the nirK, nosZ, xylanase genes. These results indicated that network interactions for most functional genes became more complex in JFL soils than in the other forest soils. On the contrary, the Average Path (GD) of JFL (3.30–8.79) was smaller than those of the other forests, suggesting that functional genes were more closely related with each other in JFL soils. Two MENs on behalf of the nifH gene were visualized using soil samples from HS and JFL, which revealed a more complex and closer MEN of JFL (Figure 1).

Table 3.
Soil microbial functional gene network properties. R2 of Power-law (R2) indicates how well a curve of network connectivity distribution is fitted with the power-law model. A higher R2 means a scale-free network. Network size indicates the number of nodes in a network. Modularity indicates how well a network can be separated into modules. Average connectivity (avgK) indicates the average connection strength between pairwise nodes. Higher avgK means a more complex network. Average Clustering coefficient (avgCC) is the average clustering coefficient of all nodes. Average path (GD) is the average shortest path between pairwise nodes. Smaller GD indicates that the nodes in the network are closer.
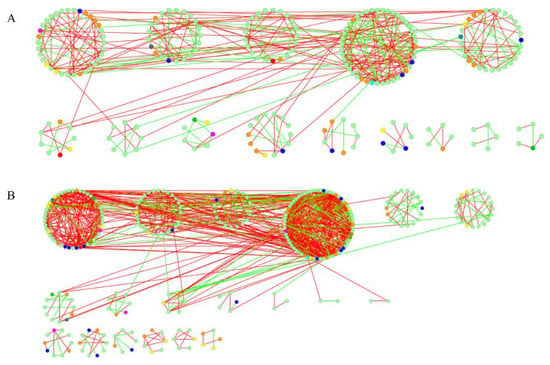
Figure 1.
Networks constructed by the RMT-based approach of nifH genes in GeoChip data in HS (A) and JFL (B) forests. Each node indicates a functional gene. Green lines indicate negative interactions between two individual nodes, while red lines indicate positive interactions.
Correlations calculated between environmental variables (Table S2) and main MEN properties were calculated (Table 4). Temperature was the only detected variable that had a significant (r = 0.62, P < 0.05) influence on the modularity of MENs. Temperature also had a significant influence on avgK (r = 0.63, P < 0.01) and avgCC (r = 0.58, P < 0.05). Soil moisture, TK, and TS also played considerable roles on avgK (r > 0.55, P < 0.05) and avgCC (r > 0.56, P < 0.05).

Table 4.
Partial mantel tests to determine relationships between environmental variables and soil microbial communities or microbial ecological network properties.
3.5. Soil Microbial Community Assembly Processes and Quantitative Spatial Turnover
Drift, selection, dispersal limitation and homogenizing dispersal were determined using a framework to quantify community assembly processes in governing spatial turnover of soil microbial communities. We found that about 67% of turnover in community composition was due to selection, 0–70% of turnover was due to dispersal limitation acting alongside drift, 0–22% of turnover was due to homogenizing dispersal, and 0-8% of turnover was due to drift acting alone (Figure 2).
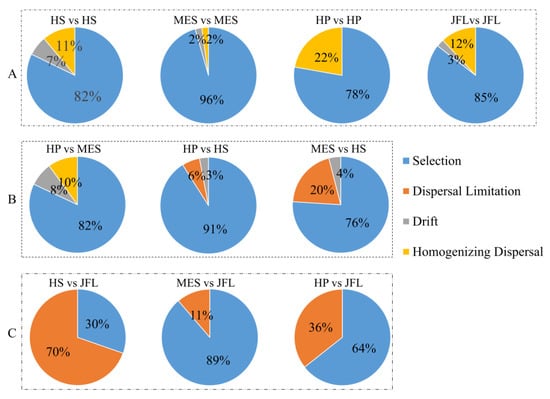
Figure 2.
Quantitative spatial turnover of soil microbial community compositions (A) within each forest, (B) between subtropical forests and (C) between subtropical and tropical forests. The percent of turnover in community compositions is governed by selection (blue fill), dispersal limitation (orange fill), drift (gray fill), and homogenizing dispersal (yellow fill).
Homogenizing dispersal had a substantial influence over community compositions within each forest soil (~2–22%) (Figure 2A); but it had little influence over community assembly between the three subtropical forests and JFL forest (0%) (Figure 2C). In contrast, dispersal limitation had a detectable influence over community assembly between the three subtropical forests and JFL soils (11–70%) (Figure 2C).
Partial Mantel tests were used to identify major environmental variables in shaping microbial communities (Table 4). Soil pH was significantly and most (r > 0.62, P < 0.5) correlated with soil microbial communities in HS. SOC and AN were significantly correlated (r > 0.31, P < 0.05) with soil microbial communities in MES. TN and TP were significantly correlated (r > 0.40, P < 0.05) with soil microbial communities in HP.
4. Discussion
Soil microbial taxonomic compositions and functional gene structures involved in carbon and nitrogen cycling in four broadleaf forests in China were determined. Initial hypotheses that soil microbial diversities would be lower in the forest with low nitrogen and phosphorus contents were supported by taxonomic composition data as well as functional gene analysis (Table 1). Environmental selection was found to dominate the shape of microbial community compositions, but dispersal limitation had a substantial influence over community composition variations of JFL and the other three forests (Figure 2). Soil microbial functional gene networks of JFL were distinctly more complex and functional genes were more closely related with each other than those of the other three forests (Table 3).
Tropical forest soils lose tremendous quantities of nitrogen to water in dissolved organic forms [34]. N2O and NO are lost to the atmosphere, while as is lost to stream water [35]. TN and AN in the JFL soils therefore were found to be nearly one third of those in the subtropical forest soils (HP, MES and HS) (Table S2), implying that JFL soils might be nitrogen limited. At the same time, environmental stresses, such as nutrient limitation, pollution, etc. can reduce microbial diversity [36]. Therefore, the relative abundance of nitrogen cycling genes in this study was lowest in JFL, which implies that nitrogen mineralization, nitrification and denitrification potential were lowest.
Phosphorus is limited in lowland tropical forest soils [37]. The JFL forest is a typical lowland tropical forest, which is low in phosphorus content (21.8% of subtropical soil TP and 41.2% of subtropical soil AP, Table S2). Therefore, the JFL forest faces both nitrogen and phosphorus limitation. TP was determined to be a significant driver of soil microbial communities, and it also significantly affected avgCC of nifH gene networks (Table 4). Lower SOC was detected in JFL (70.5% of SOC in the other three forests). It is well documented that soil conditions such as soil pH, nitrogen, phosphorus, potassium and soil organic matter are the foremost determinants of soil microbial diversities [38,39]. Although tropical forests have the most diverse animal and plant populations on Earth [40], relatively lower bacterial and fungal diversities were found in the rainforests of North East Ecuador compared with those in more temperate regions [3,41]. Similarly, observations showed the lowest soil microbial diversities in the JFL forest (Table 1), owing to the low nutrition availability in JFL (Table S2).
Selection induces differential in survival and reproductive success across individuals and species, and then constrains and differentiates microbial community composition among locations [10,32]. Selection is caused by biotic pressures like competition, predation, and mutualism among species [10], and abiotic pressures like environmental physical and chemical properties [39]. Selection often has detectable influence over microbial community assembly [32,42,43,44]. For example, Stegen et al. [32] found that selection governed about 60% of microbial community assembly, which was imposed by an unmeasured environmental variable in deeper sediments in an aquifer of Washington State. Quantitative results from this study further support the idea that selection is dominant in driving microbial community assembly, as selection comprises up to 30–96% of processes.
Dispersal leads to homogenization among communities; however, dispersal limitation constrains the movement and colonization of organisms to new locations [32]. High levels of dispersal limitation lead to spatial turnover in microbial communities, which plays a significant role in community assembly, as is shown by a growing body of literature [32,45]. This study found that dispersal limitation was distinctly larger in governing spatial turnover of soil microbial communities between the JFL forest and the other three forests (11–70%), than those among the other three forests (0–20%) (Figure 2). The spatial turnover can be largely attributed to the different distances between the JFL forest and the others. This result challenges the classic dictum that "everything is everywhere but the environment selects" by constraining microbial dispersal to long-distance locations.
MENs make it possible to compare architectural patterns of microbial communities and to explore latent mechanisms of the stability and resilience of communities [46]. In this study, we found that the functional MENs of the JFL forest were more complex, and functional genes were more closely related with each other than those of the other three soils (Table 3, Figure 1). It has been well documented that closely connected communities are less resilient and thus more susceptible to disturbance [47,48], so soil microbial communities in JFL soils would have a higher response rate to perturbations.
In conclusion, comprehensive surveys of microbial taxonomic compositions and carbon/nitrogen cycling genes in sub- and tropical broadleaf forest soils found the lowest microbial diversities and the most complex functional potential interactions in the JFL forest soils. The distinct microbial compositions can be attributed to the constraint of nitrogen and phosphorus contents. These results suggest that nitrogen and phosphorus have a significant influence on not only microbial community diversities and compositions, but also functional gene interactions. This study provides a crucial baseline of soil microbial biogeography in sub- and tropical forests, which is a prerequisite for understanding their ecological consequences.
Availability of data and materials
The high-throughput sequencing data datasets generated and/or analyzed during the current study are available in the GenBank databases by the accession number SRP062748 (http://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP062748). Microarray data can be found at NCBI’s Gene Expression Omnibus by the accession number GSE69171 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69171).
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/3/285/s1, Table S1: Major topological properties of the empirical and random MENs of nifH genes in four forests, Table S2: Plant and environmental variables in subtropical and tropical forest soils. Values are shown as mean ± standard deviation (SD), Figure S1: Nonmetric multidimensional scaling of soil microbial taxonomic compositions (A) and functional structures (B) to examine microbial distribution patterns, Figure S2: Relative abundance of main phyla in four forests, Figure S3: Relative abundance (total signal intensities of functional genes) of (A) nitrogen cycling genes and (B) part of carbon cycling genes in GeoChip data.
Author Contributions
M.Z. analyzed data and wrote paper. J.C. (Jing Cong), H.L., and D.L. collected soil samples. J.C. (Jing Cong) and Y.S. contributed to DNA extraction and plant and environmental variables measurements. D.N. implemented the R code. J.C. (Jingmin Cheng) and Q.Q. contributed to final revision of the manuscript. K.N.W. polished the language. Y.D. contributed to development and design of methodology. J.Z. contributed to formulation of overarching research goals and aims. Y.Z. designed the experiment and supervised all work. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Nature Science Foundation of China (No. 31670614, No.31370145), the Public Welfare Project of the National Scientific Research Institution (CAFRIFEEP201101), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB15010302), and National Biological Specimens and Resources Sharing Platform in Nature Reserve (2005DKA21404).
Acknowledgments
We acknowledge all authors’ contributions to this work.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| SOM | soil organic matter; |
| TN | total nitrogen; |
| TK | total potassium, |
| TP | total phosphorus, |
| AN | available nitrogen, |
| AP | available phosphorus; |
| OUT | operational taxonomic units; |
| MRPP | multiple response permutation procedure; |
| NMDS | non-metric multidimensional scaling; |
| Βnti | β-nearest taxon index; |
| MEN | molecular ecological network; |
| RMT | random matrix theory; |
| avgCC | average clustering coefficients; |
| GD | average path; |
| avgK | average degree. |
References
- Pan, Y.; Birdsey, R.A.; Fang, J.; Houghton, R.; Kauppi, P.E.; Kurz, W.A.; Phillips, O.L.; Shvidenko, A.; Lewis, S.L.; Canadell, J.G.; et al. A large and persistent carbon sink in the world’s forests. Science 2011, 333, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Schluter, D.; Pennell, M.W. Speciation gradients and the distribution of biodiversity. Nature 2017, 546, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Sadam, A.; Zambrano, M.; Valencia, R.; Bahram, M. Low diversity and high host preference of ectomycorrhizal fungi in western Amazonia, a neotropical biodiversity hotspot. ISME J. 2010, 4, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Buée, M.; Deveau, A.; Mieszkin, S.; Martin, F. Ecology of the forest microbiome: Highlights of temperate and boreal ecosystems. Soil Biol. Biochem. 2016, 103, 471–488. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Krashevska, V.; Klarner, B.; Widyastuti, R.; Maraun, M.; Scheu, S. Impact of tropical lowland rainforest conversion into rubber and oil palm plantations on soil microbial communities. Biol. Fert. Soils 2015, 51, 697–705. [Google Scholar] [CrossRef]
- Sall, S.N.; Masse, D.; Ndour, N.Y.B.; Chotte, J.-L. Does cropping modify the decomposition function and the diversity of the soil microbial community of tropical fallow soil? Appl. Soil Ecol. 2006, 31, 211–219. [Google Scholar] [CrossRef]
- Hamaoui, G.S.; Rodrigues, J.L.M.; Bohannan, B.J.M.; Tiedje, J.M.; Nüsslein, K. Land-use change drives abundance and community structure alterations of thaumarchaeal ammonia oxidizers in tropical rainforest soils in Rondônia, Brazil. Appl. Soil Ecol. 2016, 107, 48–56. [Google Scholar] [CrossRef]
- Paula, F.S.; Rodrigues, J.L.; Zhou, J.; Wu, L.; Mueller, R.C.; Mirza, B.S.; Bohannan, B.J.; Nusslein, K.; Deng, Y.; Tiedje, J.M.; et al. Land use change alters functional gene diversity, composition and abundance in Amazon forest soil microbial communities. Mol. Ecol. 2014, 23, 2988–2999. [Google Scholar] [CrossRef]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B.H. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef]
- Vellend, M. Conceptual Synthesis in Community Ecology. Q. Rev. of Biol. 2010, 85, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Santoferrara, L.F.; Grattepanche, J.D.; Katz, L.A.; McManus, G.B. Patterns and processes in microbial biogeography: do molecules and morphologies give the same answers? ISME J. 2016, 10, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.M.; Myers, J.A. Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. B: Biol. Sci. 2011, 366, 2351–2363. [Google Scholar] [CrossRef]
- Long, W.; Yang, X.; Li, D. Patterns of species diversity and soil nutrients along a chronosequence of vegetation recovery in Hainan Island, South China. Ecol. Res. 2012, 27, 561–568. [Google Scholar] [CrossRef]
- Lu, R. Soil Agricultural Chemical Analysis Methods; Agricultural Sci-Tech. Press: Beijing, China, 1999; pp. 106–135. [Google Scholar]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef]
- Zhao, M.; Xue, K.; Wang, F.; Liu, S.; Bai, S.; Sun, B.; Zhou, J.; Yang, Y. Microbial mediation of biogeochemical cycles revealed by simulation of global changes with soil transplant and cropping. ISME J. 2014, 8, 2045–2055. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Zhao, M.; Sun, B.; Wu, L.; Gao, Q.; Wang, F.; Wen, C.; Wang, M.; Liang, Y.; Hale, L.; Zhou, J. Zonal Soil Type Determines Soil Microbial Responses to Maize Cropping and Fertilization. mSystems 2016, 1, e00075-16. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wen, C.; Qin, Y.; Yin, H.; Tu, Q.; Van Nostrand, J.D.; Yuan, T.; Yuan, M.; Deng, Y.; Zhou, J. Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol. 2015, 15, 125. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Van Nostrand, J.D.; Yin, H.; Wu, L.; Yuan, T.; Zhou, J. Hybridization of Environmental Microbial Community Nucleic Acids by GeoChip. Methods Mol. Biol. 2016, 1399, 183–196. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Liu, D.; Wu, H.; Lu, X.; Fang, Y.; Cheng, W.; Luo, W.; Jiang, P.; Shi, J.; et al. Aridity threshold in controlling ecosystem nitrogen cycling in arid and semi-arid grasslands. Nat. Commun. 2014, 5, 4799. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, Y.; Wang, S.; Xu, D.; Yu, H.; Wu, L.; Lin, Q.; Hu, Y.; Li, X.; He, Z.; et al. The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 2014, 8, 430–440. [Google Scholar] [CrossRef]
- Kruskal, J.B. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 1964, 29, 1–27. [Google Scholar] [CrossRef]
- Smouse, P.E.; Long, J.C.; Sokal, R.R. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 1986, 35, 627–632. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2016. [Google Scholar]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Tu, Q.; Zhi, X. Functional molecular ecological networks. mBio 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Houlton, B.Z.; Marklein, A.R.; Bai, E. Representation of nitrogen in climate change forecasts. Nat. Clim. Chang. 2015, 5, 398–401. [Google Scholar] [CrossRef]
- Hall, S.J.; Matson, P.A. Nitrogen oxide emissions after nitrogen additions in tropical forests. Nature 1999, 400, 152–155. [Google Scholar] [CrossRef]
- Schimel, J. Ecosystem consequences of microbial diversity and community structure. In Arctic and Alpine Biodiversity: Patterns, Causes and Ecosystem Consequences; Chapin, F.S., Korner, C., Eds.; Springer: Berlin, Germany, 1995; pp. 239–254. [Google Scholar]
- Zemunik, G.; Davies, S.J.; Turner, B.L. Soil drivers of local-scale tree growth in a lowland tropical forest. Ecology 2018, 99, 2844–2852. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xia, B.; Treves, D.S.; Wu, L.Y.; Marsh, T.L.; O’Neill, R.V.; Palumbo, A.V.; Tiedje, J.M. Spatial and Resource Factors Influencing High Microbial Diversity in Soil. Appl. Environ. Microbiol. 2002, 68, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Lewis, S.L.; Lloyd, J.; Sitch, S.; Mitchard, E.T.; Laurance, W.F. Changing ecology of tropical forests: evidence and drivers. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 529–549. [Google Scholar] [CrossRef]
- Kim, M.; Kim, W.S.; Tripathi, B.M.; Adams, J. Distinct bacterial communities dominate tropical and temperate zone leaf litter. Microbiol. Ecol. 2014, 67, 837–848. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Bohannan, B.J.; Brown, J.H.; Colwell, R.K.; Fuhrman, J.A.; Green, J.L.; Horner-Devine, M.C.; Kane, M.; Krumins, J.A.; Kuske, C.R. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 4, 102–112. [Google Scholar] [CrossRef]
- Andersson, A.F.; Riemann, L.; Bertilsson, S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010, 4, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Martiny, J.B.; Eisen, J.A.; Penn, K.; Allison, S.D.; Horner-Devine, M.C. Drivers of bacterial beta-diversity depend on spatial scale. Proc. Natl. Acad. Sci. USA 2011, 108, 7850–7854. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, Y.; Wang, M.; Sun, X.; Cong, J.; Deng, Y.; Lu, H.; Yuan, T.; Van Nostrand, J.D.; Li, D.; et al. Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests. Mol. Ecol. 2015, 24, 5175–5185. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, S.; Stouffer, D.B.; Uzzi, B.; Bascompte, J. Strong contributors to network persistence are the most vulnerable to extinction. Nature 2011, 478, 233–235. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).