Soil Microbial Biomass and Community Composition Relates to Poplar Genotypes and Environmental Conditions
Abstract
1. Introduction
2. Materials and Methods
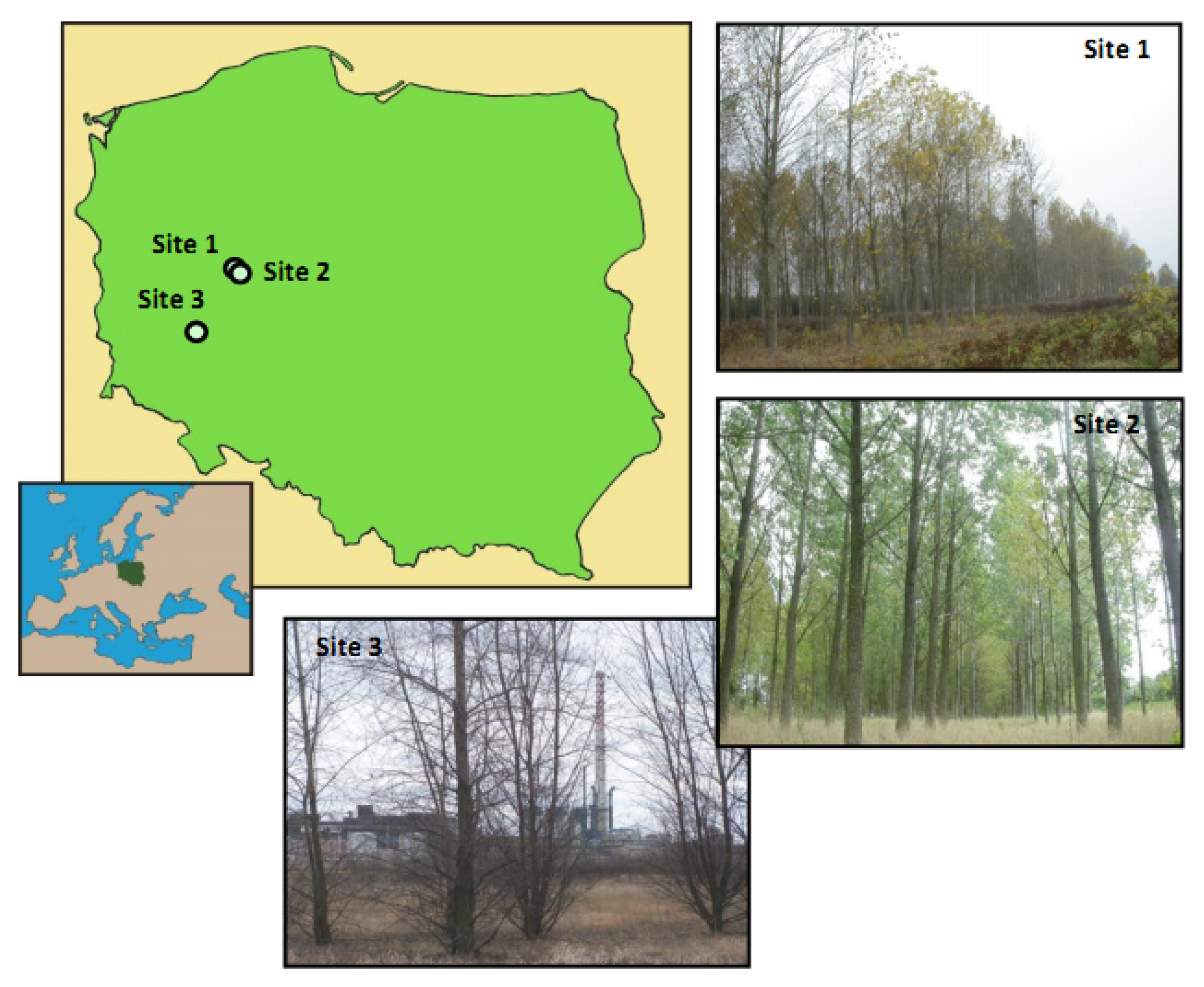
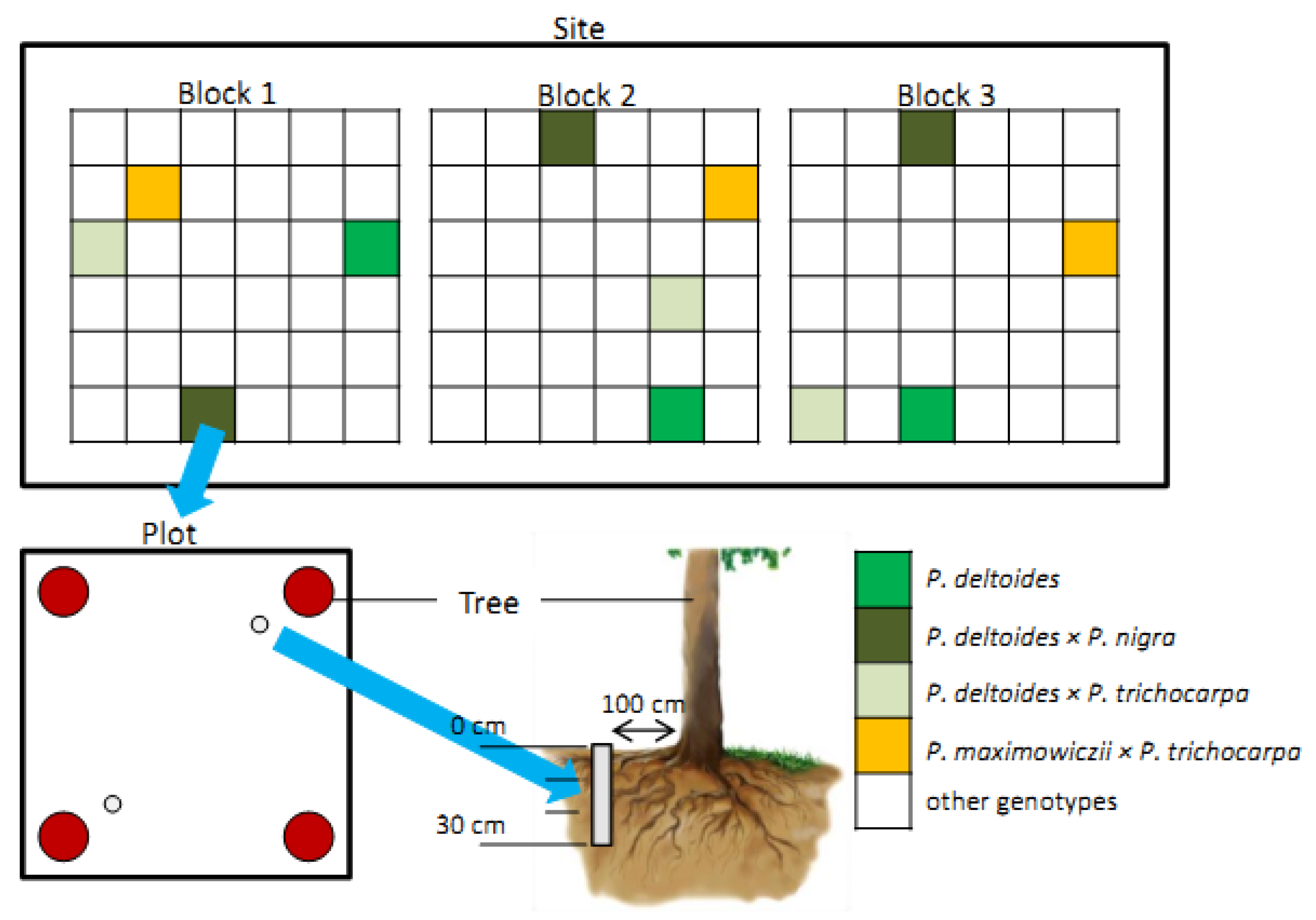
2.1. Plant Material and Study Sites
2.2. Sample Collection
2.3. Whole Cell Fatty Acid (WCFA) Analysis
2.4. Statistics
3. Results
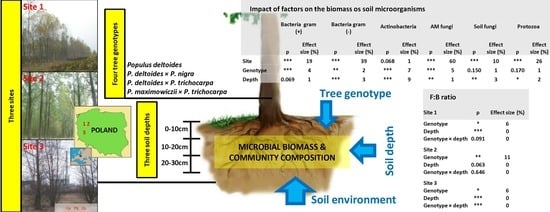
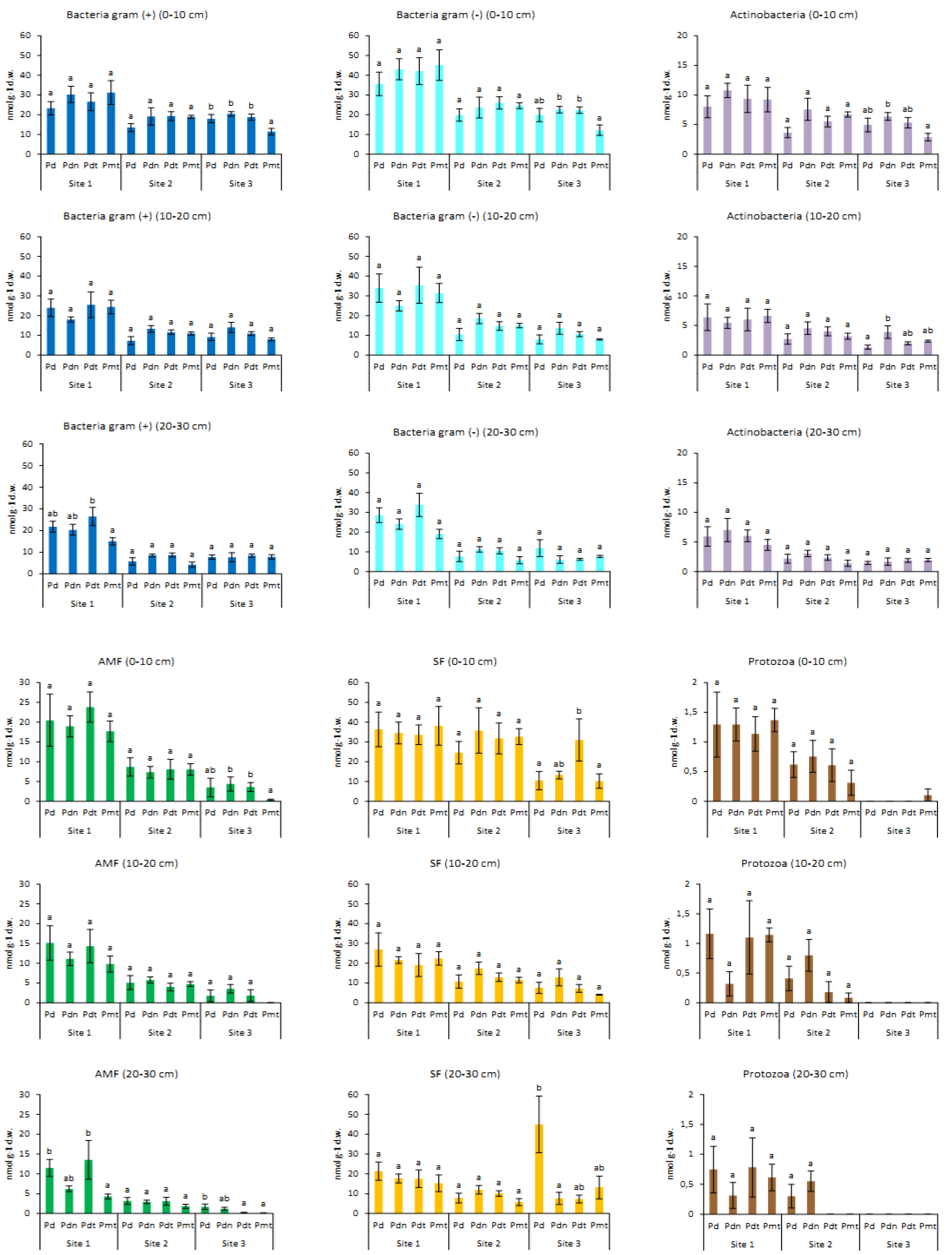
3.1. The Impact of Site, Poplar Genotype and Soil Depth on Biomass of Microorganisms
3.2. Biomass and Contribution of Microorganisms in Microbial Community
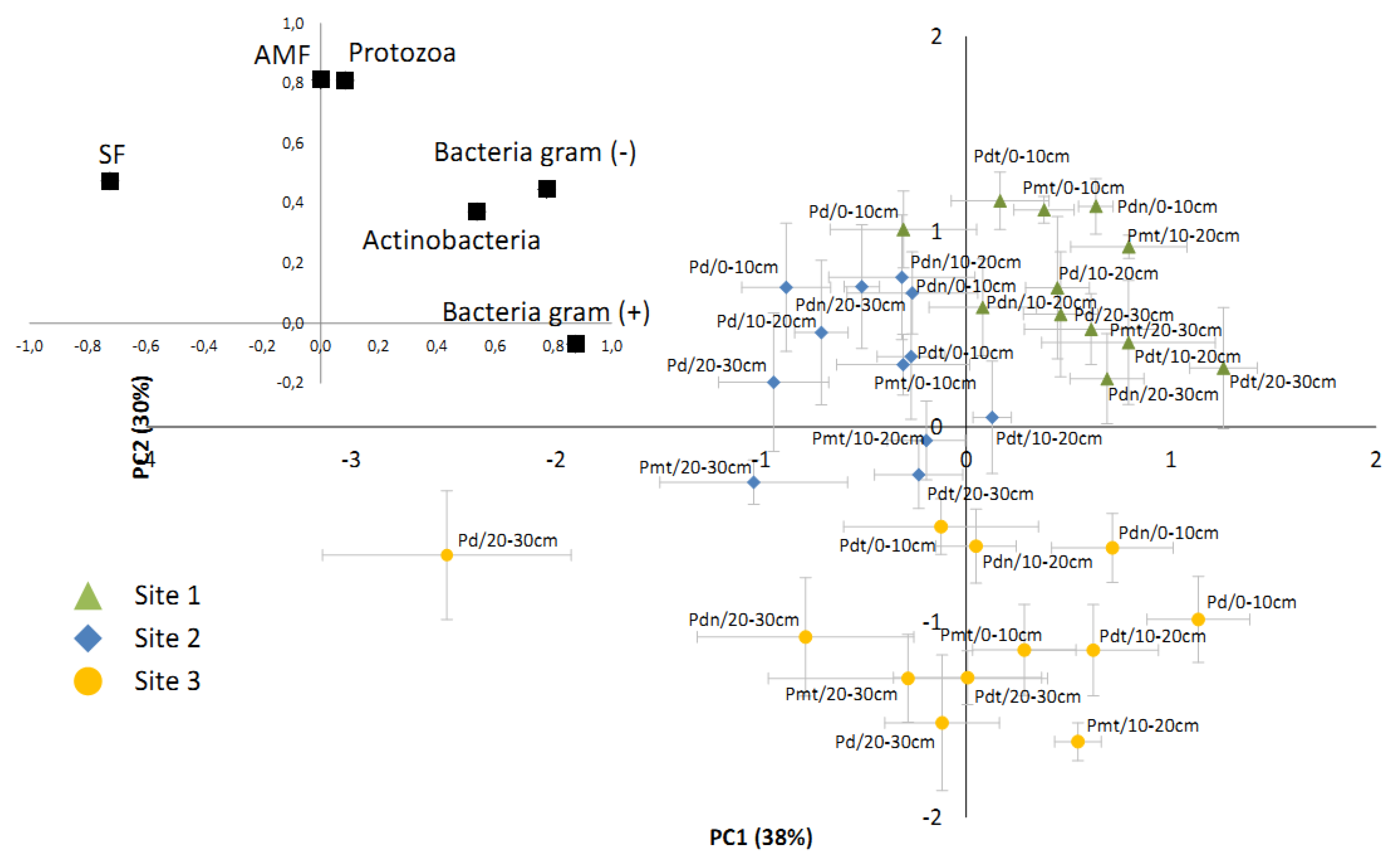
3.3. The Comparison of Microbial Communities
3.4. The Impact of Soil Factors of Microbial Community
4. Discussion
4.1. Host-Genotype Effect
4.2. Soil Depth Effect
4.3. Site Effect
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gottel, N.R.; Castro, H.F.; Kerley, M.; Yang, Z.; Pelletier, D.A.; Podar, M.; Karpinets, T.; Uberbacher, E.; Tuskan, G.A.; Vilgalys, R.; et al. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 2011, 77, 5934–5944. [Google Scholar] [CrossRef] [PubMed]
- Shakya, M.; Gottel, N.; Castro, H.; Yang, Z.K.; Gunter, L.; Labbe, J.; Muchero, W.; Bonito, G.; Vilgalys, R.; Tuskan, G.; et al. A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS ONE 2013, 8, e76382. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; Op De Beeck, M.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Chodak, M.; Gołębiewski, M.; Morawska-Płoskonka, J.; Kuduk, K.; Niklińska, M. Diversity of microorganisms from forest soils differently polluted with heavy metals. Appl. Soil Ecol. 2013, 64, 7–14. [Google Scholar] [CrossRef]
- Frey, B.; Stemmer, M.; Widmer, F.; Luster, J.; Sperisen, C. Microbial activity and community structure of a soil after heavy metal contamination in a model forest ecosystem. Soil Biol. Biochem. 2006, 38, 1745–1756. [Google Scholar] [CrossRef]
- Peacock, A.D.; Macnaughton, S.J.; Cantu, J.M.; Dale, V.H.; White, D.C. Soil microbial biomass and community composition along an anthropogenic disturbance gradient within a long-leaf pine habitat. Ecol. Indic. 2001, 1, 113–121. [Google Scholar] [CrossRef]
- Van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Stephen, J.R.; Chang, Y.J.; Gan, Y.D.; Peacock, A.D.; Pfiffner, S.M.; Barcelona, M.J.; White, D.C.; Macnaughton, S.J. Microbial characterization of a JP-4 fuel-contaminated site using a combined lipid biomarker/polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE)-based approach. Environ. Microbiol. 1999, 1, 231–241. [Google Scholar] [CrossRef]
- Zogg, G.P.; Zak, D.R.; Ringelberg, D.B.; Macdonald, N.W.; Pregitzer, K.S.; White, D.C. Compositional and functional shifts in microbial communities due to soil warming. Soil Sci. Soc. Am. J. 1997, 61, 475–481. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Droge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Lottmann, J.; O’Callaghan, M.; Baird, D.; Walter, C. Bacterial and fungal communities in the rhizosphere of field-grown genetically modified pine trees (Pinus radiata D.). Environ. Biosaf. Res. 2010, 9, 25–40. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schweitzer, J.A.; Bailey, J.K.; Fischer, D.G.; LeRoy, C.J.; Lonsdorf, E.V.; Whitham, T.G.; Hart, S.C. Plant-soil-microorganism interactions: Heritable relationship between plant genotype and associated soil microorganisms. Ecology 2008, 89, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Ellouze, W.; Hamelb, C.; Vujanovic, V.; Ganb, Y.; Bouzid, S.; St-Arnaud, M. Chickpea genotypes shape the soil microbiome and affect the establishment of the subsequent durum wheat crop in the semiarid North American Great Plains. Soil Biol. Biochem. 2013, 63, 129–141. [Google Scholar] [CrossRef]
- Ravnskov, S.; Larsen, J. Functional compatibility in cucumber mycorrhizas in terms of plant growth performance and foliar nutrient composition. Plant Biol. 2016, 18, 816–823. [Google Scholar] [CrossRef]
- Gamalero, E.; Cesaro, P.; Cicatelli, A.; Todeschini, V.; Musso, C.; Castiglione, S.; Fabiani, A.; Lingua, G. Poplar clones of different sizes, grown on a heavy metal polluted site, are associated with microbial populations of varying composition. Sci. Tot. Environ. 2012, 425, 262–270. [Google Scholar] [CrossRef]
- Fernández-González, A.J.; Villadas, P.J.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Belaj, A.; Mercado-Blanco, J.; Fernández-López, M. Defning the root endosphere and rhizosphere microbiomes from the World Olive Germplasm Collection. Sci. Rep. 2019, 9, 20423. [Google Scholar] [CrossRef]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.J.; Vilgalys, G.A.; Tuskan, G.A.; Schadt, C.W. The Populus holobiont: Dissecting the effect of plant niches and genotype on the microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef]
- Bonito, G.; Benucci, G.M.N.; Hameed, K.; Weighill, D.; Jones, P.; Chen, K.-H.; Jacobson, D.; Schadt, C.; Vilgalys, R. Fungal-bacterial networks in the Populus rhizobiome are impacted by soil properties and host genotype. Front. Microbiol. 2019, 10, 481. [Google Scholar] [CrossRef]
- Bálint, M.; Tiffin, P.; Hallström, B.; O’Hara, R.B.; Olson, M.S.; Fankhauser, J.D.; Piepenbring, M.; Schmitt, I. Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera). PLoS ONE 2013, 8, e53987. [Google Scholar] [CrossRef]
- Corredor, A.H.; Van Rees, K.; Vujanovic, V. Host genotype and health status influence on the composition of the arbuscular mycorrhizal fungi in Salix bioenergy plantations. For. Ecol. Manag. 2014, 314, 112–119. [Google Scholar] [CrossRef]
- Gehring, C.A.; Mueller, C.; Whitham, T.G. Environmental and genetic effects on the formation of ectomycorrhizal and arbuscular mycorrhizal associations in cottonwoods. Oecologia 2006, 149, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Gherghel, F.; Behringer, D.; Haubrich, S.; Schlauß, M.; Fey-Wagner, C.; Rexer, K.-H.; Janßen, A.; Kost, G. Former land use and host genotype influence the mycorrhizal colonization of poplar roots. Forests 2014, 5, 2980–2995. [Google Scholar] [CrossRef]
- Karliński, L.; Rudawska, M.; Kieliszewska-Rokicka, B.; Leski, T. Relationship between genotype and soil environment during colonization of poplar roots by mycorrhizal and endophytic fungi. Mycorrhiza 2010, 20, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Karliński, L.; Rudawska, M.; Leski, T. The influence of host genotype and soil conditions on ectomycorrhizal community of poplar clones. Eur. J. Soil Biol. 2013, 58, 51–58. [Google Scholar] [CrossRef]
- Tagu, D.; Rampant, P.F.; Lapeyrie, F.; Frey-Klett, P.; Vion, P. Variation in the ability to form ectomycorrhizas in the F1progeny of an interspecific poplar (Populus spp.) cross. Mycorrhiza 2001, 10, 237–240. [Google Scholar] [CrossRef]
- Bakker, M.G.; Manter, D.K.; Sheflin, A.M.; Weir, T.L.; Vivanco, J.M. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 2012, 360, 1–13. [Google Scholar] [CrossRef]
- Langer, I.; Santner, J.; Krpata, D.; Fitz, W.J.; Wenzel, W.W.; Schweiger, P.F. Ectomycorrhizal impact on Zn accumulation of Populus tremula L. grown in metalliferous soil with increasing levels of Zn concentration. Plant Soil 2012, 355, 283–297. [Google Scholar] [CrossRef]
- Sebastiani, L.; Scebba, F.; Tognetti, R. Heavy metal accumulation and growth responses in poplar clones Eridano (Populus deltoides × maximowiczii) and I-214 (P. euramericana) exposed to industrial waste. Environ. Exp. Bot. 2004, 52, 79–88. [Google Scholar] [CrossRef]
- Zsuffa, L.; Giordano, E.; Pryor, L.D.; Stettler, R. Trends in poplar culture: Some global and regional perspectives. In Biology of Populus; Stettler, R.F., Bradshaw, H.D., Jr., Heilman, P.E., Hinckley, T.M., Eds.; NRC Research Press: Ottawa, ON, Canada, 1996; Chapter 20; pp. 515–539. [Google Scholar]
- Smulders, M.J.M.; Cottrell, J.E.; Lefèvre, F.; van der Schoot, J.; Arens, P.; Vosman, B.; Tabbener, H.E.; Grassi, F.; Fossati, T.; Castiglione, S.; et al. Structure of the genetic diversity in black poplar (Populus nigra L.) populations across European river systems: Consequences for conservation and restoration. For. Ecol. Manag. 2008, 255, 1388–1399. [Google Scholar] [CrossRef]
- Szuba, A. Ectomycorrhiza of Populus. For. Ecol. Manag. 2015, 347, 156–169. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, J.; Pan, G.; Wang, G.; Liu, X.; Zhang, X.; Li, L.; Bian, R.; Cheng, K.; Zheng, J. A long-term hybrid poplar plantation on cropland reduces soil organic carbon mineralization and shifts microbial community abundance and composition. Appl. Soil Ecol. 2017, 111, 94–104. [Google Scholar] [CrossRef]
- Bradshaw, H.D.; Reinhart, C.; Davis, J.; Stettler, R. Emerging model systems in plant biology: Poplar (Populus) as a model forest tree. J. Plant Growth Regul. 2000, 19, 306–313. [Google Scholar] [CrossRef]
- Dillen, S.Y.; Marron, N.; Bastien, C.; Ricciotti, L.; Salani, F.; Sabatti, M.; Pinel, M.P.C.; Rae, A.M.; Taylor, G.; Ceulemans, R. Effects of environment and progeny on biomass estimations of five hybrid poplar families grown at three contrasting sites across Europe. For. Ecol. Manag. 2007, 252, 12–23. [Google Scholar] [CrossRef]
- Larchevêque, M.; Maurel, M.; Desrochers, A.; Larocque, G.R. How does drought tolerance compare between two improved hybrids of balsam poplar and an unimproved native species? Tree Physiol. 2011, 31, 240–249. [Google Scholar] [CrossRef]
- Marmiroli, M.; Pietrini, F.; Maestri, E.; Zacchini, M.; Marmiroli, N.; Massacci, A. Growth, physiological and molecular traits in Salicaceae trees investigated for phytoremediation of heavy metals and organics. Tree Physiol. 2011, 31, 1319–1334. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Use and misuse of PLFA measurement in soils. Soil Biol. Biochem. 2011, 43, 1621–1625. [Google Scholar] [CrossRef]
- Wallander, H.; Ekblad, A.; Godbold, D.L.; Johnson, D.; Bahr, A.; Baldrian, P.; Björk, R.G.; Kieliszewska-Rokicka, B.; Kjøller, R.; Kraigher, H.; et al. Evaluation of methods to estimate production, biomass and turnover of ectomycorrhizal mycelium in forests soils—A review. Soil Biol. Biochem. 2013, 57, 1034–1047. [Google Scholar] [CrossRef]
- Sasser, M. Identification of bacteria through fatty acid analysis. In Methods in Phyto-Bacteriology; Klement, Z., Rudolph, K., Sands, D.C., Eds.; Akademiai Kiado: Budapest, Hungary, 1990; pp. 199–203. [Google Scholar]
- Steger, K.; Jarvis, Å.; Smårs, S.; Sundh, I. Comparison of signature lipid methods to determine microbial community structure in compost. J. Microb. Methods 2003, 55, 371–382. [Google Scholar] [CrossRef]
- Parsley, R. Operation manual, version 6. In Microbial Identification System; MIDI Inc.: Newark, DE, USA, 1996. [Google Scholar]
- Ravnskov, S.; Jensen, B.; Knudsen, I.M.B.; Bødker, L.; Funck Jensen, D.; Karliński, L.; Larsen, J. Soil inoculation with the biocontrol agent Clonostachys rosea and the mycorrhizal fungus Glomus intraradices results in mutual inhibition, plant growth promotion and alteration of soil microbial communities. Soil Biol. Biochem. 2006, 38, 3453–3462. [Google Scholar] [CrossRef]
- Wilkinson, S.C.; Anderson, J.M.; Scardelis, S.P.; Tisiafouli, M.; Taylor, A.; Wolters, V. PLFA profiles of microbial communities in decomposing conifer litters subject to moisture stress. Soil Biol Biochem. 2002, 34, 189–200. [Google Scholar] [CrossRef]
- Zelles, L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: A review. Biol. Fertil. Soils 1999, 29, 111–129. [Google Scholar] [CrossRef]
- Frostegård, Å.; Bååth, E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertil. Soils 1996, 22, 59–65. [Google Scholar] [CrossRef]
- Graham, J.; Hodge, N.C.; Morton, J.B. Fatty acid methyl ester profiles for characterisation of glomalean fungi and their endomycorrhizae. Appl. Environ. Microbiol. 1995, 1, 58–64. [Google Scholar] [CrossRef]
- Madan, R.; Pankhurst, C.; Hawke, B.; Smith, S. Use of fatty acids for identification of AM fungi and estimation of the biomass of AM spores in soil. Soil Biol. Biochem. 2002, 34, 125–128. [Google Scholar] [CrossRef]
- Olsson, P.A. Signature fatty acids provide tools for determination of the distribution and interaction of mycorrhizal fungi in soil. FEMS Microbiol. Ecol. 1999, 29, 303–310. [Google Scholar] [CrossRef]
- Bååth, E. The use of neutral lipid fatty acids to indicate the physiological conditions of soil fungi. Microb. Ecol. 2003, 45, 373–383. [Google Scholar] [CrossRef]
- Lechevalier, H.; Lechevalier, M.P. Chemotaxonomic use of lipids—An overview. In Microbial Lipids; Ratledge, C., Wilkinson, S.G., Eds.; Academic Press Ltd.: London, UK, 1988; Volume 1. [Google Scholar]
- White, D.C.; Stair, J.O.; Ringelberg, D.B. Quantitative comparisons of in situ microbial biodiversity by signature biomarker analysis. J. Industrial Microbiol. 1996, 17, 185–196. [Google Scholar] [CrossRef]
- Snedecor, W.; Cochran, W.G. Statistical Methods, 6th ed.; The Iowa State University Press: Ames, IA, USA, 1976; pp. 327–329. [Google Scholar]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Paleont. Electron. 2001, 4, 1–9. [Google Scholar]
- Brunner, I.; Luster, J.; Günthardt-Goerg, M.S.; Frey, B. Heavy metal accumulation and phytostabilisation potential of tree fine roots in a contaminated soil. Environ. Pollut. 2008, 152, 559–568. [Google Scholar] [CrossRef]
- Bailey, V.L.; Smith, J.L.; Bolton, H., Jr. Fungal-to-bacterial biomass ratios in soils investigated for enhanced carbon sequestration. Soil Biol. Biochem. 2002, 34, 997–1007. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, J.; Yang, F.; Lei, Y.; Zhang, Q.; Cheng, X. Alternations in soil microbial community composition and biomass following agricultural land use change. Sci. Rep. 2016, 6, 36587. [Google Scholar] [CrossRef] [PubMed]
- Holland, E.A.; Coleman, D.C. Litter lacement effects on microbial and organic matter dynamics in an agroecosystem. Ecology 1987, 68, 425–433. [Google Scholar] [CrossRef]
- Joergensen, R.G.; Wichern, F. Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol. Biochem. 2008, 40, 2977–2991. [Google Scholar] [CrossRef]
- Lamit, J.L.; Holeski, L.M.; Flores-Rentería, L.; Whitham, T.G.; Gehring, C.A. Tree genotype influences ectomycorrhizal fungal community structure: Ecological and evolutionary implications. Fungal Ecol. 2016, 24, 124–134. [Google Scholar] [CrossRef]
- Holste, E.K.; Kobe, R.K.; Gehring, C.A. Plant species differ in early seedling growth and tissue nutrient responses to arbuscular and ectomycorrhizal fungi. Mycorrhiza 2017, 27, 211–223. [Google Scholar] [CrossRef]
- Ostonen, I.; Lõhmus, K.; Pajuste, K. Fine root biomass, production and its proportion of NPP in a fertile middle-aged Norway spruce forest: Comparison of soil core and ingrowth core methods. For. Ecol. Manag. 2005, 212, 264–277. [Google Scholar] [CrossRef]
- Hobley, E.U.; Wilson, B. The depth distribution of organic carbon in the soils of eastern Australia. Ecosphere 2016, 7, e01214. [Google Scholar] [CrossRef]
- Song, Y.; Deng, S.P.; Acosta-Martínez, V.; Katsalitou, E. Characterization of redox-related soil microbial communities along a river floodplain continuum by fatty acid methyl ester (FAME) and 16S rRNA genes. Appl. Soil Ecol. 2008, 40, 499–509. [Google Scholar] [CrossRef]
- Fritze, H.; Pietikäinen, T.; Pennanen, T. Distribution of microbial biomass and phospholipid fatty acids in Podzol profiles under coniferous forest. Eur. J. Soil Sci. 2000, 51, 565–573. [Google Scholar] [CrossRef]
- Blume, E.; Bischoff, M.; Reichert, J.M.; Moorman, T.; Konopka, A.; Turco, R.F. Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season. Appl. Soil Ecol. 2002, 20, 171–181. [Google Scholar] [CrossRef]
- Pennanen, T.; Frostegård, Å.; Fritze, H.; Bååth, E. Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal-polluted gradients in coniferous forests. Appl. Environ. Microbiol. 1996, 62, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Eklund, F.; Rønn, R.; Christensen, S. Distribution with depth of protozoa, bacteria and fungi in soil profiles from three Danish forest sites. Soil Biol. Biochem. 2001, 33, 475–481. [Google Scholar] [CrossRef]
- Henkes, G.J.; Kandeler, E.; Marhan, S.; Scheu, S.; Bonkowski, M. Interactions of mycorrhiza and protists in the rhizosphere systemically alter microbial community composition, plant shoot-to-root ratio and within-root system nitrogen allocation. Front. Environ. Sci. 2018, 6, 117. [Google Scholar] [CrossRef]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl. Environ. Microbiol. 1993, 59, 3606–3617. [Google Scholar] [CrossRef]
- Hagerberg, D.; Manique, N.; Brandt, K.K.; Larsen, J.; Nybroe, O.; Olsson, S. Low concentration of copper inhibits colonization of soil by the arbuscular mycorrhizal fungus Glomus intraradices and changes the microbial community structure. Microb. Ecol. 2011, 61, 844–852. [Google Scholar] [CrossRef]
- Rajapaksha, R.M.C.P.; Tobor-Kapłon, M.A.; Bååth, E. Metal toxicity affects fungal and bacterial activities in soil differently. Appl. Environ. Microbiol. 2004, 70, 2966–2973. [Google Scholar] [CrossRef]
- Duxbury, T.; Binknell, B. Metal-tolerant bacterial populations from natural and metal-polluted soils. Soil Biol. Biochem. 1983, 15, 243–250. [Google Scholar] [CrossRef]
- O’Leary, W.M.; Wilkinson, S.G. Gram-Positive Bacteria. In Microbial Lipids; Retledge, C., Wilkinson, S.G., Eds.; Academic Press Ltd.: London, UK, 1988; Volume 1, pp. 117–185. [Google Scholar]
- Gupta, K.; Chatterjee, C.; Gupta, B. Isolation and characterization of heavy metal tolerant Gram-positive bacteria with bioremedial properties from municipal waste rich soil of Kestopur canal (Kolkata), West Bengal, India. Biologia 2012, 67, 827–836. [Google Scholar] [CrossRef]
- Pennanen, T.; Strömmer, R.; Markkola, A.; Fritze, H. Microbial and plant community structure across a primary succession gradient. Scand. J. For. Res. 2001, 16, 37–43. [Google Scholar] [CrossRef]
- Buyer, J.S.; Teasdale, J.R.; Roberts, D.P.; Zasada, I.A.; Maul, J.E. Factors affecting soil microbial community structure in tomato cropping system. Soil Biol. Biochem. 2010, 42, 831–841. [Google Scholar] [CrossRef]
- Jordan, M.J.; Lechevalier, M.P. Effects zinc-smelter emissions on forest soil microflora. Can. J. Mirobiol. 1975, 21, 1855–1865. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghe, B.A.T.D.; Parkinson, D. Actinomycetes as antagonists of litter decomposer fungi. Appl. Soil Ecol. 2008, 38, 109–118. [Google Scholar] [CrossRef]
- Gomiero, A.; Dagnino, A.; Nasci, C.; Viarengo, A. The use of protozoa in ecotoxicology: Application of multiple endpoint tests of the ciliate E. Crassus for the evaluation of sediment quality in coastal marine ecosystems. Sci. Tot. Environ. 2013, 442, 534–544. [Google Scholar] [CrossRef]
- Li, Y.-D.; Chen, Y.; Wang, L.; Yao, L.; Pan, X.-M.; Lee, D.-J. Pollution tolerant protozoa in polluted wetland. Biores. Technol. 2017, 240, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ananyeva, N.D.; Susyan, E.A.; Chernova, O.V.; Chernov, I.Y.; Makarova, O.L. The ratio of fungi and bacteria in the biomass of different types of soil determined by selective inhibition. Microbiology 2006, 75, 702–707. [Google Scholar] [CrossRef]


| Site 1 | Site 2 | Site 3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0–10 cm | 10–20 cm | 20–30 cm | * | 0–10 cm | 10–20 cm | 20–30 cm | * | 0–10 cm | 10–20 cm | 20–30 cm | * | |
| pH[H2O] | 7.6 | 7.3 | 7.6 | c | 7.1 | 7.1 | 7.1 | b | 6.8 | 6.3 | 6.2 | a |
| pH[KCl] | 7.2 | 7.2 | 7.5 | c | 6.8 | 6.8 | 6.8 | b | 6.1 | 5.4 | 5.2 | a |
| Cu | 7.88 | 9.1 | 4.17 | a | 5.08 | 8.41 | 4.97 | a | 2137.4 | 1295.2 | 748.8 | b |
| Pb | 8.47 | 10.78 | 9.47 | a | 9.33 | 10.02 | 6.69 | a | 2154.35 | 518.8 | 515.55 | b |
| Zn | 20.90 | 18.40 | 18.90 | a | 30.30 | 32.80 | 25.90 | b | 94.20 | 71.80 | 114.2 | c |
| Cd | 0.29 | 0.28 | 0.26 | a | 0.32 | 0.31 | 0.30 | b | 0.47 | 0.43 | 0.43 | c |
| N-NO3 | 24.69 | 8.75 | 8.51 | b | 20.59 | 8.66 | 5.76 | b | 10.29 | 0.95 | 1.34 | a |
| N-NH3 | 2.29 | 0.98 | 0.17 | 2.3 | 2.2 | 1.66 | 1.97 | 1.22 | 1.32 | |||
| P | 492 | 358 | 397.5 | a | 446 | 438 | 324.5 | a | 870 | 488.5 | 512.5 | b |
| K | 1073 | 820.5 | 750 | a | 1617 | 1616.5 | 1631.5 | b | 3361 | 3641.5 | 4333 | c |
| Ca | 2521.5 | 2414 | 3955.5 | 2932 | 1736 | 1597.5 | 4220 | 2526.5 | 2316 | |||
| C % | 1.41 | 1.08 | 1.09 | b | 0.94 | 0.63 | 0.48 | a | 2.22 | 1.92 | 1.51 | c |
| N % | 0.13 | 0.1 | 0.11 | b | 0.07 | 0.04 | 0.03 | a | 0.19 | 0.1 | 0.12 | c |
| C/N | 10.97 | 11.23 | 10.18 | 14.09 | 14.95 | 17.02 | 11.46 | 19.09 | 12.48 | |||
| Bacteria Gram (+) | Bacteria Gram (−) | Actinobacteria | AMF | SF | Protozoa | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | p | ω2 | F | p | ω2 | F | p | ω2 | F | p | ω2 | F | p | ω2 | F | p | ω2 | |
| Overall | ||||||||||||||||||
| Site | 33.7 | *** | 19 | 92.0 | *** | 39 | 2.7 | 0.068 | 1 | 230.6 | *** | 60 | 17.6 | *** | 10 | 43.1 | *** | 26 |
| Genotype | 5.7 | *** | 4 | 4.8 | ** | 2 | 7.3 | *** | 7 | 13.4 | *** | 5 | 1.8 | 0.150 | 1 | 1.7 | 0.170 | 1 |
| Depth | 2.7 | 0.069 | 1 | 8.2 | *** | 3 | 13.0 | *** | 9 | 6.7 | ** | 1 | 5.9 | ** | 3 | 3.6 | * | 2 |
| Site × genotype | 0.5 | 0.835 | 0 | 0.5 | 0.824 | 0 | 0.5 | 0.831 | 0 | 7.1 | *** | 5 | 1.4 | 0.204 | 1 | 5.5 | *** | 8 |
| Site × depth | 7.7 | *** | 8 | 10.2 | *** | 8 | 1.1 | 0.362 | 0.1 | 2.4 | * | 1 | 6.8 | *** | 7 | 0.6 | 0.665 | 0 |
| Genotype × depth | 2.7 | * | 3 | 1.3 | 0.277 | 0.3 | 0.8 | 0.536 | 0 | 1.2 | 0.292 | 0.2 | 3.1 | ** | 4 | 0.4 | 0.905 | 0 |
| Site × genotype × depth | 1.9 | * | 3 | 2.0 | * | 3 | 1.6 | 0.086 | 3 | 0.5 | 0.911 | 0 | 2.5 | ** | 6 | 0.7 | 0.770 | 0 |
| At each of the sites | ||||||||||||||||||
| Site 1 | ||||||||||||||||||
| Genotype | 1.3 | 0.280 | 1 | 1.1 | 0.373 | 0.2 | 2.3 | 0.086 | 5 | 4.7 | ** | 11 | 3.9 | * | 10 | 2.4 | 0.073 | 6 |
| Depth | 5.4 | ** | 10 | 4.4 | * | 8 | 3.5 | * | 7 | 10.5 | *** | 19 | 5.1 | ** | 9 | 2.4 | 0.101 | 4 |
| Genotype × depth | 2.0 | 0.0743 | 7 | 1.5 | 0.188 | 4 | 0.1 | 0.994 | 0 | 0.5 | 0.816 | 0 | 1.1 | 0.369 | 1 | 0.4 | 0.897 | 0 |
| Site 2 | ||||||||||||||||||
| Genotype | 5.1 | ** | 15 | 2.3 | 0.087 | 5 | 2.0 | 0.125 | 4 | 2.0 | 0.125 | 19 | 1.1 | 0.361 | 0 | 6.3 | *** | 18 |
| Depth | 1.0 | 0.382 | 0 | 4.4 | * | 8 | 3.4 | * | 6 | 3.4 | * | 0 | 8.4 | *** | 18 | 0.8 | 0.433 | 0 |
| Genotype × depth | 0.7 | 0.646 | 0 | 0.7 | 0.624 | 0 | 1.0 | 0.456 | 0 | 1.0 | 0.456 | 0 | 0.4 | 0.895 | 0 | 0.7 | 0.623 | 0 |
| Site 3 | ||||||||||||||||||
| Genotype | 1.3 | 0.298 | 1 | 2.1 | 0.109 | 3 | 4.0 | * | 8 | 12.6 | *** | 32 | 1.3 | 0.299 | 1 | 1.0 | 0.399 | 0 |
| Depth | 9.4 | *** | 16 | 17.7 | *** | 28 | 8.4 | *** | 14 | 2.1 | 0.1340 | 2 | 6.4 | ** | 11 | 1.0 | 0.3739 | 0 |
| Genotype × depth | 3.0 | * | 12 | 2.9 | * | 9 | 3.0 | * | 11 | 1.1 | 0.362 | 1 | 3.4 | ** | 15 | 1.0 | 0.434 | 0 |
| AMF:SF | F:B | Bacteria Gram (+):Gram (−) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| F | p | ω2 | F | p | ω2 | F | p | ω2 | |
| Overall | |||||||||
| Site | 152.4 | *** | 50.7 | 17.4 | *** | 9 | 34.0 | *** | 23.2 |
| Genotype | 9.0 | *** | 4.0 | 7.7 | *** | 5 | 0.7 | 0.558 | 0 |
| Depth | 3.9 | * | 1.0 | 7.4 | *** | 3 | 2.8 | 0.062 | 1.3 |
| Site × genotype | 9.3 | *** | 8.4 | 1.5 | 0.181 | 1 | 0.6 | 0.744 | 0 |
| Site × depth | 3.2 | * | 1.5 | 9.6 | *** | 9 | 2.2 | 0.069 | 1.7 |
| Genotype × depth | 0.995 | 0.995 | 0 | 4.5 | *** | 6 | 0.8 | 0.544 | 0 |
| Site × genotype × depth | 0.867 | 0.867 | 0 | 33.5 | *** | 8 | 0.8 | 0.627 | 0 |
| At each of the sites | |||||||||
| Site 1 | |||||||||
| Genotype | 4.9 | ** | 14.3 | 2.9 | * | 6 | 0.1 | 0.971 | 0 |
| Depth | 2.1 | 0.130 | 2.7 | 10.5 | *** | 0 | 0.5 | 0.629 | 0 |
| Genotype × depth | 0.3 | 0.924 | 0 | 1.9 | 0.091 | 0 | 1.3 | 0.272 | 2.6 |
| Site 2 | |||||||||
| Genotype | 4.0 | * | 11.1 | 4.1 | ** | 11 | 0.1 | 0.938 | 0 |
| Depth | 3.8 | * | 6.7 | 2.9 | 0.063 | 0 | 3.2 | * | 6.1 |
| Genotype × depth | 0.3 | 0.950 | 0 | 0.7 | 0.646 | 0 | 0.9 | 0.522 | 0 |
| Site 3 | |||||||||
| Genotype | 14.4 | *** | 34.7 | 3.5 | * | 6 | 1.0 | 0.415 | 0 |
| Depth | 4.2 | ** | 5.6 | 10.2 | *** | 0 | 2.3 | 0.111 | 3.5 |
| Genotype × depth | 0.5 | 0.774 | 0 | 4.9 | *** | 0 | 0.7 | 0.627 | 0 |
| Bacteria Gram (+) | Bacteria Gram (−) | Actinobacteria | AMF | SF | Protozoa | |
|---|---|---|---|---|---|---|
| pH[H2O] | 0.29 | 0.78 | 0.32 | 0.85 | 0.27 | 0.77 |
| 0.081 | *** | 0.056 | *** | 0.110 | *** | |
| pH[KCl] | 0.34 | 0.81 | 0.27 | 0.81 | 0.23 | 0.73 |
| * | *** | 0.110 | *** | 0.169 | *** | |
| Cu | 0.15 | −0.29 | −0.07 | −0.54 | −0.51 | −0.51 |
| 0.379 | 0.086 | 0.692 | *** | ** | ** | |
| Pb | 0.34 | −0.14 | −0.11 | −0.62 | −0.65 | −0.57 |
| * | 0.402 | 0.520 | *** | *** | *** | |
| Zn | −0.34 | −0.73 | −0.14 | −0.80 | −0.24 | −0.70 |
| * | *** | 0.409 | *** | 0.160 | *** | |
| Cd | −0.25 | −0.69 | −0.04 | −0.77 | −0.23 | −0.67 |
| 0.145 | *** | 0.800 | *** | 0.180 | *** | |
| N-NO3 | 0.06 | 0.43 | 0.59 | 0.57 | 0.39 | 0.58 |
| 0.714 | ** | *** | *** | * | *** | |
| N-NH3 | −0.52 | −0.25 | 0.38 | 0.15 | 0.51 | 0.21 |
| ** | 0.134 | * | 0.385 | ** | 0.219 | |
| P | 0.03 | −0.30 | 0.23 | −0.47 | −0.27 | −0.37 |
| 0.876 | 0.075 | 0.170 | ** | 0.115 | * | |
| K | −0.36 | −0.84 | −0.26 | −0.80 | −0.19 | −0.73 |
| * | *** | 0.121 | *** | 0.271 | *** | |
| Ca | 0.54 | 0.30 | 0.42 | −0.17 | −0.29 | −0.07 |
| *** | 0.081 | 0.011 | 0.328 | 0.085 | 0.667 | |
| C | 0.44 | −0.07 | 0.12 | −0.49 | −0.59 | −0.44 |
| ** | 0.683 | 0.485 | ** | *** | ** | |
| N | 0.44 | 0.18 | 0.28 | −0.15 | −0.37 | −0.12 |
| ** | 0.294 | 0.093 | 0.399 | * | 0.494 | |
| C/N | −0.48 | −0.67 | −0.32 | −0.46 | 0.01 | −0.43 |
| ** | *** | 0.054 | ** | 0.933 | ** | |
| Soil temperature | −0.26 | −0.75 | −0.22 | −0.87 | −0.31 | −0.78 |
| 0.128 | *** | 0.199 | *** | 0.068 | *** | |
| Soil humidity | 0.43 | −0.13 | −0.08 | −0.55 | −0.65 | −0.52 |
| ** | 0.446 | 0.662 | *** | *** | ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karliński, L.; Ravnskov, S.; Rudawska, M. Soil Microbial Biomass and Community Composition Relates to Poplar Genotypes and Environmental Conditions. Forests 2020, 11, 262. https://doi.org/10.3390/f11030262
Karliński L, Ravnskov S, Rudawska M. Soil Microbial Biomass and Community Composition Relates to Poplar Genotypes and Environmental Conditions. Forests. 2020; 11(3):262. https://doi.org/10.3390/f11030262
Chicago/Turabian StyleKarliński, Leszek, Sabine Ravnskov, and Maria Rudawska. 2020. "Soil Microbial Biomass and Community Composition Relates to Poplar Genotypes and Environmental Conditions" Forests 11, no. 3: 262. https://doi.org/10.3390/f11030262
APA StyleKarliński, L., Ravnskov, S., & Rudawska, M. (2020). Soil Microbial Biomass and Community Composition Relates to Poplar Genotypes and Environmental Conditions. Forests, 11(3), 262. https://doi.org/10.3390/f11030262