Genome-Wide Identification of the Auxin Response Factor (ARF) Gene Family and Their Expression Analysis during Flower Development of Osmanthus fragrans
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of ARF Genes in Osmanthus fragrans
2.2. Analysis of Gene Structures, Conserved Domains and Subcellular Localizations
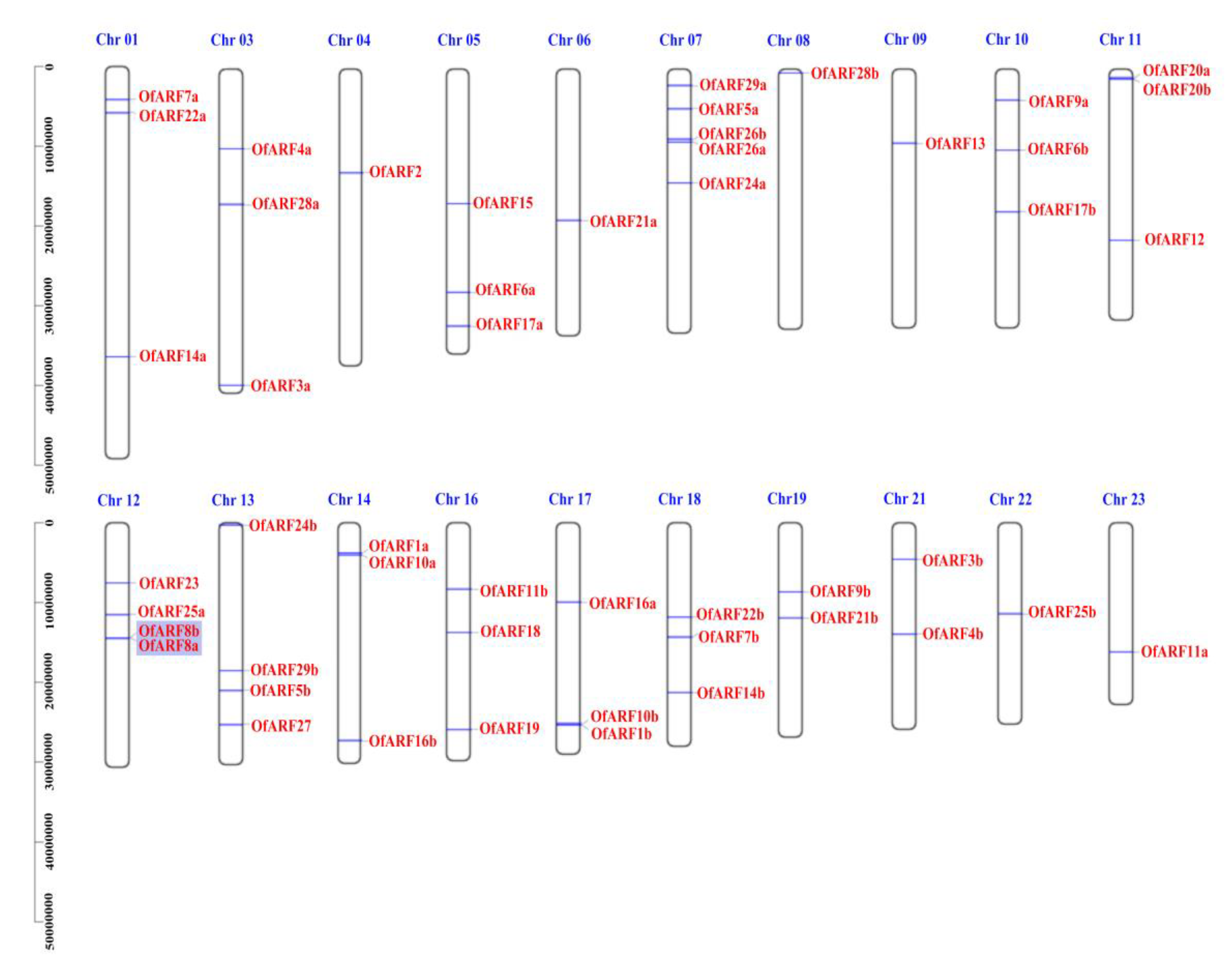
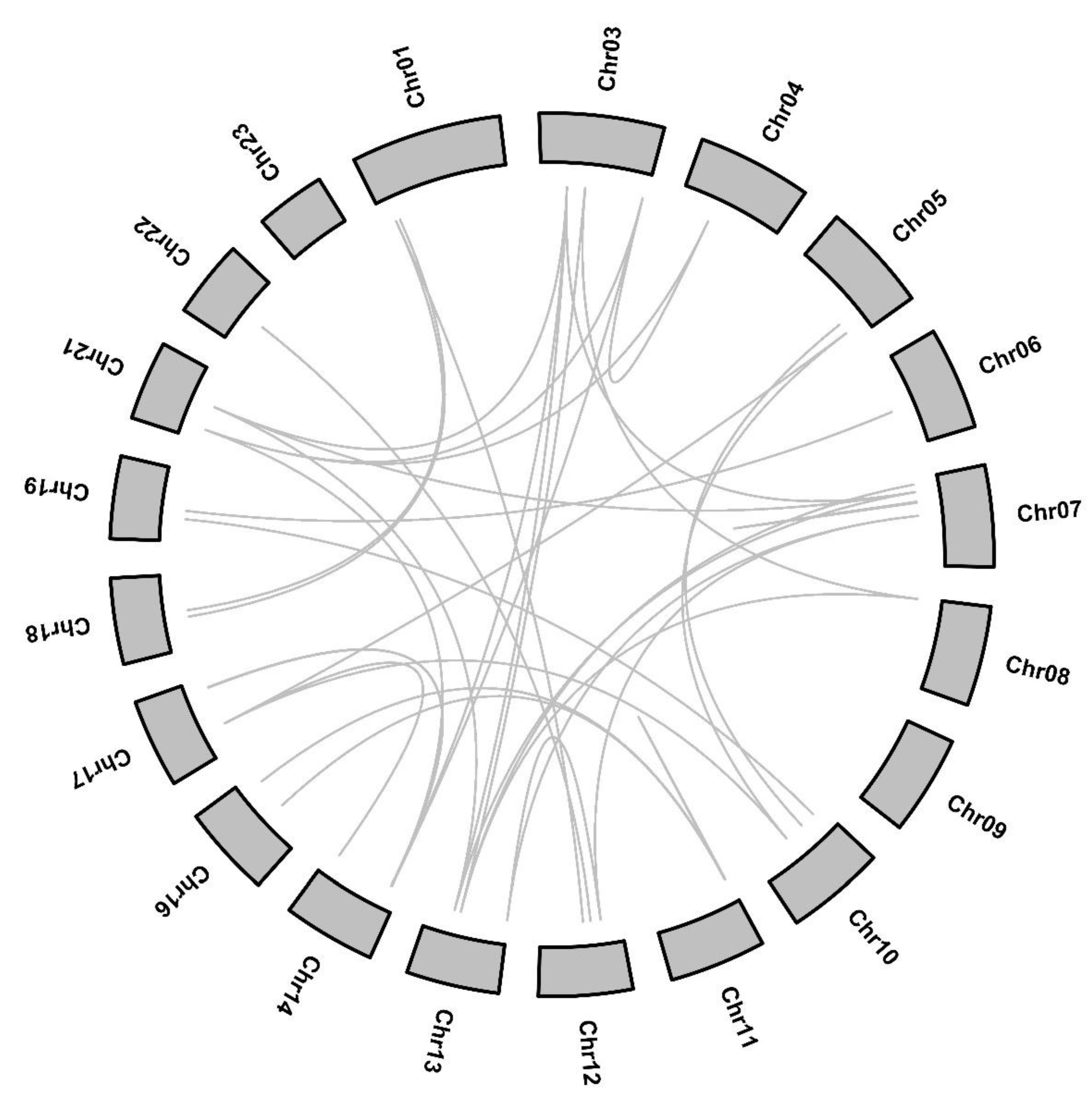
2.3. Chromosomal Localization, Gene Duplication and Evolution Analysis of OfARF Genes
2.4. Conserved Motifs and Promoter Cis-acting Regulatory DNA Elements Analysis
2.5. Phylogenetic Analysis of OfARF Genes
2.6. Plant Material Collection, RNA extraction and cDNA Synthesis
2.7. Expression Analysis of OfARF Genes
2.8. IAA Content Measurement
3. Results
3.1. Identification of the OfARF Gene Family
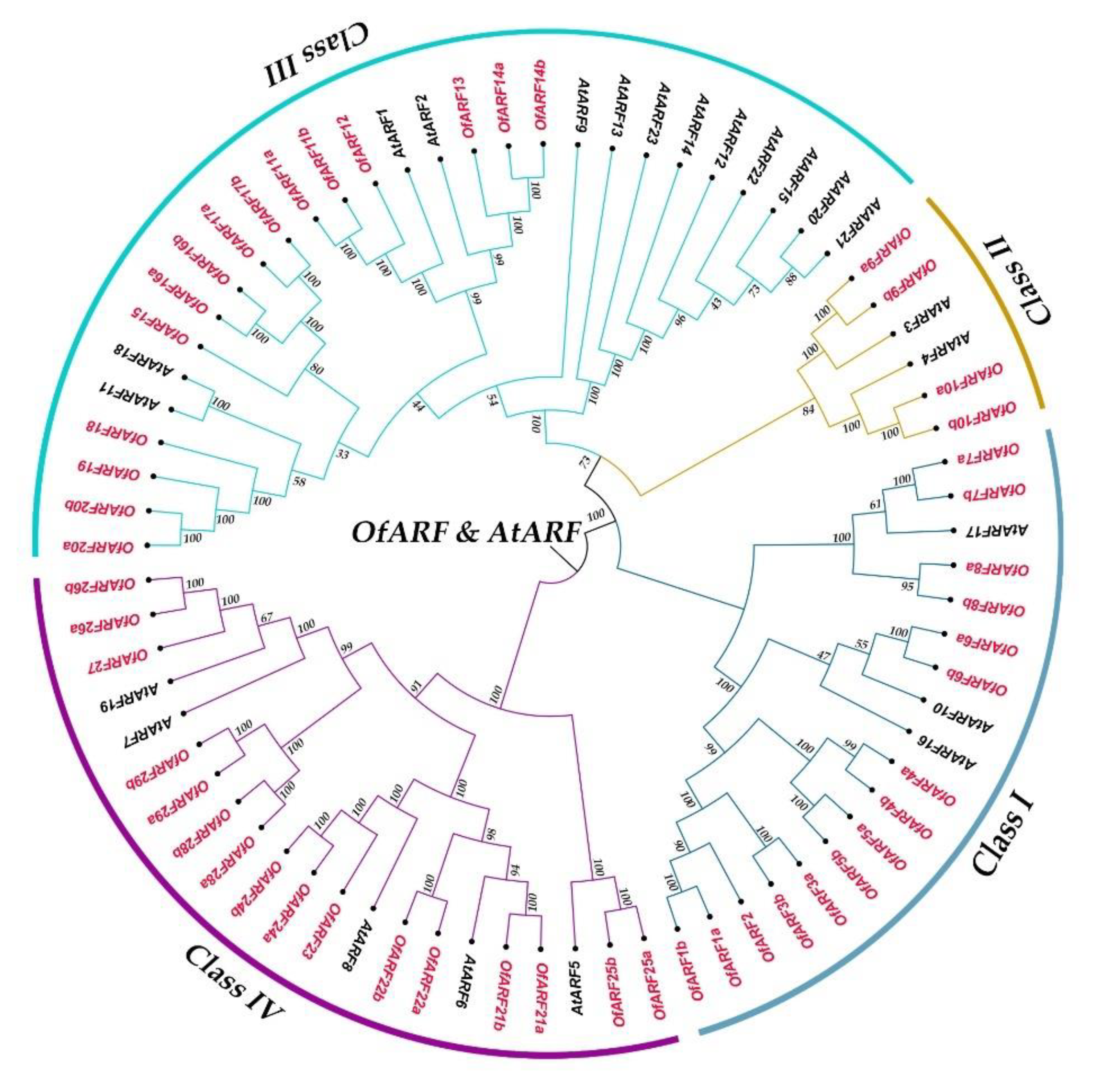
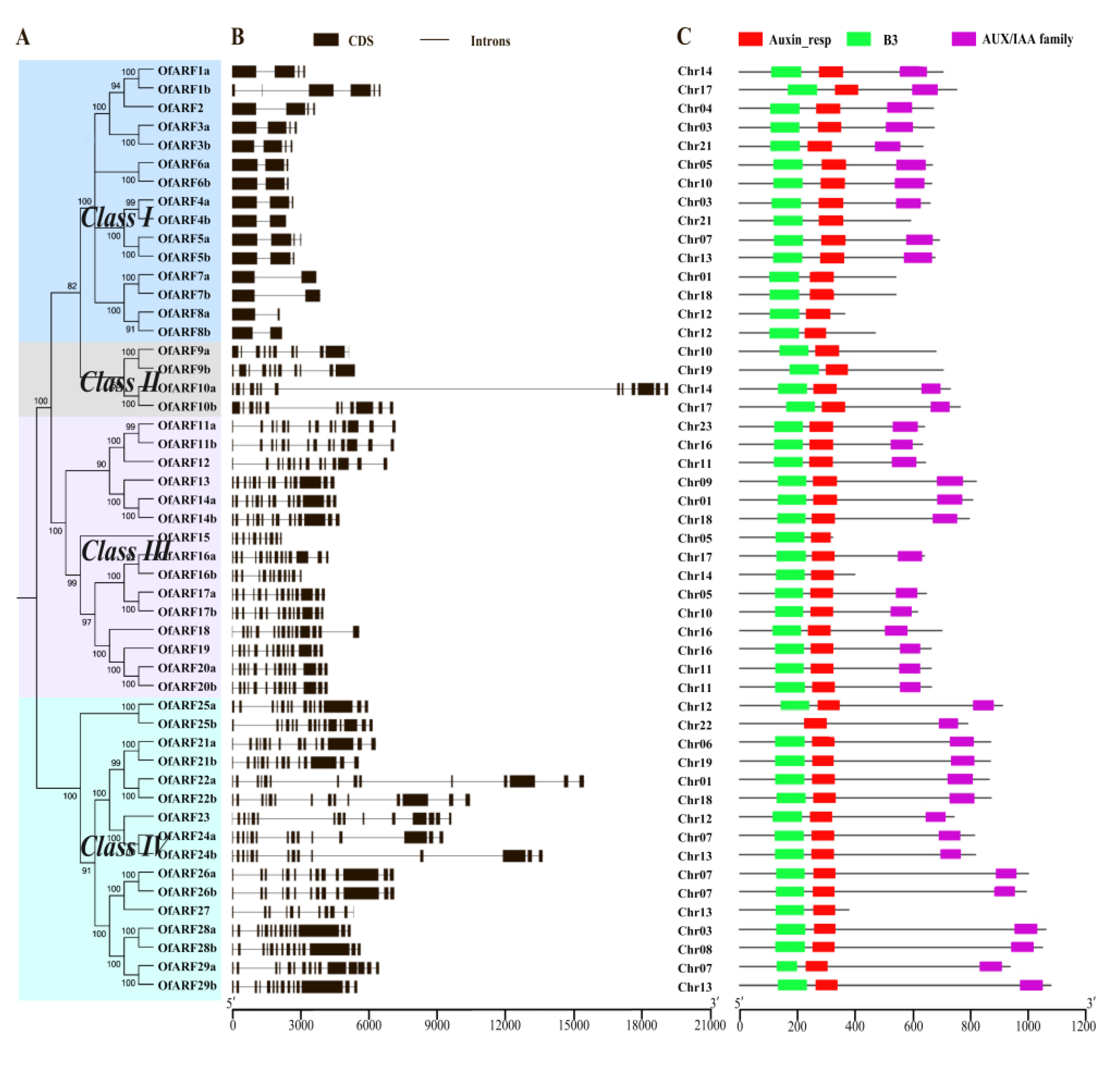
3.2. Phylogenetic Analysis and Gene Structure of OfARFs
3.3. Analysis of Conserved Domains of Osmanthus fragrans Auxin Response Factor (OfARF) Proteins
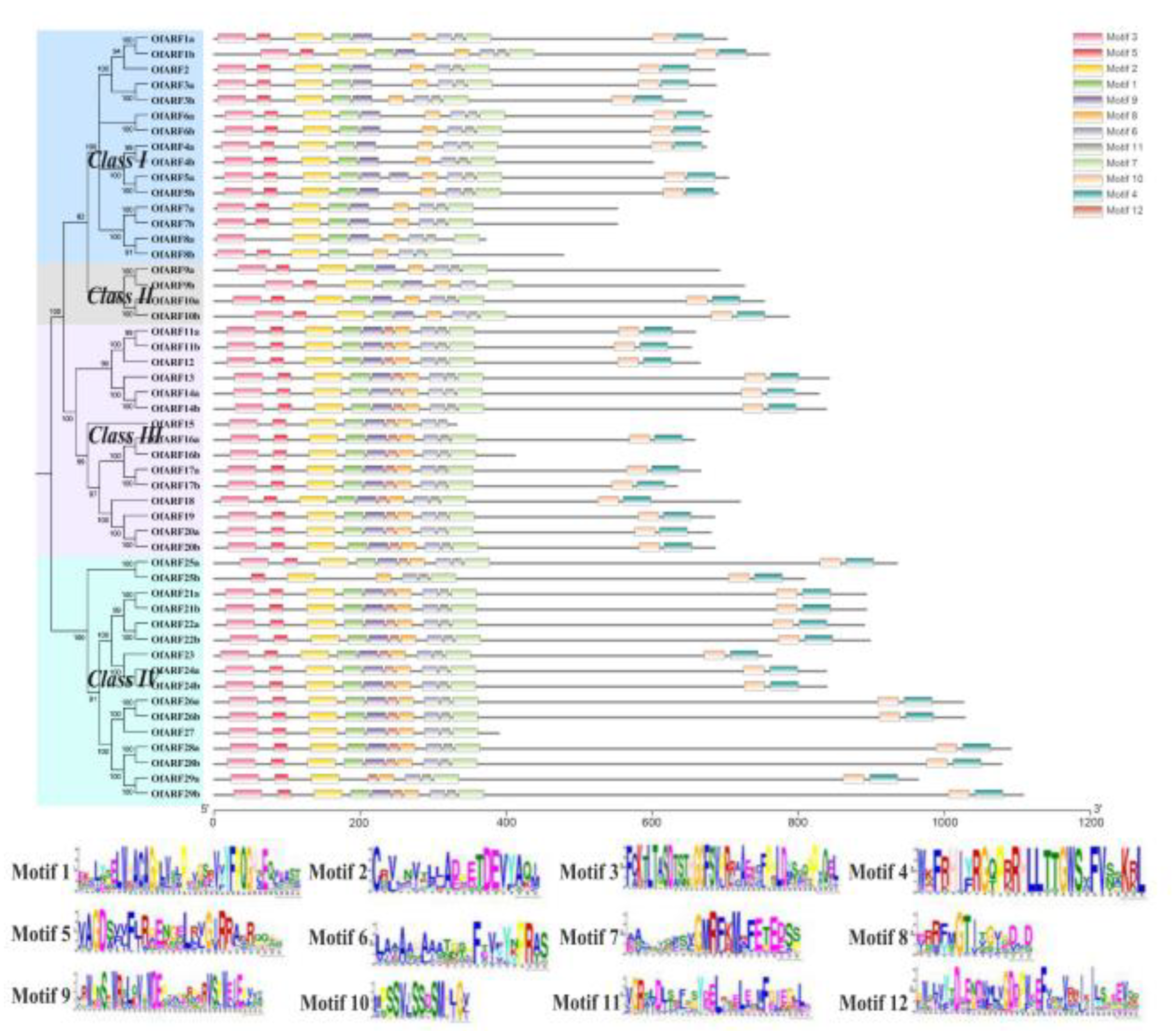
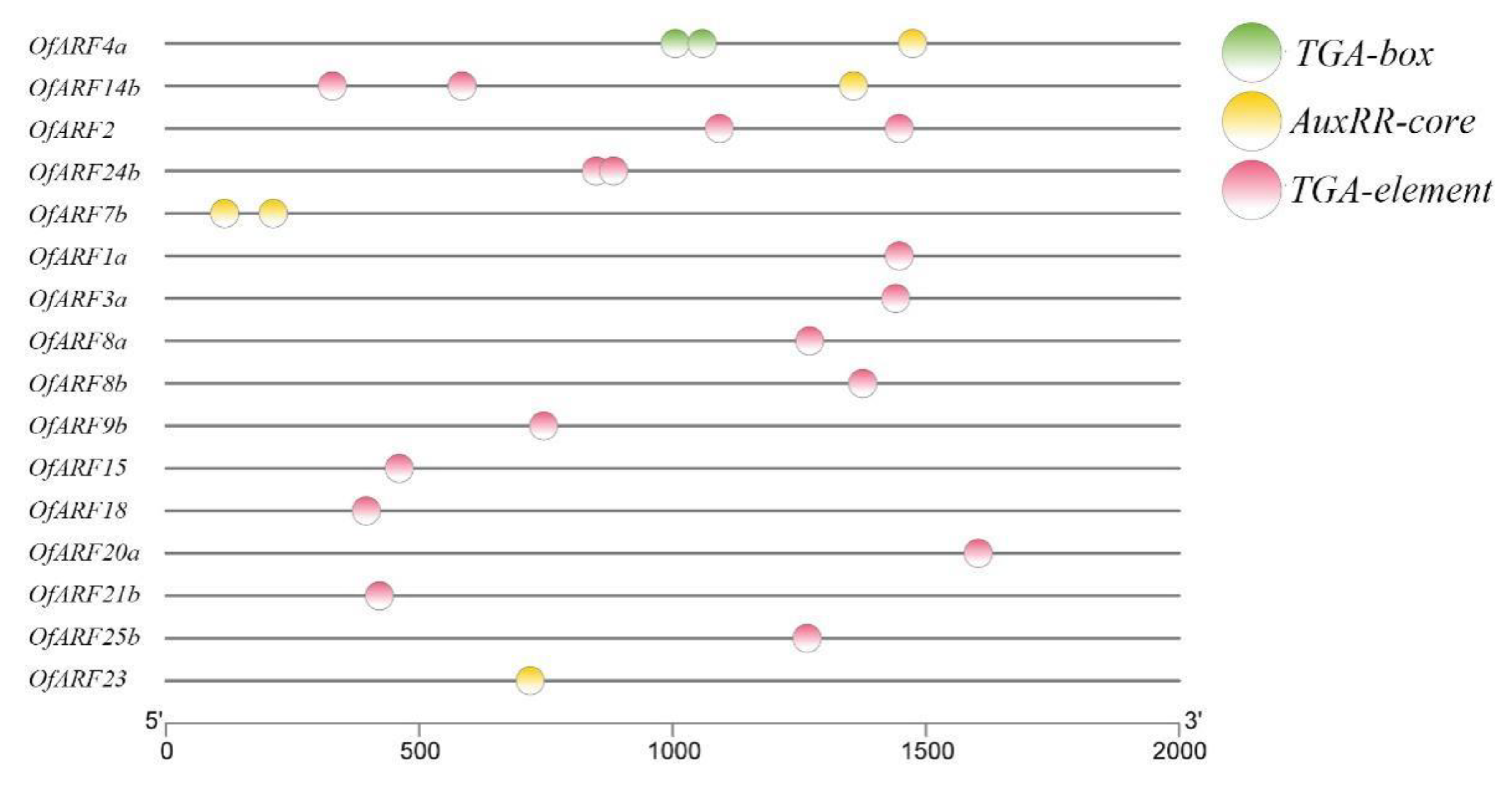
3.4. Motif Characterization and Promoter Analysis of Osmanthus fragrans Auxin Response Factor (OfARF) Proteins
3.5. Subcellular Localization Analysis of OfARFs
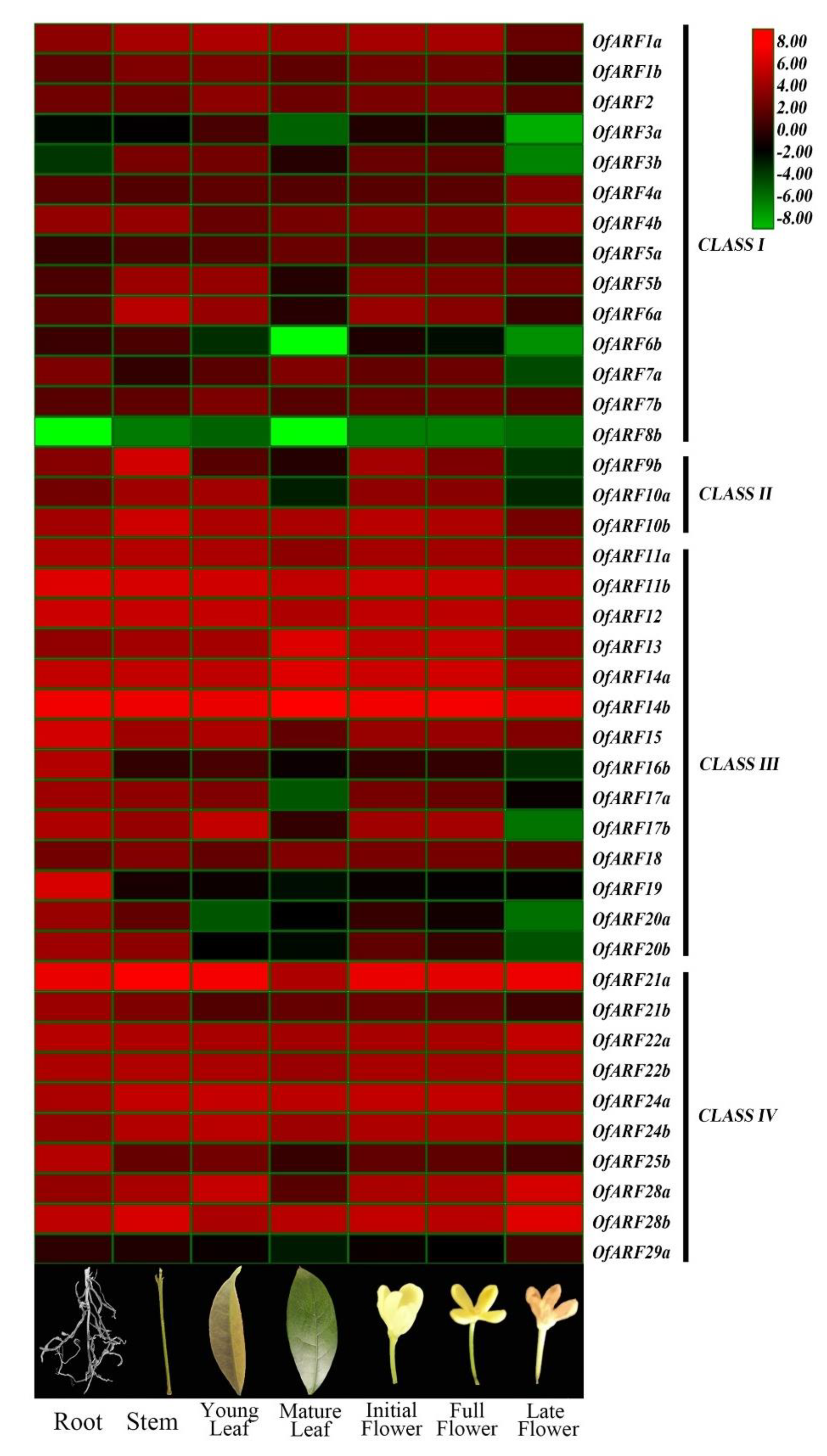
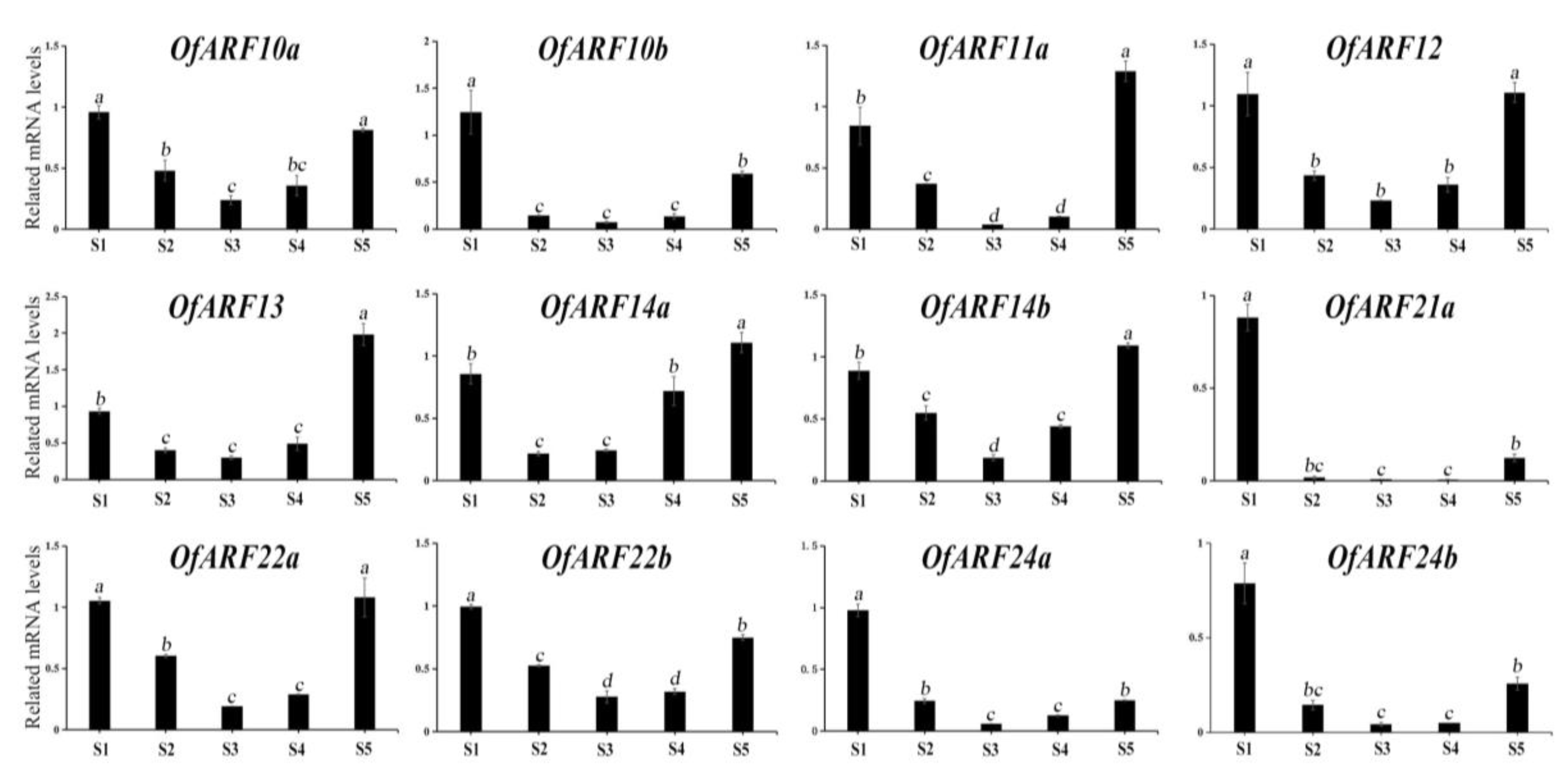
3.6. Expression of OfARF Genes in Different Tissues and Various Flower Development Stages of O. fragrans
3.7. Endogenous Indoleacetic Acid (IAA) Concentration Measurement
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, X.L.; Li, H.Y.; Yue, Y.Z.; Ding, W.J.; Xu, C.; Shi, T.T.; Chen, G.W.; Wang, L.G. Transcriptomic analysis of the candidate genes related to aroma formation in Osmanthus fragrans. Molecules 2018, 23, 1604. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.A.; Guo, X.F.; Kong, X.S.; Zhang, G.H.; Bao, M.Z. Respiration Rate and Endogenous Hormone Levels in Relation to the Flower Development of Tree Peonies. Acta Hortic. Sin. 2011, 38, 78–82. [Google Scholar]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin response factors, output control in auxin biology. J. Exp. Bot. 2017, 69, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.F.; Zhao, Y.D. A role for auxin in flower development. J. Integr. Plant Biol. 2007, 49, 99–104. [Google Scholar] [CrossRef]
- Naramoto, S. Polar transport in plants mediated by membrane transporters: Focus on mechanisms of polar auxin transport. Curr. Opin. Plant Biol. 2017, 40, 8–14. [Google Scholar] [CrossRef]
- Warren, W.J.; Warren, W.P. Mechanisms of Auxin Regulation of Structural and Physiological Polarity in Plants, Tissues, Cells and Embryos. Funct. Plant Biol. 1993, 20, 555–571. [Google Scholar] [CrossRef]
- Xu, D.Y.; Miao, J.H.; Yumoto, E.; Yokota, T.; Asahina, M.; Watahiki, M. YUCCA9-Mediated Auxin Biosynthesis and Polar Auxin Transport Synergistically Regulate Regeneration of Root Systems Following Root Cutting. Plant Cell Physiol. 2017, 58, 1710–1723. [Google Scholar] [CrossRef]
- Abel, S.; Theologis, A. Early genes and auxin action. Plant Physiol. 1996, 111, 9–17. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef] [PubMed]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef]
- Goh, T.; Kasahara, H.; Mimura, T.; Kamiya, Y.; Fukaki, H. Multiple AUX/IAA-ARF modules regulate lateral root formation: The role of Arabidopsis SHY2/IAA3-mediated auxin signaling. Philos. Trans. R. Soc. B 2012, 367, 1461–1468. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef]
- Okushima, Y.; Overvoorde, P.; Arima, K.; Alonso, J.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional Genomic Analysis of the AUXIN RESPONSE FACTOR Gene Family Members in Arabidopsis thaliana: Unique and Overlapping Functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef]
- Mu, H.N.; Sun, T.Z.; Xu, C.; Wang, L.G.; Yue, Y.Z.; Li, H.Y.; Yang, X. Identification and validation of reference genes for gene expression studies in sweet osmanthus (Osmanthus fragrans). J. Genet. 2017, 96, 273–281. [Google Scholar] [CrossRef]
- Ni, X.; Song, W.T.; Zhang, H.C.; Yang, X.L.; Wang, L.G. Effects of mulching on soil properties and growth of tea olive (Osmanthus fragrans). PLoS ONE 2016, 11, e0158228. [Google Scholar] [CrossRef]
- Yang, X.L.; Ding, W.J.; Yue, Y.Z.; Xu, C.; Wang, X.; Wang, L.G. Cloning and expression analysis of three critical Triterpenoid pathway genes in Osmanthus fragrans. Electron. J. Biotechn. 2018, 36, 1–8. [Google Scholar] [CrossRef]
- Mu, H.N.; Li, H.G.; Wang, L.G.; Yang, X.L.; Sun, T.Z.; Xu, C. Transcriptome sequencing and analysis of sweet osmanthus (Osmanthus fragrans Lour.). Genes Genom. 2014, 36, 777–788. [Google Scholar] [CrossRef]
- Xu, C.; Li, H.G.; Yang, X.L.; Gu, C.S.; Mu, H.N.; Yue, Y.Z. Cloning and expression analysis of MEP pathway enzyme-encoding genes in Osmanthus fragrans. Genes 2016, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.X.; Hou, D.; Zhang, C.; Bao, Z.Y.; Zhao, H.B.; Hu, S.Q. The emission of the floral scent of four Osmanthus fragrans cultivars in response to different temperatures. Molecules 2017, 22, 430. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.G.; Zhang, C.; Dong, B.; Fu, J.X.; Hu, S.Q.; Zhao, H.B. Carotenoid accumulation and its contribution to flower coloration of Osmanthus fragrans. Front. Plant Sci. 2018, 9, 1499–1513. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.G.; Fu, J.X.; Bao, Z.Y.; Zhao, H.B. Transcriptomic analysis and carotenogenic gene expression related to petal coloration in Osmanthus fragrans ‘Yanhong Gui’. Trees Struct. Funct. 2016, 30, 1207–1223. [Google Scholar] [CrossRef]
- Zhou, F.; Peng, J.Y.; Zhao, Y.J.; Huang, W.S.; Jiang, Y.R.; Li, M.Q.; Wu, X.; Lu, B. Varietal classification and antioxidant activity prediction of Osmanthus fragrans Lour. flowers using UPLC-PDA/QTOF-MS and multivariable analysis. Food Chem. 2017, 217, 490–497. [Google Scholar] [CrossRef]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178–189. [Google Scholar] [CrossRef]
- Zhuang, H.X.; Ning, C.; Dong, C.J.; Shang, Q.M. Genome-wide identification and expression of ARF gene family during adventitious root development in hot pepper (Capsicum annuum). Hortic. Plant J. 2017, 4, 22–35. [Google Scholar] [CrossRef]
- Wu, J.; Wang, F.; Cheng, L.; Kong, F.; Peng, Z.; Liu, S.; Yu, X.; Lu, G. Identification, isolation and expression analysis of Auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 2011, 30, 2059–2073. [Google Scholar] [CrossRef]
- Song, J.; Gao, Z.H.; Huo, X.M.; Sun, H.L.; Xu, Y.S.; Shi, T.; Ni, Z. Genome-wide identification of the auxin response factor (ARF) gene family and expression analysis of its role associated with pistil development in Japanese apricot (Prunus mume Sieb.et Zucc). Acta Physiol. Plant 2015, 37, 145–158. [Google Scholar] [CrossRef]
- Li, S.B.; OuYang, W.Z.; Hou, X.J.; Xie, L.L.; Hu, C.G.; Zhang, J.Z. Genome-wide identification, isolation and expression analysis of Auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front. Plant Sci. 2015, 6, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, U.C.; Difazio, S.P.; Brunner, A.M.; Tuskan, G.A. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007, 7, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Yue, Y.Z.; Li, H.Y.; Ding, W.J.; Chen, G.W.; Shi, T.T.; Chen, J.; Park, M.S.; Chen, F.; Wang, L. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic. Res. 2018, 5, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Li, J.S.; Dai, X.H.; Zhao, Y.D. A role for Auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 2006, 140, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.C.; Sun, M.H.; Xu, R.R.; Shu, H.R.; Wang, J.W.; Zhang, S.Z. Genome-wide identification and expression analysis of the ARF gene family in apple. J. Genet. 2014, 93, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server, interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.N.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD, a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, 225–229. [Google Scholar] [CrossRef]
- Shen, C.; Wang, S.; Bai, Y.; Wu, Y.; Zhang, S.; Chen, M.; Chen, M.; Guilfoyle, T.J.; Wu, P.; Qi, Y. Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). J. Exp. Bot. 2012, 61, 3971–3981. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2012, 40, 290–301. [Google Scholar] [CrossRef]
- Zouine, M.; Fu, Y.; Chateigner-Boutin, A.L.; Mila, I.; Frasse, P.; Wang, H.; Audran, C.; Roustan, J.P.; Bouzayen, M. Characterization of the tomato ARF gene family uncovers a multilevels post-transcriptional regulation including alternative splicing. PLoS ONE 2014, 9, e84203. [Google Scholar] [CrossRef]
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.K.; Lu, C.H. CELLO2GO: A Web Server for Protein subCELlular LOcalization Prediction with Functional Gene Ontology Annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2014, 45, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0 toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, 1040–1045. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Yue, Y.Z.; Yin, C.Q.; Guo, R.; Peng, H.; Yang, Z.N.; Liu, G.F.; Bao, M.; Hu, H. An anther-specific gene PhGRP is regulated by PhMYC2 and causes male sterility when overexpressed in Petunia anthers. Plant Cell Rep. 2017, 36, 1401–1415. [Google Scholar] [CrossRef]
- Yue, Y.Z.; Tian, S.Z.; Wang, Y.; Ma, H.; Liu, S.Y.; Wang, Y.Q.; Hu, H. Transcriptomic and GC-MS metabolomic analyses reveal the sink strength changes during Petunia anther development. Int. J. Mol. Sci. 2018, 19, 955. [Google Scholar] [CrossRef]
- Zhang, C.; Fu, J.X.; Wang, Y.G.; Bao, Z.Y.; Zhao, H.B. Identification of suitable reference genes for gene expression normalization in the quantitative real-time PCR analysis of Sweet Osmanthus (Osmanthus fragrans Lour.). PLoS ONE 2015, 10, e136355. [Google Scholar] [CrossRef]
- Zhang, F.J.; Jin, Y.J.; Xu, X.Y.; Lu, R.C.; Chen, H.J. Study on the Extraction, Purification and Quantification of Jasmonic Acid, Abscisic acid and Indole-3-acetic Acid in Plants. Phytochem. Anal. 2008, 19, 560–567. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of Auxin-Regulated Gene Expression in Plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef]
- Zhao, Y.D. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 2, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.D.; Yuan, C.C.; Li, H.L.; Lin, W.H.; Yang, Y.J.; Shen, C.J.; Zheng, X. Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genom. 2015, 16, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Liscum, E.; Reed, J.W. Genetics of Aux/IAA action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Exon No. a | Len b (aa) | MW c (kDa) | pI d | Instability Index | GRAVY e | Conserved Domain | Homologous |
|---|---|---|---|---|---|---|---|---|---|
| OfARF1a | Contig151.75 | 4 | 702 | 77.19 | 7.29 | 48.43 | −0.305 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF1b | Contig73.170 | 6 | 760 | 83.87 | 7.82 | 46.64 | −0.289 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF2 | Contig328.11 | 4 | 685 | 74.8 | 5.81 | 40.99 | −0.305 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF3a | Contig281.61 | 4 | 687 | 75.86 | 7.91 | 50.00 | −0.322 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF3b | Contig592.12 | 5 | 646 | 71.52 | 6.55 | 46.00 | −0.248 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF4a | Contig37.40 | 3 | 674 | 74.86 | 6.43 | 47.95 | −0.370 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF4b | Contig295.42 | 2 | 601 | 66.52 | 6.36 | 42.06 | −0.428 | B3, ARF | AtARF16 |
| OfARF5a | Contig275.68 | 4 | 704 | 77.28 | 6.48 | 44.24 | −0.283 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF5b | Contig10.361 | 3 | 690 | 76.3 | 6.69 | 47.23 | −0.332 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF6a | Contig86.172 | 3 | 681 | 75.78 | 6.02 | 48.21 | −0.374 | B3, ARF, Aux/IAA | AtARF16 |
| OfARF6b | Contig12.215 | 2 | 677 | 75.43 | 7.25 | 43.42 | −0.407 | B3, ARF, Aux/IAA | AtARF10 |
| OfARF7a | Contig84.135 | 2 | 552 | 60.82 | 5.25 | 47.13 | −0.355 | B3, ARF | AtARF17 |
| OfARF7b | Contig77.21 | 2 | 552 | 60.85 | 5.95 | 46.36 | −0.281 | B3, ARF | AtARF17 |
| OfARF8a | Contig38.67 | 2 | 372 | 42.26 | 6.8 | 41.46 | −0.257 | B3, ARF | AtARF17 |
| OfARF8b | Contig38.68 | 2 | 478 | 53.89 | 6.06 | 42.24 | −0.343 | B3, ARF | AtARF17 |
| OfARF9a | Contig26.229 | 11 | 692 | 75.99 | 6.33 | 60.34 | −0.451 | B3, ARF | AtARF3 |
| OfARF9b | Contig172.11 | 11 | 726 | 79.71 | 6.92 | 48.75 | −0.367 | B3, ARF | AtARF3 |
| OfARF10a | Contig151.50 | 13 | 753 | 84.46 | 6.05 | 51.77 | −0.432 | B3, ARF, Aux/IAA | AtARF4 |
| OfARF10b | Contig73.186 | 12 | 787 | 87.64 | 5.97 | 48.99 | −0.409 | B3, ARF, Aux/IAA | AtARF4 |
| OfARF11a | Contig324.17 | 14 | 659 | 73.98 | 5.84 | 65.97 | −0.551 | B3, ARF, Aux/IAA | AtARF1 |
| OfARF11b | Contig97.3 | 14 | 653 | 73.33 | 5.76 | 64.99 | −0.525 | B3, ARF, Aux/IAA | AtARF1 |
| OfARF12 | Contig194.92 | 14 | 665 | 74.53 | 6.08 | 61.36 | −0.520 | B3, ARF, Aux/IAA | AtARF1 |
| OfARF13 | Contig76.252 | 14 | 842 | 94.36 | 6.28 | 56.97 | −0.568 | B3, ARF, Aux/IAA | AtARF2 |
| OfARF14a | Contig6.38 | 14 | 828 | 92.59 | 6.06 | 61.49 | −0.549 | B3, ARF, Aux/IAA | AtARF2 |
| OfARF14b | Contig2.130 | 14 | 838 | 93.5 | 6.02 | 64.87 | −0.609 | B3, ARF, Aux/IAA | AtARF2 |
| OfARF15 | Contig105.88 | 10 | 332 | 37.46 | 7.26 | 55.57 | −0.436 | B3, ARF | AtARF9 |
| OfARF16a | Contig317.63 | 14 | 658 | 74.16 | 6.7 | 57.20 | −0.550 | B3, ARF, Aux/IAA | AtARF9 |
| OfARF16b | Contig153.78 | 12 | 411 | 46.67 | 6.77 | 60.92 | −0.372 | B3, ARF | AtARF9 |
| OfARF17a | Contig54.9 | 14 | 666 | 74.97 | 6.67 | 56.90 | −0.414 | B3, ARF, Aux/IAA | AtARF9 |
| OfARF17b | Contig21.49 | 14 | 634 | 71.54 | 6.33 | 49.95 | −0.476 | B3, ARF, Aux/IAA | AtARF9 |
| OfARF18 | Contig354.37 | 15 | 720 | 81.77 | 9.05 | 50.43 | −0.551 | B3, ARF, Aux/IAA | AtARF18 |
| OfARF19 | Contig389.17 | 14 | 685 | 76.95 | 6.06 | 52.79 | −0.436 | B3, ARF, Aux/IAA | AtARF18 |
| OfARF20a | Contig267.9 | 14 | 680 | 76.3 | 6.26 | 55.17 | −0.516 | B3, ARF, Aux/IAA | AtARF18 |
| OfARF20b | Contig390.3 | 14 | 686 | 77 | 6.19 | 55.97 | −0.514 | B3, ARF, Aux/IAA | AtARF18 |
| OfARF21a | Contig314.6 | 14 | 893 | 98.79 | 6.11 | 67.35 | −0.362 | B3, ARF, Aux/IAA | AtARF6 |
| OfARF21b | Contig236.41 | 14 | 893 | 98.5 | 5.74 | 65.08 | −0.339 | B3, ARF, Aux/IAA | AtARF6 |
| OfARF22a | Contig109.86 | 14 | 890 | 98.99 | 6.2 | 67.24 | −0.454 | B3, ARF, Aux/IAA | AtARF6 |
| OfARF22b | Contig154.52 | 14 | 898 | 99.9 | 6.07 | 68.19 | −0.462 | B3, ARF, Aux/IAA | AtARF6 |
| OfARF23 | Contig72.89 | 15 | 763 | 86.2 | 6.16 | 61.71 | −0.456 | B3, ARF, Aux/IAA | AtARF8 |
| OfARF24a | Contig189.73 | 14 | 838 | 93.16 | 6.04 | 57.06 | −0.445 | B3, ARF, Aux/IAA | AtARF8 |
| OfARF24b | Contig165.100 | 14 | 839 | 93.65 | 6.04 | 57.64 | −0.478 | B3, ARF, Aux/IAA | AtARF8 |
| OfARF25a | Contig158.77 | 14 | 935 | 103.58 | 5.35 | 56.60 | −0.440 | B3, ARF, Aux/IAA | AtARF5 |
| OfARF25b | Contig16.261 | 15 | 809 | 90.34 | 5.22 | 56.51 | −0.292 | ARF, Aux/IAA | AtARF5 |
| OfARF26a | Contig183.139 | 13 | 1026 | 113.47 | 6.16 | 61.78 | −0.567 | B3, ARF, Aux/IAA | AtARF7 |
| OfARF26b | Contig259.21 | 13 | 1028 | 113.72 | 6.16 | 62.17 | −0.573 | B3, ARF, Aux/IAA | AtARF7 |
| OfARF27 | Contig248.99 | 11 | 390 | 43.46 | 8.26 | 64.10 | −0.282 | B3, ARF | AtARF7 |
| OfARF28a | Contig312.37 | 14 | 1091 | 120.3 | 6.07 | 56.21 | −0.516 | B3, ARF, Aux/IAA | AtARF7 |
| OfARF28b | Contig116.35 | 14 | 1078 | 119.52 | 6.17 | 56.52 | −0.512 | B3, ARF, Aux/IAA | AtARF7 |
| OfARF29a | Contig132.116 | 15 | 964 | 106.71 | 6.44 | 58.78 | −0.555 | B3, ARF, Aux/IAA | AtARF7 |
| OfARF29b | Contig8.81 | 14 | 1108 | 122.67 | 6.3 | 57.79 | −0.553 | B3, ARF, Aux/IAA | AtARF7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Yue, Y.; Li, L.; Li, Y.; Li, H.; Ding, W.; Shi, T.; Yang, X.; Wang, L. Genome-Wide Identification of the Auxin Response Factor (ARF) Gene Family and Their Expression Analysis during Flower Development of Osmanthus fragrans. Forests 2020, 11, 245. https://doi.org/10.3390/f11020245
Chen G, Yue Y, Li L, Li Y, Li H, Ding W, Shi T, Yang X, Wang L. Genome-Wide Identification of the Auxin Response Factor (ARF) Gene Family and Their Expression Analysis during Flower Development of Osmanthus fragrans. Forests. 2020; 11(2):245. https://doi.org/10.3390/f11020245
Chicago/Turabian StyleChen, Gongwei, Yuanzheng Yue, Ling Li, Yuli Li, Haiyan Li, Wenjie Ding, Tingting Shi, Xiulian Yang, and Lianggui Wang. 2020. "Genome-Wide Identification of the Auxin Response Factor (ARF) Gene Family and Their Expression Analysis during Flower Development of Osmanthus fragrans" Forests 11, no. 2: 245. https://doi.org/10.3390/f11020245
APA StyleChen, G., Yue, Y., Li, L., Li, Y., Li, H., Ding, W., Shi, T., Yang, X., & Wang, L. (2020). Genome-Wide Identification of the Auxin Response Factor (ARF) Gene Family and Their Expression Analysis during Flower Development of Osmanthus fragrans. Forests, 11(2), 245. https://doi.org/10.3390/f11020245




