Abstract
The rubber tree (Hevea brasiliensis) is a widely cultivated crop in tropical acidic soil that is tolerant to high concentration of aluminum and the aluminum-activated malate transporter (ALMT) plays an important role in plant aluminum detoxification. However, the effects of ALMT on rubber tree aluminum tolerance, growth performance, and latex production are unclear. In this study, 17 HbALMT genes were identified from the genome of rubber trees. The physiological and biochemical characteristics, phylogenetic relationships, gene structures, conserved motifs, cis-elements of promoter, and expression patterns of the identified HbALMT genes were studied. Phylogenetic relationships indicated that these genes were divided into four clusters and genes in the same cluster have similar gene structures and conserved motifs. The promoters of HbALMT genes contain many cis-elements associated with biotic stress and abiotic stress. Quantitative real-time PCR analysis revealed HbALMTs showed various expression patterns in different tissues, indicating the functional diversity of HbALMT genes in different tissues of rubber trees. Transcriptome analysis and qRT-PCR assay showed that most of the HbALMT genes responded to aluminum stress, and among the 17 HbALMTs, HbALMT1, HbALMT2, HbALMT13, and HbALMT15 displayed higher expression levels in roots after two or five days of Al treatments, indicating their potential involvement in aluminum detoxification. Taken together, this study laid a foundation for further understanding the molecular evolution of the ALMT genes and their involvement in rubber tree aluminum adaption.
1. Introduction
Soil acidification is a major stress that affects about 50% of potentially arable lands throughout the world, particularly in tropical and subtropical zones [1,2]. Soil acidity is being gradually exacerbated by an increase in environmental problems, including acid rain and inappropriate farming practices [3]. The primary cause of poor acid soil fertility is the toxicity of metal elements and the loss of nutrients, among which aluminum (Al) toxicity is the most prominent constraint to worldwide crop production in acidic soils [4]. When the soil is acidic (pH < 5), Al3+ cation is released from the aluminosilicate clay complexes and absorbed by the plant roots [5]. The Al3+ cation would combine with plant roots, forming a “short, thick and unbranched” root system, which would inhibit root normal growth and physiological function, and thereby restrict the yield of crops [6].
In the process of long-term evolution, many plant species in acidic soils have developed a series of tolerance mechanisms to cope with and resist Al toxicity [7]. Gramineous plants, such as maize and wheat, show signs of inhibiting root growth within minutes or hours of being exposed to Al stress, even at micromolar concentration levels [3,8]. However, rice which is the most Al-tolerant cereal crop shows 2-6 times more resistance to Al than maize, wheat, and sorghum [9]. In addition, some plant species, such as buckwheat, tea plant, and hydrangea (Hydrangea macrophylla) can also accumulate high concentrations of Al without showing toxic symptoms [10,11,12]. In previous studies, we found that rubber tree saplings could tolerate 100–200 mmol/L of Al at pH 4.2, which is far higher than other crops [13]. Broader cultivation of these Al-resistant crops will help promote the development of sustainable agriculture. Nevertheless, the underlying mechanism, especially the molecular mechanism of plant Al tolerance, is still unclear.
The physiological mechanism of plant resistance to Al toxicity can be divided into two categories that include external exclusion (exclusion of Al from the root apex) and internal tolerance (intracellular tolerance of Al transported into the plant symplasm) [14]. A large number of studies have shown that organic acid secretion is the most important external exclusion mechanism for plants to adapt to the Al toxicity [4,14]. Organic acid anions such as malate, citrate, and oxalate reduce Al toxicity by forming a stable chelate with harmful Al3+ cations preventing Al from entering the symplast [7,15]. However, how organic acids secrete from roots to the root rhizosphere soil was still unknown until the identification of Al-activated malate transporter (TaALMT1) from wheat [16].
The aluminum-activated malate transporter (ALMT) gene encodes anion transporter/channel and it can transport organic and inorganic anions through the tonoplast and plasma membrane in cells [15,17]. Genes encoding malate transporters have been found in many plant species, such as Arabidopsis (AtALMT1), rape (BnALMT1, BnALMT2), rye (ScALMT1), and soybean (GmALMT1) [18,19,20,21]. Furthermore, more studies showed that members of the ALMT family were not only associated with Al resistance but also participated in other physiological processes, including stomatal movement, cellular signaling, cell expansion, nutrients acquisition, fruit flavor regulation, and GABA (γ–aminobutyric acid) signaling [22,23,24]. Nevertheless, the ALMT family member has not been reported in the rubber trees.
The rubber tree (Hevea brasiliensis) is one of the most important cash trees widely cultivated in tropical regions. It is native to the Amazon forest and is the sole commercially cultivated source of natural rubber (cis-1, 4-polyisoprene) which originates from the milky latex flowing out once rubber trees are tapped [25]. Rubber trees are cultivated in about 95,000 hectares in 45 countries [26]. They are the main income source of smallholders in many countries, especially Southeast Asia. In these regions, soil acidification and aluminum toxicity has been a prominent challenge for rubber tree cultivation [27]. However, the physiological effect of Al toxicity on rubber trees is unclear.
We previously reported that Al toxicity affected rubber tree growth performance and that it was probably associated with rubber tree tapping panel dryness and decreasing yields [13]. Even though the rubber tree is an Al tolerant plant, its Al tolerant mechanism has not been studied. It was reported that ALMTs played vital roles in Al detoxification of many plants [15,17]. Therefore, identification of ALMTs from rubber tree genome will be helpful for the comprehensive understanding of the Al tolerant mechanism and for the development of Al-resistant cultivars. To identify the rubber tree ALMTs and investigate their involvement in rubber tree aluminum detoxification, the whole family of ALMT genes was identified from the published rubber tree genomes—their phylogenetic classification, gene structures, tissue specific expression, and expression subjected to various Al concentrations were analyzed.
2. Materials and Methods
2.1. Plant Materials and Treatments
The most widely planted high-yielding Reyan 7-33-97 rubber tree cultivar was used as the plant material. The two-whorled-leaf old tissue culture rubber tree saplings with similar height, stem diameter, number of leaves, and growth stage, were cultured in the Hoagland’s solution (2.8 mg/L H3BO3, 3.4 mg/L MnSO4·H2O, 0.1 mg/L CuSO4·5H2O, 0.22 mg/L ZnSO4·7H2O, 0.1 mg/L (NH4)6·7H2O, 20 mg/L Na2Fe-EDTA, 0.94 g/L Ca(NO3)2·4H2O, 0.52 g/L MgSO4·7H2O, 0.66 g/L KNO3, 0.12 g/L NH4H2PO4) in a growth chamber. The growth chamber was set to 28 °C/day (16 h) and 25 °C/night (8 h) cycle and the saplings were treated with Hoagland’s solution (pH = 4.2) supplemented with or without 200 mM of AlCl3 for 0 (CK), 2 or 5 d, respectively. The nutrient solution was changed every two days. The roots from 4 saplings for each treatment were collected as a pool for RNA extraction.
2.2. Identification, Sequence Alignments, and Phylogenetic Analysis of Rubber Tree ALMT Genes
The whole genome sequence of rubber trees was downloaded from National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/). All the amino acid sequences of ALMTs in Arabidopsis were obtained from TAIR database (http://www.arabidopsis.org/) and were used as queries for BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The default parameters (e-value ≤ 1e-5) against rubber tree genome database were used. The HMM profile of the predicted rubber tree ALMT protein domain (PF11744) was confirmed by HMMER3.0 software using AtALMTs as the queries and the deduced HbALMT proteins were verified using conserved domain database (CDD, https://www.ncbi.nlm.nih.gov/cdd). Multiple sequence alignment was executed in ClustalX with the default parameters (gap opening, 10; gap extension, 0.2; delay divergent sequences (%), 30; DNA Transition weight, 0.5; use negative matrix, off) [28]. Furthermore, to facilitate the evolutionary analysis of HbALMTs, a similar method was also used to identify homologs from Populus and other representative Euphorbiaceae family plant genomes such as cassava, Jatropha curcas, and Ricinus communis, which were obtained from Phytozome v12 (https://phytozome.jgi.doe.gov/pz/portal.html) or NCBI (http://www.ncbi.nlm.nih.gov/) (see Figure 1 and Supplementary Table S1). Phylogenetic analysis was performed using MEGA 7.0 software with the neighbor-joining method by setting bootstrap value to 1000 replicates.
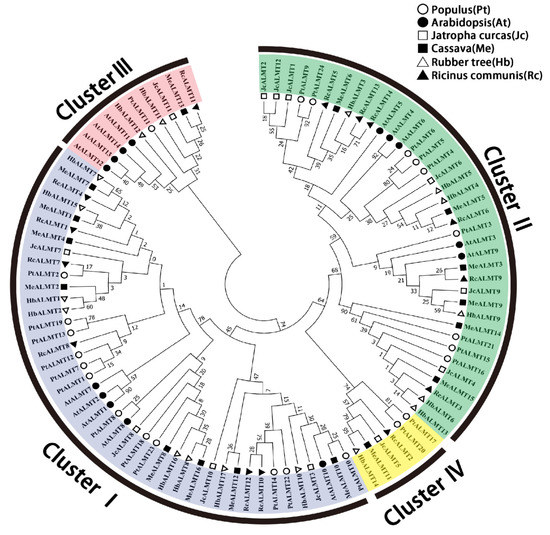
Figure 1.
Phylogenetic analysis of ALMTs from Populus, Arabidopsis, Jatropha curcas, cassava, rubber tree, and Ricinus communis using the complete protein sequences. The phylogenetic tree was constructed by MEGA 7.0 using the neighbor-joining method with 1000 bootstrap replicates and displayed by iTOL v4 online software. The four clusters of HbALMT proteins are distinguished by different colors.
2.3. Protein Properties, Gene Structure, Conserved Motif, and Promoter Analysis
Using online ExPASy software (https://web.expasy.org/protparam/), the basic information about ALMT proteins, including the number of amino acid, molecular weight (MW), and isoelectric point (pI) was collected and predicted. The transmembrane region was predicted by using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). The subcellular location was predicted with ProtComp v.9.0 (http://www.softberry.com/berry.phtml). The ALMT gene structures were drawn using Gene Structure Display Sever V2.0 (http://gsds.cbi.pku.edu.cn/) [29] and TBtools [30]. Motifs in ALMT proteins were identified by MEME program (http://meme-suite.org/tools/meme) with the motif width from 6 to 50 and maximum number of different motifs to be 15. The upstream sequences (2000 bp) of the HbALMT coding region were submitted to Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to detect their cis-elements and 7 resultant cis-elements (i.e., ABRE, involved in the ABA responsiveness; ERE, ethylene-responsive element; LTRE, low temperature-responsive element; GARE, gibberellin-responsive element; MBS, MYB binding site involved in drought-inducibility; TC-rich repeats, involved in defense and stress responsiveness; W-box, binding site of WRKY transcription factor in defense response) related to stress response were further analyzed.
2.4. cDAN Library Construction and Transcriptome Sequencing
The total RNA of each sample was extracted with a plant RNA extraction kit (TIANGEN, China) according to the manufacturer’s instructions. The residual genomic DNA was removed by treating RNA samples with RNase-free DNase I (TIANGEN, China). Subsequently, RNA purity and concentration were verified with a spectrophotometer (NANODROP 2000c, ThermoScientific, USA). The cDNA library was conducted based on the Illumina manufacturer’s instructions [31,32]. The cDNA library was sequenced using Illumina HiseqTM 2000 platform as operated by Guangdong Longsee Biomedical Corporation (Guangzhou, China). Clean reads were obtained by after filtering the adapter and low-quality reads using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Trinity program was used for the de novo transcriptome assembly of clean reads into unigenes [33]. The expression of unigenes was calculated and normalized to fragments per kilobase of transcript per million fragments mapped (FPKM) values [34]. The expression of unigenes was calculated and normalized to FPKM values. To annotate the assembled unigenes, sequences were subjected to BLASTx (http://www. ncbi.nlm.nih.gov/BLAST/) searching of four databases (e-value < 1e-5), including the NCBI non-redundant protein (Nr) database (http://www.ncbi.nlm.nih.gov), the Swiss-Prot protein database (http://www.expasy.ch/sprot), the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg), and the COG database (http://www.ncbi.nlm.nih.gov/COG).
2.5. Real-time Quantitative PCR (qRT-PCR) Analysis
The cDNA templates were synthesized by reverse transcribing the isolated RNAs with a RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania) according to the manufacturer’s protocol. Real-time qPCR reaction was performed utilizing the CFX96 real-time PCR system (Bio-Rad, Hercules, CA, USA) and SYBR®Green Premix kit (Takara, Dalian, China). Each qRT-PCR reaction volume contained 10 μL of 2 × SYBR Premix ExTaqTM, 0.4 μL of each primer (10 μM), 1 μL of cDNA, and 8.2 μL of RNase-free water. The qRT-PCR reaction program was set as follows: 95 °C for 3 min, followed by 45 cycles of 95 °C for 10 s and 60 °C for 20 s, and 30 s of extension at 72 °C. The rubber tree UBC4 (GenBank: HQ323249) gene, which was proved to be the most stable housekeeping gene in response to Al stress, was selected as the internal reference gene [35]. The qRT-PCR data was analyzed with 2−ΔΔCT method [36]. Each treatment was performed with at least four biological replicates and three technical replicates. All the gene specific primers were designed by using Primer 5.0 software and listed in Supplementary Table S2.
2.6. Statistical Analysis
Statistical analyses were performed using the SAS 9.1 software (SAS Institute Inc., Cary, NC, USA) and the values were presented as mean ± standard deviation (SD). Different letters represent significant differences (P < 0.05) determined by a Duncan one-way analysis of variance.
3. Results
3.1. Identification and Characterization of ALMT Gene Family in Rubber Trees
A total of 17 members of the ALMT homologous genes were identified in the rubber tree genome by using BLAST and HMMER, which were named as HbALMT1-HbALMT17 according to the phylogenetic relationship with Arabidopsis. The basic properties of HbALMTs that include gene name, gene ID, the lengths of genomic and coding region, amino acid numbers, molecular weight (MW), and theoretical isoelectric point (pI) were listed in Table 1. The lengths of CDS ranged from 1266 (HbALMT14) to 1788 bp (HbALMT4), coding 421 (HbALMT14) to 625 (HbALMT4) amino acids; the molecular weights were varied from 46.47 to 69.81 kDa and the theoretical isoelectric point (pI) was 5.22 (HbALMT5)-8.89 (HbALMT15). All identified HbALMTs were predicted to contain 5 to 7 transmembrane regions in the N-terminus of the ALMT proteins (Table 1, Supplementary File S1). Subcellular localization prediction showed that 11 HbALMTs were localized on the plasma membrane and 6 HbALMTs were located on the endoplasmic reticulum. The HbALMTs protein sequences were aligned by ClustalX, and sequences of the 17 identified HbALMTs proteins were conserved (Supplementary File S2). The sequence similarity of HbALMTs at the amino acid level was between 28.82% and 95.19%, and HbALMT11 and HbALMT12 showed the highest sequence similarity (95.19%) (Supplementary Table S3). Conserved domain analysis demonstrated all identified 17 HbALMTs harbored ALMT domains, which was a characteristic of the ALMT family (Supplementary Table S4).

Table 1.
The basic data of aluminum-activated malate transporter (ALMT) family members in rubber tree.
3.2. Phylogenetic Analysis of HbALMTs
As shown in Figure 1, phylogenetic analysis of the 17 HbALMTs, 14 AtALMTs, 24 PtALMTs, 16 MeALMTs, 12 JcALMTs, and 14 RcALMTs was conducted to demonstrate the evolutionary relationship and putative functions of HbALMT genes. The results showed that 97 ALMTs could be classified into four clusters. Most of the ALMTs, including 14 HbALMTs, were split into Cluster I and Cluster II; the same results were found in Arabidopsis. For rubber trees, Cluster I contained HbALMT1,-2,-7,-8,-10,-15,-16,-17; Cluster II included HbALMT3,-4,-5,-6,-9,-13; Cluster III consisted of HbALMT11,-12 and Cluster IV harbored HbALMT14. Furthermore, Cluster IV was specific to woody plants. It was found that ALMTs in Cluster IV were more closely related to ALMTs from different species than other ALMTs from the same species, which implied a relatively high homology between the same ALMT cluster across various species.
3.3. Gene Structure of HbALMTs
All the identified HbALMT genes were further explored to detect their exon-intron structures. As shown in Figure 2, all identified HbALMT genes possessed five to eight exons. Most of them contained six exons, while only HbALMT14 contained five exons, HbALMT1 and HbALMT5 contained seven exons, and HbALMT6 and HbALMT17 contained eight exons. In addition, genes within the same cluster usually displayed similar structure—Cluster I contained six exons except HbALMT1 and HbALMT16, Cluster III contained six exons, and Cluster IV contained five exons.
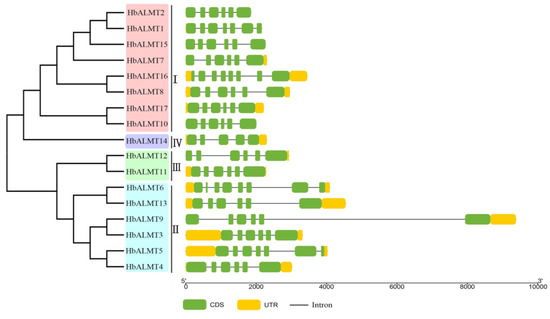
Figure 2.
Gene structures of HbALMTs according to their phylogenetic relationship. The phylogenetic tree was constructed by MEGA 7.0 using the neighbor-joining method with 1000 bootstrap replicates. Gene structure analyses of the HbALMT genes were performed with the online website GSDS and TBtools software. Green boxes indicate exons; orange boxes indicate untranslated regions; black line indicates introns.
3.4. Conserved Motif Analysis of HbALMTs
We obtained 15 conserved motifs for the 17 HbALMT proteins through the online MEME website search (Figure 3). The lengths of those conserved motifs varied between 15 and 50 amino acids. Annotating the obtained motifs using InterProScan online server, the results showed that motif 1–10 were harbored in the ALMT domain (Pfam No. PF11744) and were closely related to Al-activated malate transport and Al3+ resistance (Supplementary Table S4). As shown in Figure 3, all the identified HbALMTs contained motif 2–9, suggesting that the majority of Al-activated malate transporter motifs existed in all HbALMTs. Furthermore, most HbALMT proteins shared similar motifs features within subfamilies, which further supported the phylogenetic classification of HbALMTs.
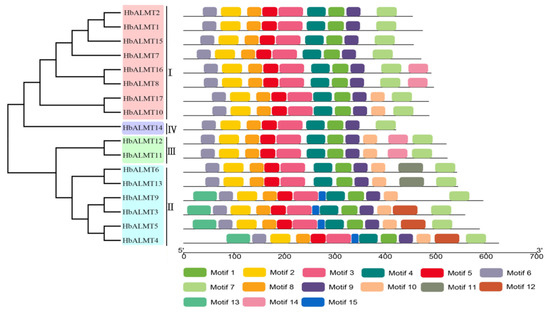
Figure 3.
Conserved motifs of HbALMT proteins based on their phylogenetic relationship. The phylogenetic tree was constructed by MEGA 7.0 using the neighbor-joining method with 1000 bootstrap replicates. The conserved motifs of the HbALMT proteins were detected using online MEME program and drawn using TBtools software. Motifs 1–15 were displayed in different colored boxes. The sequence information of each motif was supplied with Supplementary Table S5. The length of protein could be estimated using the scale at the bottom.
3.5. The Cis-elements Prediction of HbALMT Promoters
Cis-elements of HbALMT promoters analysis indicated that all of the HbALMT promoters except HbALMT7 and HbALMT13 contained one or more cis-elements related to plant hormone responses (Figure 4). For example, 58.8% of HbALMT genes contained ABRE, 64.7% contained ERE, and two genes (HbALMT10,-17) harbored GARE. Additionally, the W box and TC-rich repeats appeared in eight HbALMT genes, the MBS cis-element was located in seven HbALMT genes, and LTRE was found in three HbALMTs (HbALMT1,-5,-13). All HbALMTs contained at least one of the tested elements in their promoter region, indicating HbALMT genes may be involved in plant hormone and stress responses.
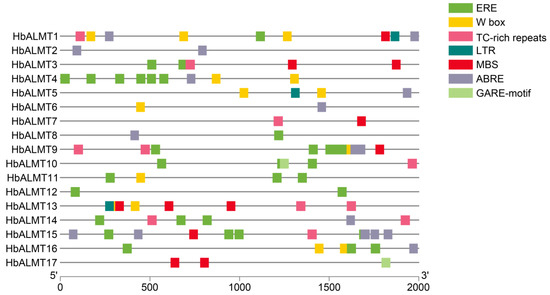
Figure 4.
Predicted cis-elements in HbALMT promoters. The 2000 bp upstream HbALMT gene sequences were chosen to detect promoter cis-elements of HbALMTs by PLANTCARE database. ABRE, involved in the ABA responsiveness; ERE, ethylene-responsive element; LTRE, low temperature-responsive element; GARE, gibberellin-responsive element; MBS, MYB binding site involved in drought-inducibility; TC-rich repeats, involved in defense and stress responsiveness; W-box, binding site of WRKY transcription factor in defense response. Different color boxes represent cis-elements.
3.6. Expression Profiles of HbALMTs in Different Tissues
The tissue specific expression of HbALMT genes in different tissues, i.e., root, stem tip, leaf, bark, latex of rubber tree, was analyzed by using qRT-PCR. As shown in Figure 5, the 17 HbALMT genes were expressed in all tested tissues, although the transcript abundance of some genes in the tissues was very low. The seven HbALMTs (HbALMT1,-2,-3,-6,-12,-13,-16) displayed quite similar expression profiles among the tested tissues—the expression levels of HbALMTs were shown to be most abundant in stem tips, moderate in root and leaf, and considerably low in latex. Some HbALMTs, i.e., HbALMT4,-5,-7,-8,-9,-10,-14 were significantly more expressed in roots and stem tips than other tissues, whereas 14 HbALMTs (HbALMT1,-2,-3,-4,-6,-7,-8,-9,-10,-11,-12,-13,-14,-16) had a lower expression level in bark and latex than other tissues. Notably, HbALMT5 and HbALMT9 showed lower transcript abundance than the other genes in all tissues. In addition, some genes were highly expressed in specific tissues. For example, HbALMT10 showed the highest expression pattern in roots and leaves compared to other genes and HbALMT4 and HbALMT15 displayed the highest expression pattern in bark and stem tips, respectively. These results indicated that HbALMTs were involved in various aspects of physiological and developmental processes of rubber tree.
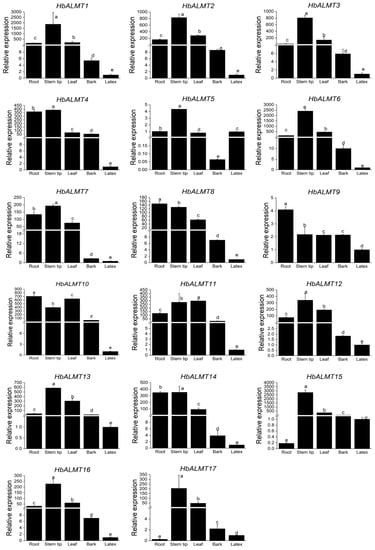
Figure 5.
Expression patterns of HbALMT genes in different rubber tree tissues. Total RNA was extracted from roots, stem tips, leaves, bark, and latex according to the manufacturer’s instructions. The relative expression of HbALMTs in different tissues was quantified by qRT-PCR with four biological replicates and three technical replicates. The rubber tree UBC4 gene (GenBank: HQ323249) was used as the internal control. To calculate the relative expression level of each gene in different tissues, the transcript level in latex was used to normalize the transcript levels in other tissues. Values were means ± SD of four biological replicates. Different lowercase letters above the bars represent significant differences (P < 0.05) determined by a Duncan one-way analysis of variance. All primer sequences were listed in Supplementary Table S2.
3.7. The Expression Pattern of HbALMTs in Response to Aluminum Stress
RNA-seq was used to investigate the transcript levels of each HbALMT gene under Al stress. As shown in Figure 6, a significant difference in the transcript abundance of HbALMT gene was found when the rubber trees were subjected to various Al treatment days. Most of the HbALMT genes were sensitive to Al stress, and only one gene (HbALMT6) was insensitive. Nearly half of the HbALMTs were upregulated after Al treatment for two days or five days. In order to further confirm the involvement of HbALMTs in rubber tree Al detoxification and validate the RNA-seq results, all the 17 identiified HbALMTs were selected for qRT-PCR assay. Many genes, such as HbALMT1,-2,-5,-11,-12,-13,-15 showed higher expression levels at Al treatments after two or five days in comparison with control. Among these, HbALMT1, -15 significantly upregulated at two days after Al treatment and further upregulated at five days after Al treatment, HbALMT2,-5,-13 significantly upregulated only at five days after Al treatment, and HbALMT11 significantly upregulated at two days after Al treatment but significantly down-regulated at five days after Al treatment. By contrast, other HbALMT genes, including HbALMT3,-7,-8,-9,-10,-14,-16,-17 were inhibited by Al treatments. These results indicate that all HbALMT genes, except HbALMT6, are involved in Al stress response. The expression profiles of HbALMTs obtained by qRT-PCR were generally consistent with the RNA-seq expression data, indicating the reliability of RNA-seq data.
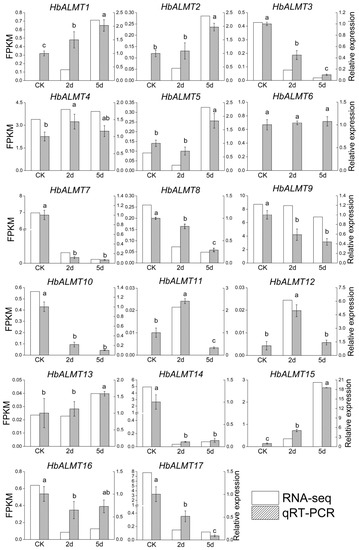
Figure 6.
Expression patterns of the 17 HbALMTs responding to Al treatment. Same plants and samples were used for qRT-PCR and RNA-seq measurements with four biological replicates and three technical replicates. The relative expression of HbALMTs in the rubber tree root under Al treatment was quantified by qRT-PCR, and the rubber tree UBC4 gene (GenBank: HQ323249) was used as the internal control. The average of Ct value for the UBC4 gene was 24.36 under Al treatment. The data are given as mean ± SD (n = 3). Different lowercase letters above the bars represent significant differences (P < 0.05) determined by a Duncan one-way analysis of variance. All primer sequences are listed in Supplementary Table S2.
4. Discussion
The ALMT genes are widespread in higher plants. The first plant ALMT gene, TaALMT1, was identified and characterized in wheat-root protoplasts from the aluminum tolerant line. TaALMT1 is an Al3+-activated malate transporter mediating malate transport from root cells into rhizosphere, which will chelate the Al3+ in acid soil reducing Al3+ damage to cell walls and membranes [16,37]. Subsequently, ALMT genes were identified from many plants and proved to be related to many physiological processes in plant, such as Al detoxification, mineral nutrition, stomatal movement, fruit acidity, seed development, and other abiotic stress [24,38,39,40]. It was found that there were 14 ALMT members in Arabidopsis thaliana, 16 members in Fragaria vesca, 9 members in rice, 27 members in Pyrus bretschneideri, 25 members in Malus domestica, 25 members in Prunus mume [22,23,38,41]. However, little is known to the ALMT genes in rubber trees. In the present study, we identified 17 HbALMTs, 24 PtALMTs, 16 MeALMTs, 12 JcALMTs, and 14 RcALMTs from the rubber tree, Polulus, cassava, Jatropha curcas, and Ricinus communis genomes by BLASTP search and HMMER analysis, respectively. The number of ALMT genes in H. brasiliensis was close to cassava, Jatropha curcas and Ricinus communis, which belong to Euphorbiaceae. Chromosomal location and gene duplication could provide more information for exploring the expansion and function of a gene family, which could occur via whole-genome duplication (WGD), segmental duplication, tandem duplication, and transposed duplication [42,43]. Nevertheless, due to the rubber tree draft genomes not being anchored to their chromosomes and the lack of a high-density genetic map in rubber tree, it is not able to determine the chromosomal localization and gene duplication type.
Phylogenetic evolutionary analysis plays a vital role in realizing the function of gene families. In earlier studies, 14 ALMT genes in Arabidopsis were classified into three clusters [41]; 34 soybean ALMT genes that belonged to two main groups were further divided into six subgroups [44]; nine ALMT genes in rice were clustered into five clades [45]; 25 ALMT genes in Malus domestica were classified into seven subgroups [38]; 13 grape ALMT genes were clustered into three clades [46]. These results indicated a large phylogenetic evolutionary classification difference among various plant species. In the present study, the phylogenetic tree showed that 17 HbALMTs were classified into four clusters. Cluster I was the largest cluster containing eight members, followed by Cluster II containing six members, Cluster III consists of two members, while Cluster IV harbored only one member. Unlike the evolutionary relationship classification of Arabidopsis ALMT genes, Cluster IV in the rubber tree ALMT genes evolutionary relationship classification was a unique class—similar results were found in Populus and other Euphorbiaceae plants such as cassava, Jatropha curcas, and Ricinus communis.
In the phylogenetic evolutionary analysis, most ALMT genes were grouped into Cluster I, including 8 HbALMTs and 10 previously identified ALMTs, such as AtALMT1 and OsALMT1. All the eight HbALMTs harbored five or six transmembrane helix domains within the N-terminal half of the proteins and were localized on the plasma membrane. Additionally, most HbALMTs contained six exons. Arabidopsis and soybean ALMT proteins also possess similar structures [41,44]. Ligaba et al. [47] found that the transmembrane region of the N-domain in ALMTs was mainly responsible for protein transport activity, while the amino acids in the C-domain were mainly related to Al resistance. In previous study, two common motifs were found in the N-domain of the Chinese white pear ALMT gene, and there is a common motif in the C-domain [22]. In rubber tree, common motif 2 and 6 were detected in the transmembrane region of the N-domain, while motif 7 was found in the C-domain. Therefore, we inferred that motif 2 and 6 might be involved in protein transport activity and motif 7 may be associated with Al tolerance. Furthermore, HbALMT3,-4,-5,-6,-9,-13 in Cluster II were localized on the endoplasmic reticulum, indicating they may have similar functions. These results further support the classification of HbALMTs.
Gene expression analysis plays an important role in exploring gene function. The expression patterns of ALMT genes in different tissues have been reported in rice and soybean. In rice, OsALMT1,-2,-4 were expressed in leaves and roots, while OsALMT7,-9 were only expressed in roots [45]; gene specific expression patterns of 34 ALMTs in soybean were investigated in different tissues (tap roots, lateral roots, leaves, and flowers) under phosphate starvation treatment, the results showed that the expression levels of 26 GmALMTs in roots, leaves, and flowers were regulated [44]. In the present study, the expression patterns of 17 HbALMTs in five different tissues, including roots, stem tips, leaves, barks, and latex, were analyzed under normal growth conditions. As shown in Figure 5, all HbALMTs were presented in all tested tissues; HbALMT8,-9,-10 were more highly expressed in roots than other tissues, indicating that HbALMT8,-9,-10 may be involved in Al detoxification.
The function of ALMT genes have been characterized from many species. For example, the homologs of TaALMT1, including AtALMT1 (Arabidopsis), GmALMT1 (soybean), BnALMT1/2 (rape), ScALMT1 (rye), MsALMT1 (Medicago sativa), BoALMT1 (cabbage), and HlALMT1 (Holcus lanatus) shared similar functions, which were involved in Al resistance by mediating malate secretion from root tip [18,19,21,48,49,50,51]. Additional identified ALMT genes were associated with other physiological functions. For example, AtALMT12 was a guard cell R-type anion channel responsible for stomatal movement [52]; OsALMT4 protein was localized on the plasma membrane, and the overexpression of OsALMT4 could affect malate efflux and the distribution of certain minerals [45]; OsALMT7 plays an important role in maintaining the development of differentiated panicles [37]. All these studies demonstrated that members of the ALMT gene family exhibited multiple physiological functions in plants. However, little is known about the role of ALMT genes in the Al tolerance of rubber trees.
Aluminum toxicity is a major factor inhibiting crop growth on tropical acidic soil [2]. Since the rubber tree is mainly distributed in tropical regions, soil acidification and aluminum toxicity is a prominent challenge for rubber tree cultivation [27]. Nevertheless, studies on the effects of aluminum toxicity on rubber tree growth and latex production are very limited and a mechanism of rubber tree tolerates to Al has not been reported. In light of this, we are more focused on the potential roles of HbALMTs in rubber tree Al resistance. ALMT genes involved in Al resistance have been reported in a variety of plants, such as wheat, Arabidopsis, rape, cabbage, soybean, rye, and Medicago sativa [16,19,21,41,48,49,51]. In Arabidopsis, AtALMT1 was induced under Al stress and enhanced Al tolerance through mediating root malate efflux [18]. BnALMT1 and BnALMT2 were expressed in rape roots and associated with Al resistance [19]. Overexpression of MsALMT1 could effectively improve the Al resistance of transgenic tobacco plants [51]. BoALMT1 was an Al-resistant gene in cabbage increasing the secretion of malate to resist Al stress [48]. These studies laid a foundation for further investigation of the Al-tolerant genes in rubber trees. Based on the previous studies, we explored the expression levels of HbALMT genes. It was found that except for HbALMT6, the expression levels of the other 16 HbALMT genes were all changed under Al stress. Moreover, the homologs of AtALMT1, HbALMT1,-2,-15 in Cluster I and HbALMT13 in Cluster II were significantly upregulated after Al treatment, signifying that these genes might play a role in Al detoxification. Besides, Arabidopsis WRKY46 was a negative regulator of AtALMT1, mutation of WRKY46 increased malate secretion and decreased Al accumulation in root apices, and thus conferred the Al tolerance for Arabidopsis [53]. In the present study, W box cis-element was detected in the promoter regions of HbALMT1,-4,-6,-5,-9,-11,-13,-16. The WRKY gene family has also been identified in rubber tree [54], which provides an insight into investigating the role of rubber tree WRKY gene in Al stress response.
This study conducted a comprehensive genomic analysis of the rubber tree ALMT gene family and provided the first step in selecting candidate Al-tolerant genes for further study. However, the effects of HbALMT genes on rubber tree growth and latex production are still unclear. Further investigations are required to study the function of ALMT genes in rubber tree.
5. Conclusions
A total of 17 HbALMT genes were identified from the rubber tree genome. All identified HbALMTs can be divided into four clusters according to the phylogenetic relationship and contain similar gene structures and conserved motifs among each cluster. Cis-elements related to plant response to biological stress and abiotic stress were found in HbALMT promoters, suggesting that these genes may play a role in regulating plant development and defending against biological and abiotic stress. HbALMT genes displayed various expression levels in different rubber tree tissues based on qRT-PCR assay. RNA-seq analysis and qRT-PCR assay showed that 16 ALMT genes responded to Al stress except HbALMT6, among which four genes i.e., HbALMT1,-2,-13,-15 were significantly upregulated after Al stress. The study provides a guide for the selection of HbALMT gene for cloning and functional analysis so as to unveil its roles in regulating rubber tree aluminum tolerance and natural rubber biosynthesis.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/2/142/s1, File S1: The secondary domains of HbALMTs proteins. File S2: The protein sequence alignment of HbALMTs. Table S1: The basic data of ALMT family members in Arabidopsis, Populus, cassava, Jatropha curcas and Ricinus communis. Table S2: Primers used in qRT-PCR analysis. Table S3: Percent identity of the identified HbALMT genes at the amino acid level. Table S4: Conserved domain analysis of the rubber tree ALMT family. Table S5: Putative motifs of HbALMT proteins.
Author Contributions
X.M. and F.A. conceived and designed the experiments; X.M., F.A., L.W. and D.G. performed the experiments; X.M., F.A., G.X., and Z.L. analyzed the data; X.M. and F.A. wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31670633), Hainan Provincial Basic and Applied-basic Research Program (Natural Science) for High-level Talents (2019RC326), the Earmarked Fund for China Agriculture Research System (CARS-33-ZP1) and the Innovation Research Fund for Hainan Provincial Graduate Student (Hys2019-149).
Conflicts of Interest
The authors declare that they have no competing interests for this study.
References
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.T.; Vitousek, P.M.; Zhang, F.S. Significant acidification in major Chinese croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef]
- Magalhaes, J.V. Aluminum tolerance genes are conserved between monocots and dicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9749–9750. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, J.; Song, X.; Zhang, Z.; Jiang, Y.; Zhu, Y.; Zhao, H.; Ni, D. An RNA-Seq transcriptome analysis revealing novel insights into aluminum tolerance and accumulation in tea plant. Planta 2017, 11, 1–103. [Google Scholar] [CrossRef]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Kochian, L.V.; Piñeros, M.A.; Hoekenga, O.A. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 2005, 274, 175–195. [Google Scholar] [CrossRef]
- Ma, J.F.; Furukawa, J. Recent progress in the research of external Al detoxification in higher plants: A minireview. J. Inorg. Biochem. 2003, 97, 46–51. [Google Scholar] [CrossRef]
- Ma, J.F. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Inter. Rev. Cytol. 2007, 264, 225–252. [Google Scholar]
- Horst, W.J.; Wang, Y.; Eticha, D. The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: A review. Ann. Bot. 2010, 106, 185–197. [Google Scholar] [CrossRef]
- Famoso, A.N.; Clark, R.T.; Shaff, J.E.; Craft, E.; McCouch, S.R.; Kochian, L.V. Development of a novel aluminum tolerance phenotyping platform used for comparisons of cereal aluminum tolerance and investigations into rice aluminum tolerance mechanisms. Plant Physiol. 2010, 153, 1678–1691. [Google Scholar] [CrossRef]
- Ma, J.F.; Hiradate, S.; Matsumoto, H. High aluminum resistance in buckwheat (II. oxalic acid detoxifies aluminum internally). Plant Physiol. 1998, 117, 753–759. [Google Scholar] [CrossRef]
- Hajiboland, R.; Bahrami Rad, S.; Barceló, J.; Poschenrieder, C. Mechanisms of aluminum-induced growth stimulation in tea (Camellia sinensis). J. Plant Nutr. Soil Sci. 2013, 176, 616–625. [Google Scholar] [CrossRef]
- Ma, J.F.; Hiradate, S.; Nomoto, K.; Iwashita, T.; Matsumoto, H. Internal detoxification mechanism of Al in Hydrangea (identification of Al form in the leaves). Plant Physiol. 1997, 113, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Li, C.Z.; Zhang, T.T.; Wang, L.F.; Wang, J.K.; Xie, G.S. Effects of aluminum toxicity on physiological and leaf chlorophyll fluorescent characteristics of rubber tree seedlings. Ying Yong Sheng Tai Xue Bao 2018, 29, 4191–4198. [Google Scholar]
- Kochian, L.V.; Pineros, M.A.; Liu, J.; Magalhaes, J.V. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Ann. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.; Dreyer, I.; Kochian, L.; Piñeros, M.A. The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front. Plant Sci. 2016, 7, 1488. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Yamamoto, Y.; Ezaki, B.; Katsuhara, M.; Ahn, S.J.; Ryan, P.R.; Delhaize, E.; Matsumoto, H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004, 37, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, M. The ALMT gene family performs multiple functions in plants. Agronomy 2018, 8, 20. [Google Scholar] [CrossRef]
- Hoekenga, O.A.; Maron, L.G.; Piñeros, M.A.; Cançado, G.M.A.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743. [Google Scholar] [CrossRef]
- Ligaba, A.; Katsuhara, M.; Ryan, P.R.; Shibasaka, M.; Matsumoto, H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006, 142, 1294–1303. [Google Scholar] [CrossRef]
- Collins, N.C.; Shirley, N.J.; Saeed, M.; Pallotta, M.; Gustafson, J.P. An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L.). Genetics 2008, 179, 669–682. [Google Scholar] [CrossRef]
- Liang, C.; Piñeros, M.A.; Tian, J.; Yao, Z.; Sun, L.; Liu, J.; Shaff, J.; Coluccio, A.; Kochian, L.V.; Liao, H. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef] [PubMed]
- Linlin, X.; Xin, Q.; Mingyue, Z.; Shaoling, Z. Genome-Wide analysis of aluminum-activated malate transporter family genes in six rosaceae species, and expression analysis and functional characterization on malate accumulation in Chinese white pear. Plant Sci. 2018, 274, 451–465. [Google Scholar] [CrossRef]
- Palmer, A.J.; Baker, A.; Muench, S.P. The varied functions of aluminium-activated malate transporters–much more than aluminium resistance. Biochem. Soc. Trans. 2016, 44, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Tyerman, S.D.; Xu, B.; Bose, J.; Kaur, S.; Conn, V.; Domingos, P.; Ullah, S.; Wege, S.; Shabala, S.; et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat. Commun. 2015, 6, 7879. [Google Scholar] [CrossRef]
- Huang, Y.; Fang, Y.; Long, X.; Liu, L.; Wang, J.; Zhu, J.; Ma, Y.; Qin, Y.; Qi, J.; Hu, X.; et al. Characterization of the rubber tree metallothionein family reveals a role in mitigating the effects of reactive oxygen species associated with physiological stress. Tree Physiol. 2018, 38, 911–924. [Google Scholar] [CrossRef]
- Chen, S.; Peng, S.; Huang, G.; Wu, K.; Fu, X.; Chen, Z. Association of decreased expression of a Myb transcription factor with the TPD (tapping panel dryness) syndrome in Hevea brasiliensis. Plant Mol. Biol. 2003, 51, 51–58. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, G.-L.; Zhao, Y.-G.; Zhao, W.-J.; Qi, Z.P. Chemical degradation of a Ferralsol (Oxisol) under intensive rubber (Hevea brasiliensis) farming in tropical China. Soil Tillage Res. 2007, 93, 109–116. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2.3.1–2.3.22. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar]
- Liu, W.; Xiong, C.; Yan, L.; Zhang, Z.; Ma, L.; Wang, Y.; Liu, Y.; Liu, Z. Transcriptome analyses reveal candidate genes potentially involved in Al stress response in Alfalfa. Front. Plant Sci. 2017, 8, 26. [Google Scholar] [CrossRef]
- Ma, Z.; Lin, S. Transcriptomic revelation of phenolic compounds involved in aluminum toxicity responses in roots of Cunninghamia lanceolata (Lamb.) Hook. Genes 2019, 10, 835. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Ma, X.W.; An, F.; Liu, Z.F.; Xie, G.S. Screening of reference genes for quantitative real-time PCR of rubber saplings under aluminum stress. Chin. J. Trop. Crop. in press.
- Livak, K.J.; Schmittgen, T.D.J.m. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Heng, Y.; Wu, C.; Long, Y.; Luo, S.; Ma, J.; Chen, J.; Liu, J.; Zhang, H.; Ren, Y.; Wang, M.; et al. OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. Plant Cell 2018, 30, 889–906. [Google Scholar] [CrossRef]
- Ma, B.; Yuan, Y.; Gao, M.; Qi, T.; Li, M.; Ma, F. Genome-wide identification, molecular evolution, and expression divergence of aluminum-activated malate transporters in apples. Int. J. Mol. Sci. 2018, 19, 2807. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Kamran, M.; Sullivan, W.; Chirkova, L.; Okamoto, M.; Degryse, F.; McLaughlin, M.; Gilliham, M.; Tyerman, S.D. Aluminum-activated malate transporters can facilitate GABA transport. Plant Cell 2018, 30, 1147–1164. [Google Scholar] [CrossRef]
- Eisenach, C.; Baetz, U.; Huck, N.V.; Zhang, J.; De Angeli, A.; Beckers, G.J.; Martinoia, E. ABA-induced stomatal closure involves ALMT4, a phosphorylation-dependent vacuolar anion channel of Arabidopsis. Plant Cell 2017, 29, 2552–2569. [Google Scholar] [CrossRef]
- Kovermann, P.; Meyer, S.; Hörtensteiner, S.; Picco, C.; Ravera, S.; Lee, Y.; Martinoia, E.; Scholz-Starke, J. The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 2007, 52, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Zhao, Y.; Jin, J.; Wu, S.; Deng, X.; Chen, Y.; Tian, W.M. Genome-Wide identification and characterization of the JAZ gene family in rubber tree (Hevea brasiliensis). Front. Genet. 2019, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Yang, J. Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. Ind. Crop. Prod. 2019, 134, 271–283. [Google Scholar] [CrossRef]
- Peng, W.; Wu, W.; Peng, J.; Li, J.; Lin, Y.; Wang, Y.; Tian, J.; Sun, L.; Liang, C.; Liao, H. Characterization of the soybean GmALMT family genes and the function of GmALMT5 in response to phosphate starvation. J. Integr. Plant Biol. 2018, 60, 216–231. [Google Scholar]
- Liu, J.; Zhou, M.; Delhaize, E.; Ryan, P.R. Altered expression of a malate-permeable anion channel, OsALMT4, disrupts mineral nutrition. Plant Physiol. 2017, 175, 1745–1759. [Google Scholar] [CrossRef]
- De Angeli, A.; Baetz, U.; Francisco, R.; Zhang, J.; Chaves, M.M.; Regalado, A. The vacuolar channel VvALMT9 mediates malate and tartrate accumulation in berries of Vitis vinifera. Planta 2013, 238, 283–291. [Google Scholar] [CrossRef]
- Ligaba, A.; Dreyer, I.; Margaryan, A.; Schneider, D.J.; Kochian, L.; Piñeros, M. Functional, structural and phylogenetic analysis of domains underlying the Al sensitivity of the aluminum-activated malate/anion transporter, TaALMT1. Plant J. 2013, 76, 766–780. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, X.X.; Wang, J.; Qi, C.; Wang, X.; Wang, G.; Li, M.; Li, X.; Guo, Y.D. BoALMT1, an Al-induced malate transporter in cabbage, enhances aluminum tolerance in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 2156. [Google Scholar] [CrossRef]
- Fontecha, G.; Silva-Navas, J.; Benito, C.; Mestres, M.A.; Espino, F.J.; Hernandez-Riquer, M.V.; Gallego, F.J. Candidate gene identification of an aluminum-activated organic acid transporter gene at the Alt4 locus for aluminum tolerance in rye (Secale cereale L.). Theor. Appl. Genet. 2007, 114, 249–260. [Google Scholar] [CrossRef]
- Chen, Z.C.; Yokosho, K.; Kashino, M.; Zhao, F.-J.; Yamaji, N. Adaptation to acidic soil is achieved by increased numbers of cis -acting elements regulating ALMT1 expression in Holcus lanatus. Plant J. 2013, 76, 10–23. [Google Scholar]
- Chen, Q.; Wu, K.H.; Wang, P.; Yi, J.; Li, K.Z.; Yu, Y.X.; Chen, L.M. Overexpression of MsALMT1, from the aluminum-sensitive Medicago sativa, enhances malate exudation and aluminum resistance in tobacco. Plant Mol. Biol. Rep. 2012, 31, 769–774. [Google Scholar] [CrossRef]
- Meyer, S.; Mumm, P.; Imes, D.; Endler, A.; Weder, B.; Al-Rasheid, K.A.; Geiger, D.; Marten, I.; Martinoia, E.; Hedrich, R. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 2010, 63, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Li, G.X.; Zheng, S.J. WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 2013, 76, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Guo, N.; Yang, Z.P.; Tang, X.; Peng, S.Q. Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 2014, 104, 14–23. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).