Transcriptome Sequencing of Different Avocado Ecotypes: de novo Transcriptome Assembly, Annotation, Identification and Validation of EST-SSR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and RNA Extraction
2.2. Library Construction and Transcriptome Sequencing
2.3. Transcriptome Assembly, Annotation, and Coding Sequence Prediction
2.4. Mining for Transcription Factor Families
2.5. EST-SSR Mining
2.6. Identification of Differentially Expressed Genes (DEGs)
2.7. Identification of EST-SSR Markers
2.8. Data Analysis
3. Results
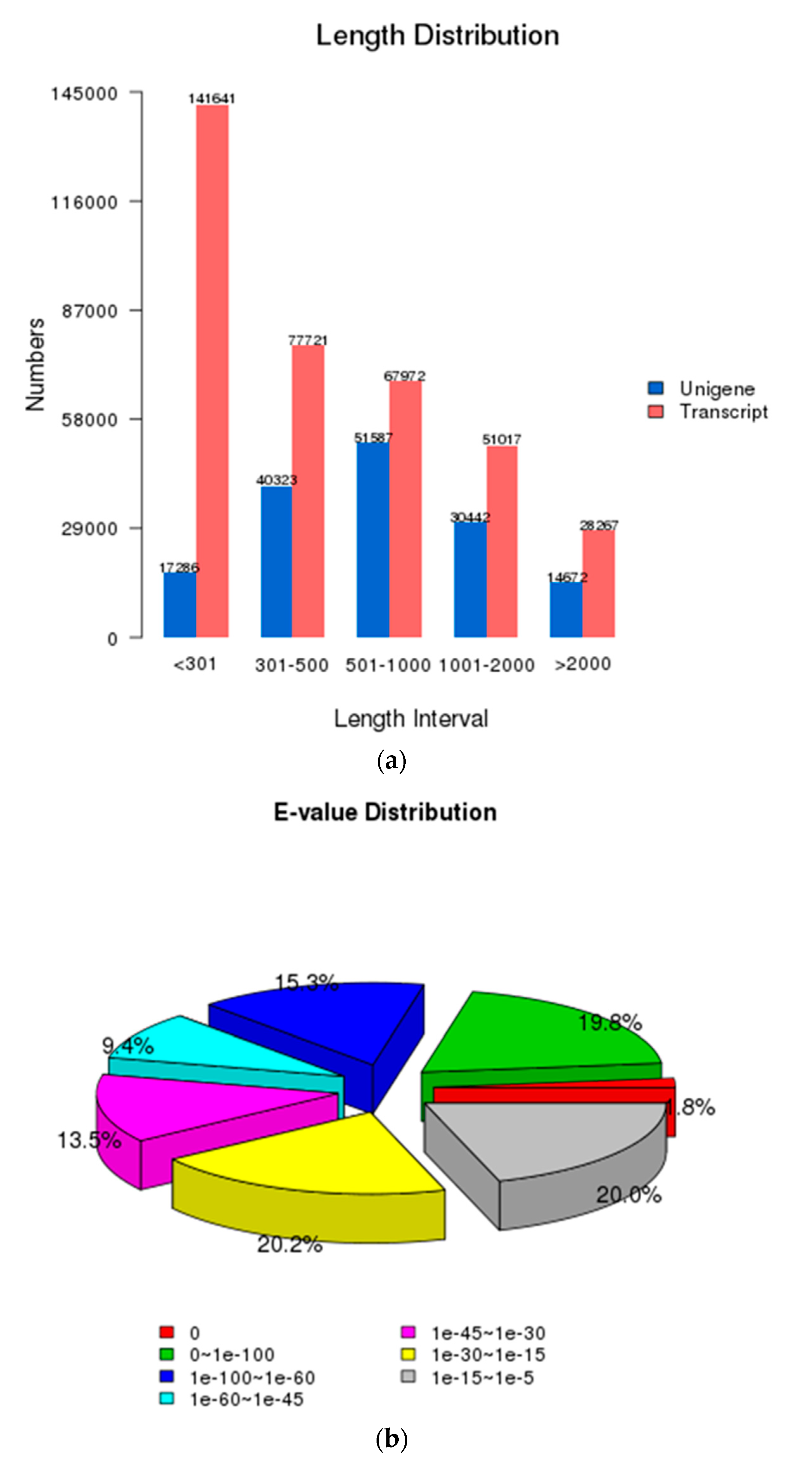
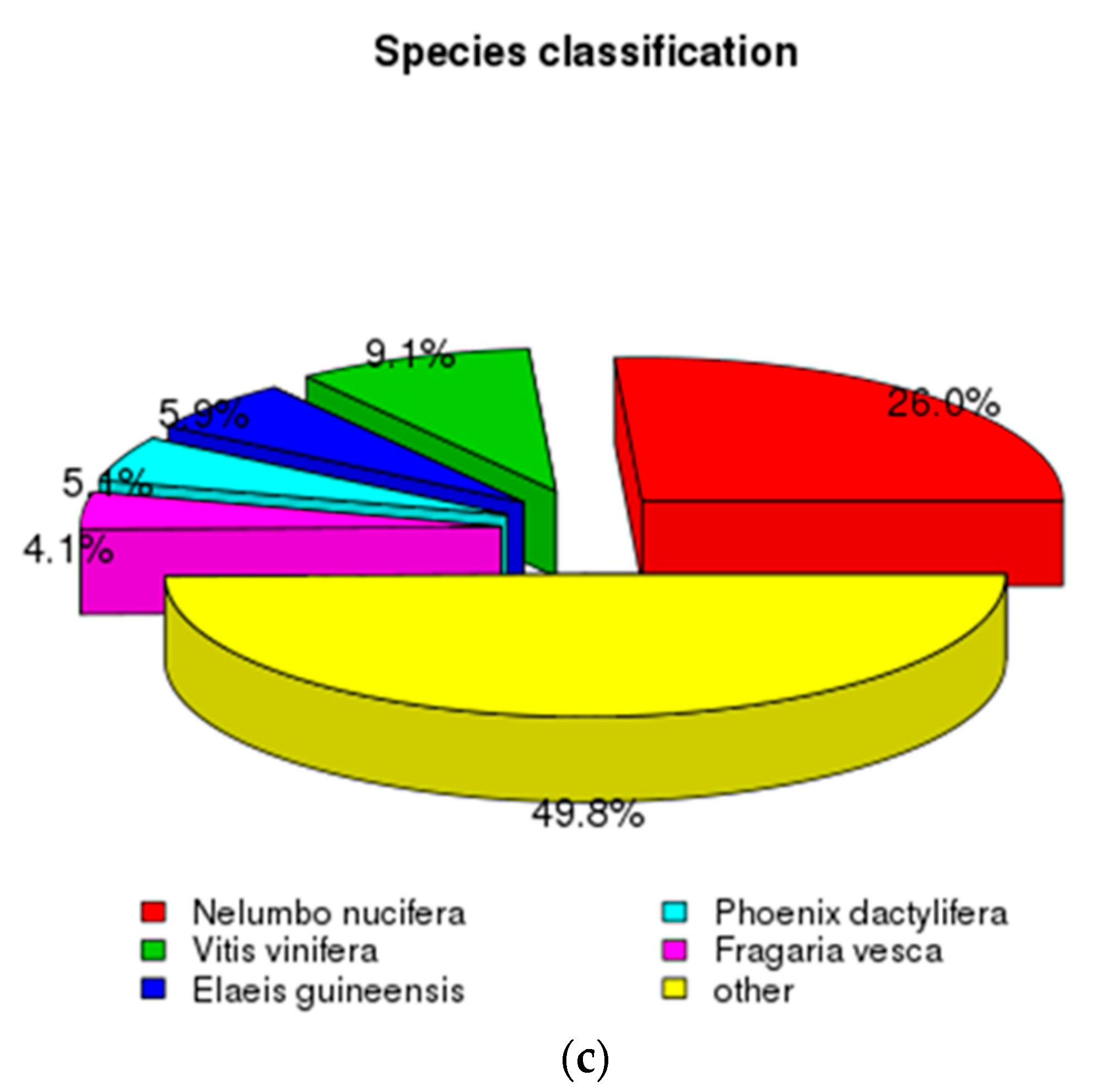
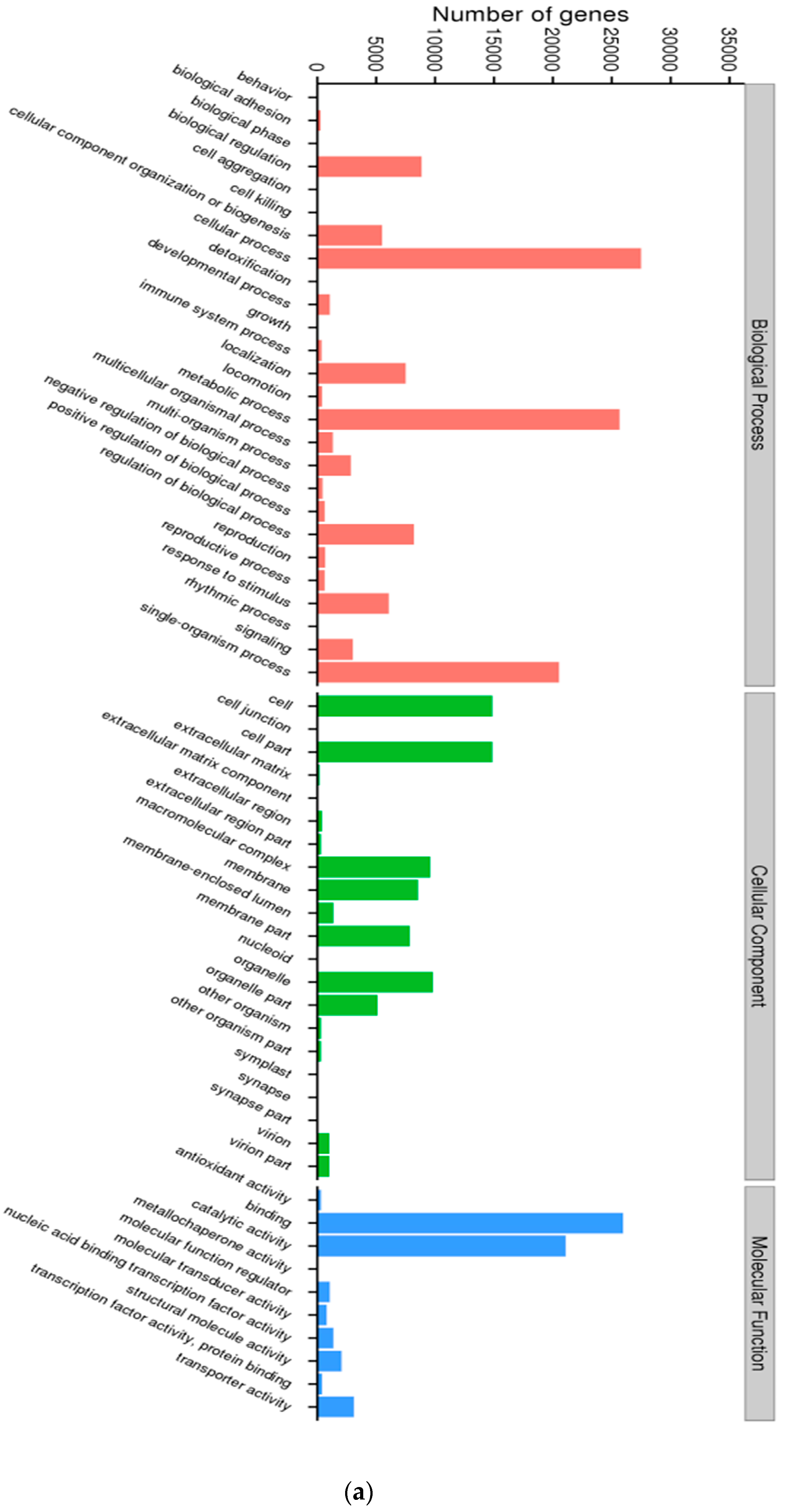
3.1. Characterization of Transcriptome Sequencing Results and Sequence Annotation
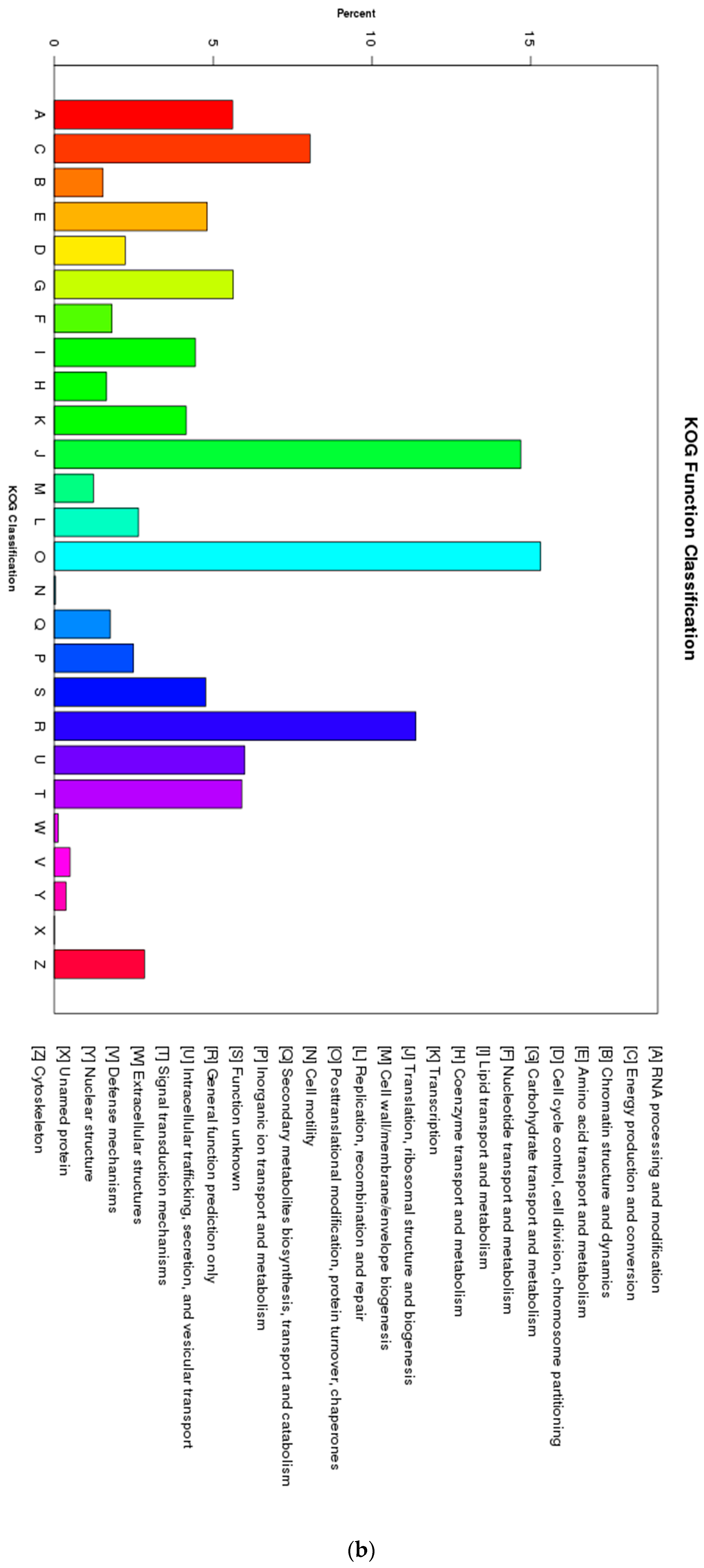
3.2. Coding Sequence Prediction and Mining for Transcription Factor Families
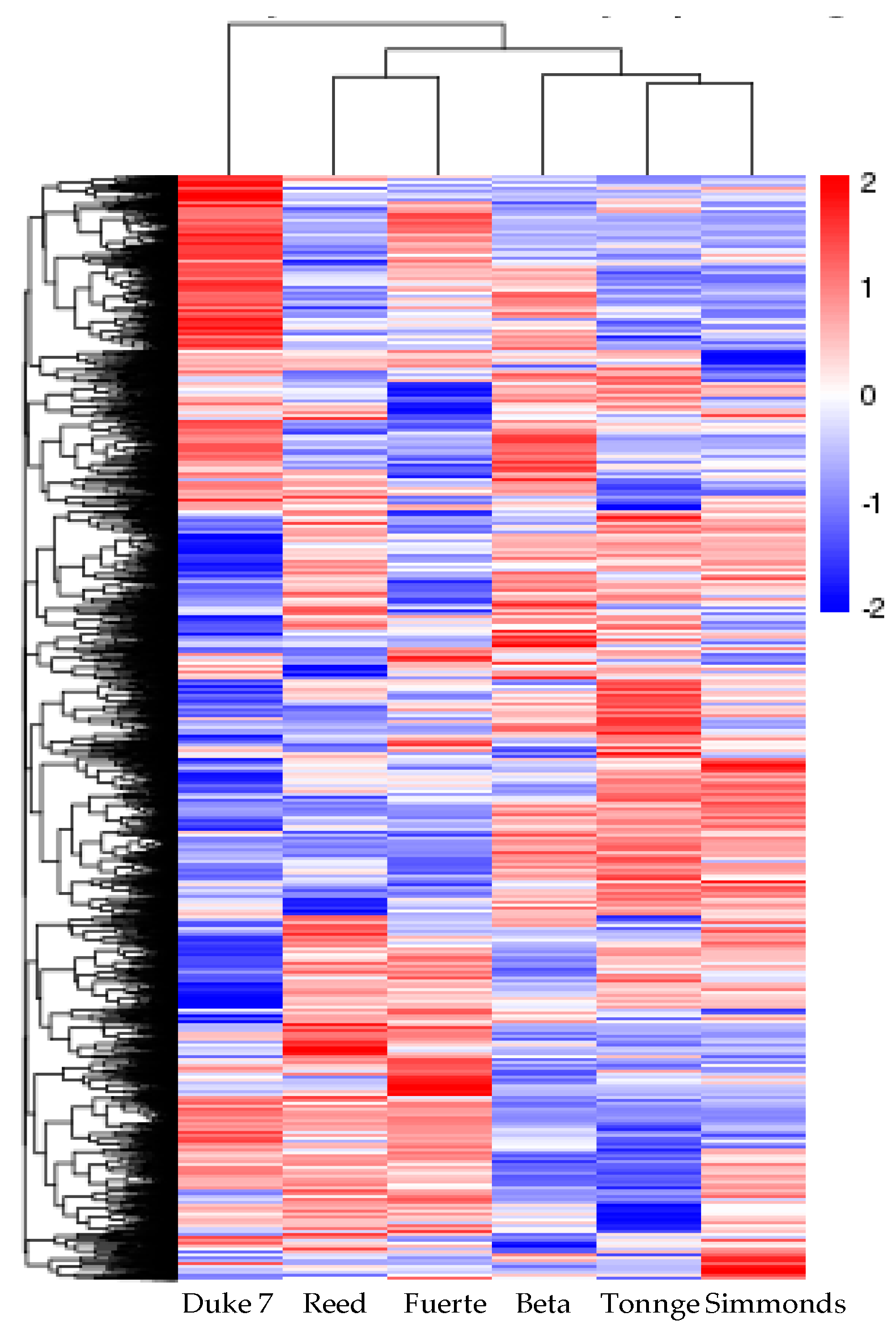
3.3. Analysis of DEGs
3.4. Frequency and Distribution of Different Types of EST-SSR Loci
3.5. Development of Polymorphism EST-SSR Markers and Genetic Diversity
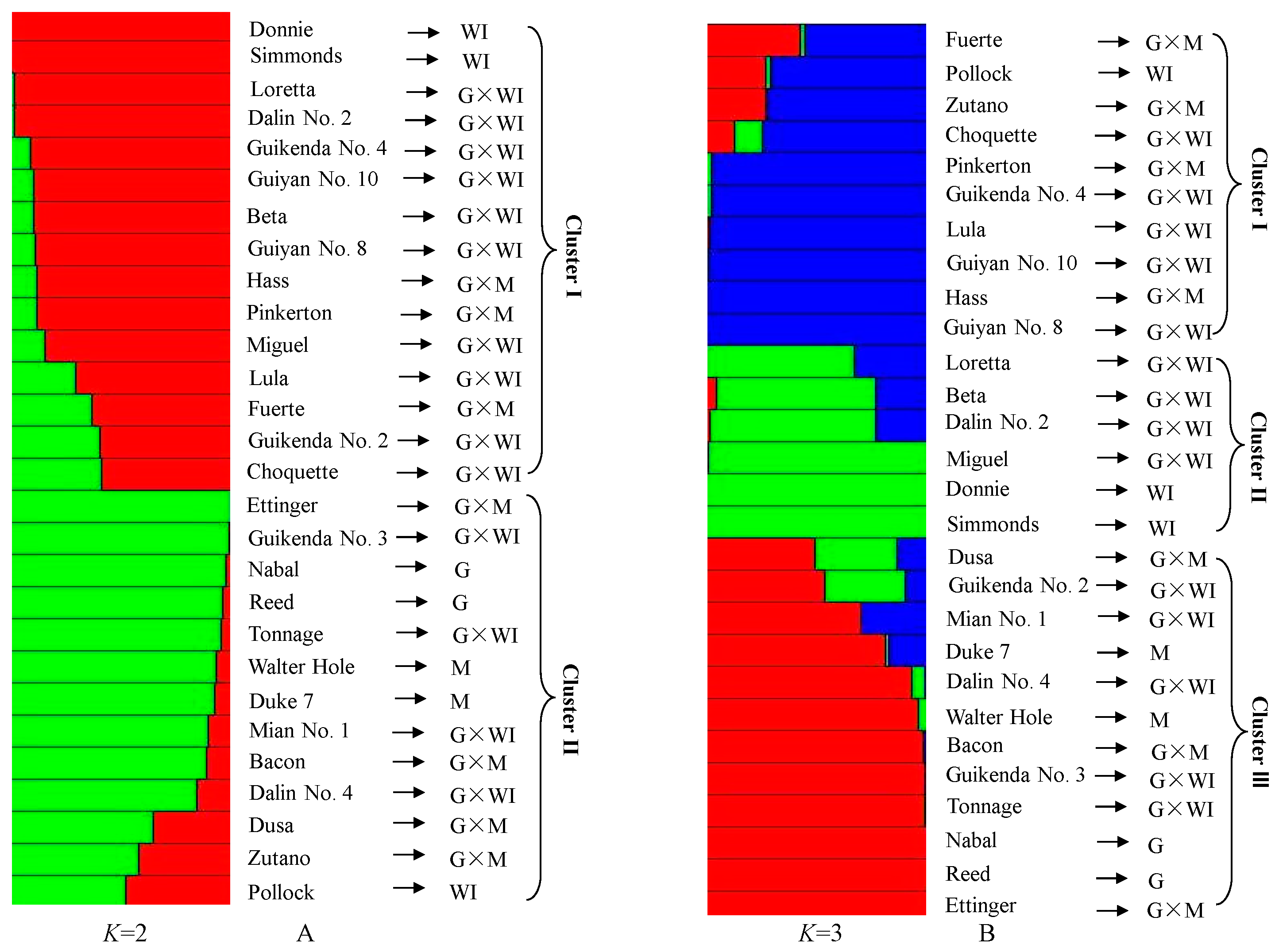
3.6. Genetic Relationship Analysis Based on Polymorphic EST-SSRs from Transcriptome Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schaffer, B.; Wolstenholme, B.N.; Whiley, A.W. The Avocado: Botany, Production and Uses, 2nd ed.; CPI Group (UK) Ltd.: Croydon, UK, 2012. [Google Scholar]
- Chanderbali, A.S.; Albert, V.A.; Ashworth, V.E.T.M.; Clegg, M.T.; Litz, R.E.; Soltis, D.E.; Soltis, P.S. Persea americana (avocado): Bringing ancient flowers to fruit in the genomics era. BioEssays 2008, 304, 386–396. [Google Scholar] [CrossRef]
- Chanderbali, A.S.; Albert, V.A.; Leebens-Mackd, J.; Altmane, N.S.; Soltis, D.E.; Soltis, P.S. Transcriptional signatures of ancient floral developmental genetics in avocado (Persea americana; Lauraceae). Proc. Natl. Acad. Sci. USA 2009, 106, 8929–8934. [Google Scholar] [CrossRef]
- Galindo-Tovar, M.E.; Ogata-Aguilar, N.; Arzate-Fernandez, A.M. Some aspects of avocado (Persea americana Mill.) diversity and domestication in Mesoamerica. Genet. Resour. Crop Evol. 2008, 55, 441–450. [Google Scholar] [CrossRef]
- Gross-German, E.; Viruel, M.A. Molecular characterization of avocado germplasm with a new set of SSR and EST-SSR markers: Genetic diversity, population structure, and identification of race-specific markers in a group of cultivated genotypes. Tree Genet. Genomes 2013, 9, 539–555. [Google Scholar] [CrossRef]
- Ibarra-Laclette, E.; Méndez-Bravo, A.; Pérez-Torres, C.A.; Albert, V.A.; Mockaitis, K.; Kilaru, A.; López-Gómez, R.; Cervantes-Luevano, J.I.; Herrera-Estrell, L. Deep sequencing of the Mexican avocado transcriptome, an ancient angiosperm with a high content of fatty acids. BMC Genom. 2015, 16, 599. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, A.; Cao, X.; Dabbs, P.B.; Sung, H.J.; Rahman, M.M.; Thrower, N.; Zynda, G.; Podicheti, R.; Ibarra-Laclette, E.; Herrera-Estrella, L.; et al. Oil biosynthesis in a basal angiosperm: Transcriptome analysis of Persea Americana mesocarp. BMC Plant Biol. 2015, 15, 203. [Google Scholar] [CrossRef]
- Vergara-Pulgar, C.; Rothkegel, K.; González-Agüero, M.; Pedreschi, R.; Campos-Vargas, R.; Defilippi, B.G.; Meneses, C. De novo assembly of Persea americana cv.’Hass’ transcriptome during fruit development. BMC Genom. 2019, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. 2019. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 27 April 2019).
- Dreher, M.L.; Davenport, A.J. Hass avocado composition and potential health effects. Crit. Rev. Food Sci. 2013, 53, 738–750. [Google Scholar] [CrossRef]
- Galvão, M.D.S.; Narain, N.; Nigam, N. Influence of different cultivars on oil quality and chemical characteristics of avocado fruit. Food Sci. Technol. 2014, 34, 539–546. [Google Scholar] [CrossRef]
- Ge, Y.; Si, X.Y.; Cao, J.Q.; Zhou, Z.X.; Wang, W.L.; Ma, W.H. Morphological characteristics, nutritional quality, and bioactive constituents in fruits of two avocado (Persea americana) varieties from hainan province, China. J. Agric. Sci. 2017, 9, 8–17. [Google Scholar] [CrossRef]
- Fiedler, J.; Bufler, G.; Bangerth, F. Genetic relationships of avocado (Persea americana Mill.) using RAPD markers. Euphytica 1998, 101, 249–255. [Google Scholar] [CrossRef]
- Mhameed, S.; Sharon, D.; Kaufman, D.; Lahav, E.; Hillel, J.; Degani, C.; Lavi, U. Genetic relationships within avocado (Persea americana Mill.) cultivars and between Persea species. Theor. Appl. Genet. 1997, 94, 279–286. [Google Scholar] [CrossRef]
- Furnier, G.R.; Cummings, M.P.; Clegg, M.T. Evolution of the avocados as revealed by DNA restriction site variation. J. Hered. 1990, 81, 183–188. [Google Scholar] [CrossRef]
- Davis, J.; Henderson, D.; Kobayashi, M. Genealogical relationships among cultivated avocado as revealed through RFLP analysis. J. Hered. 1998, 89, 319–323. [Google Scholar] [CrossRef]
- Ashworth, V.E.T.M.; Clegg, M.T. Microsatellite markers in avocado (Persea americana Mill.). genealogical relationships among cultivated avocado genotypes. J. Hered. 2003, 94, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Schnell, R.J.; Brown, J.S.; Olano, C.T.; Power, E.J.; Krol, C.A.; Kuhn, D.N.; Motamayor, J.C. Evaluation of avocado germplasm using microsatellite markers. J. Am. Soc. Hortic. Sci. 2003, 128, 881–889. [Google Scholar] [CrossRef]
- Chen, H.; Morrel, P.L.; Ashwoth, V.E.T.M.; De la Cruz, M.; Clegg, M.T. Nucleotide diversity and linkage disequilibrium in wild avocado (Persea americana Mill.). J. Hered. 2008, 99, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Morrel, P.L.; Ashwoth, V.E.T.M.; De la Cruz, M.; Clegg, M.T. Tracing the geographic origins of mayor avocado cultivars. J. Hered. 2009, 100, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Zhang, T.; Wu, B.; Tan, L.; Ma, F.N.; Zou, M.H.; Chen, H.H.; Pei, J.L.; Liu, Y.Z.; Chen, Z.H.; et al. Genome-wide assessment of avocado germplasm determined from specific length amplified fragment sequencing and transcriptomes: Population structure, genetic diversity, identification, and application of race-specific markers. Genes 2019, 10, 215. [Google Scholar] [CrossRef]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Development and linkage mapping of unigene-derived microsatellite markers in Brassica rapa L. Breed. Sci. 2011, 61, 160–167. [Google Scholar] [CrossRef]
- Hou, M.Y.; Mu, G.J.; Zhang, Y.J.; Cui, S.L.; Yang, X.L.; Liu, L.F. Evaluation of total flavonoid content and analysis of related EST-SSR in Chinese peanut germplasm. Crop Breed. Appl. Biotechnol. 2017, 17, 221–227. [Google Scholar] [CrossRef][Green Version]
- Azevedo, A.O.N.; Azevedo, C.D.O.; Santos, P.H.A.D.; Ramos, H.C.C.; Boechat, M.S.B.; Arêdes, F.A.S.; Ramos, S.R.R.; Mirizola, L.A.; Perera, L.; Aragão, W.M.; et al. Selection of legitimate dwarf coconut hybrid seedlings using DNA fingerprinting. Crop Breed. Appl. Biotechnol. 2018, 18, 409–416. [Google Scholar] [CrossRef]
- Ferreira, F.; Scapim, C.A.; Maldonado, C.; Mora, F. SSR-based genetic analysis of sweet corn inbred lines using artificial neural networks. Crop Breed. Appl. Biotechnol. 2018, 18, 309–313. [Google Scholar] [CrossRef]
- Penin, A.A.; Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D. Comparative analysis of developmental transcriptome maps of Arabidopsis thaliana and Solanum lycopersicum. Genes 2019, 10, 50. [Google Scholar] [CrossRef]
- Qiao, F.; Cong, H.Q.; Jiang, X.F.; Wang, R.X.; Yin, J.M.; Qian, D.; Wang, Z.N. De novo characterization of a cephalotaxus hainanensis transcriptome and genes related to paclitaxel biosynthesis. PLoS ONE 2014, 9, e106900. [Google Scholar] [CrossRef]
- Weisberg, A.J.; Kim, G.; Westwood, J.H.; Jelesko, J.G. Sequencing and de novo assembly of the toxicodendron radicans (Poison Ivy) transcriptome. Genes 2017, 8, 317. [Google Scholar] [CrossRef]
- Wu, J.; Cai, C.F.; Cheng, F.Y.; Cui, H.L.; Zhou, H. Characterization and development of EST-SSR markers in tree peony using transcriptome sequences. Mol. Breed. 2014, 34, 1853–1866. [Google Scholar] [CrossRef]
- Chopra, R.; Burow, G.; Hayes, C.; Emendack, Y.; Xin, Z.G.; Burke, J. Transcriptome profiling and validation of gene based single nucleotide polymorphisms (SNPs) in sorghum genotypes with contrasting responses to cold stress. BMC Genom. 2015, 16, 1040. [Google Scholar] [CrossRef]
- Thumilan, B.M.; Sajeevan, R.S.; Madhuri, B.J.T.; Nataraja, K.N.; Sreeman, S.M. Development and characterization of genic SSR markers from Indian mulberry transcriptome and their transferability to related species of Moraceae. PLoS ONE 2016, 11, e0162909. [Google Scholar] [CrossRef]
- Chen, C.L.; Xu, M.L.; Wang, C.P.; Qiao, G.X.; Wang, W.W.; Tan, Z.Y.; Wu, T.T.; Zhang, Z.S. Characterization of the Lycium barbarum fruit transcriptome and development of EST-SSR markers. PLoS ONE 2017, 12, e0187738. [Google Scholar] [CrossRef]
- Xu, D.; Chen, H.; Aci, M.; Pan, Y.; Shangguan, Y.; Ma, J.; Li, L.; Qian, G.; Wang, Q.X. De Novo assembly, characterization and development of EST-SSRs from Bletilla striata transcriptomes profiled throughout the whole growing period. PLoS ONE 2018, 13, e0205954. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.K.; Nath, U.K.; Howlader, J.; Bagchi, M.; Natarajan, S.; Kayum, M.A.; Kim, H.T.; Park, J.I.; Kang, J.G.; Nou, I.S. Exploration and exploitation of novel SSR markers for candidate transcription factor genes in Lilium species. Genes 2018, 9, 97. [Google Scholar] [CrossRef]
- Ma, S.L.Y.; Dong, W.S.; Lyu, T.; Lyu, Y.M. An RNA sequencing transcriptome analysis and development of EST-SSR markers in Chinese hawthorn through Illumina sequencing. Forests 2019, 10, 82. [Google Scholar] [CrossRef]
- Chen, Z.H.; Ge, Y.; Sun, P.G.; Sun, C.J.; Wu, Q. The effects of propagating material and culture conditions on the propagation coefficient of avocado seedlings. China Fruit 2017, 5, 61–64. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Perez-Rodriguez, P.; Riano-Pachon, D.M.; Correa, L.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, 822–827. [Google Scholar] [CrossRef]
- Krawczak, M.; Nikolaus, S.; von Eberstein, H.; Croucher, P.J.; El Mokhtari, N.E.; Schreiber, S. PopGen: Population based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006, 9, 55–61. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Du, M.; Li, N.; Niu, B.; Liu, Y.; You, D.; Jiang, D.; Ruan, C.Q.; Qin, Z.Q.; Song, T.W.; Wang, W.T. De novo transcriptome analysis of Bagarius yarrelli (Siluriformes: Sisoridae) and the search for potential SSR markers using RNA-Seq. PLoS ONE 2018, 13, e0190343. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.M.; Hong, Z.; Yang, Z.J.; Zhang, N.N.; Liu, X.J.; Xu, D.P. De Novo transcriptome analysis of Dalbergia odorifera T. Chen (Fabaceae) and transferability of SSR markers developed from the transcriptome. Forests 2019, 10, 98. [Google Scholar] [CrossRef]
- Li, W.; Zhang, C.P.; Jiang, X.Q.; Liu, Q.C.; Liu, Q.H.; Wang, K.L. De Novo transcriptomic analysis and development of EST–SSRs for Styrax japonicas. Forests 2018, 9, 748. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Fang, B.P.; Chen, J.Y.; Zhang, X.J.; Luo, Z.X.; Huang, L.F.; Chen, X.L.; Li, Y.L. De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweetpotato (Ipomoea batatas). BMC Genom. 2010, 11, 726. [Google Scholar] [CrossRef]
- Sathyanarayana, N.; Pittala, R.K.; Tripathi, P.K.; Chopra, R.; Singh, H.R.; Belamkar, V.; Bhardwaj, P.K.; Doyle, J.J.; Egan, A.N. Transcriptomic resources for the medicinal legume Mucuna pruriens: de novo Transcriptome assembly, annotation, identification and validation of EST-SSR markers. BMC Genom. 2017, 18, 409. [Google Scholar] [CrossRef]
- Hu, C.; Yang, H.X.; Jiang, K.; Wang, L.; Yang, B.; Hsireh, T.; Lan, S.; Huang, W.C. Development of polymorphic microsatellite markers by using de novo transcriptome assembly of Calanthe masuca and C. sinica (Orchidaceae). BMC Genom. 2018, 19, 800. [Google Scholar] [CrossRef] [PubMed]
- Mora-Ortiz, M.; Swain, M.T.; Vickers, M.J.; Hegarty, M.J.; Kelly, R.; Smith, L.M.J.; Skot, L. De-novo transcriptome assembly for gene identification, analysis, annotation, and molecular marker discovery in Onobrychism viciifolia. BMC Genom. 2016, 17, 756. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guo, H.; Wang, Y.; Zong, J.; Chen, J.; Li, D.D.; Li, L.; Wang, J.J.; Liu, J.X. High-throughput SSR marker development and its application in a centipedegrass (Eremochloa ophiuroides (Munro) Hack.) genetic diversity analysis. PLoS ONE 2018, 13, e0202605. [Google Scholar] [CrossRef]
- Mun, J.H.; Kim, D.J.; Choi, H.K.; Gish, J.; Debellé, F.; Mudge, J.; Denny, R.; Endré, G.; Saurat, O.; Dudez, A.M. Distribution of microsatellites in the genome of Medicago truncatula: A resource of genetic markers that integrate genetic and physical maps. Genetics 2006, 172, 2541–2555. [Google Scholar] [CrossRef]
- Chen, H.; Wang, L.; Wang, S.; Liu, C.; Blair, M.W.; Cheng, X. Transcriptome Sequencing of Mung Bean (Vigna radiate L.) Genes and the identification of EST-SSR markers. PLoS ONE 2015, 10, e0120273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yuan, D.; Yu, S.; Li, Z.; Cao, Y.; Miao, Z.; Qian, H.; Tang, K. Preference of simple sequence repeats in coding and non-coding regions of Arabidopsis thaliana. Bioinformatics 2004, 20, 1081–1086. [Google Scholar] [CrossRef]
- Alcaraz, M.L.; Hormaza, J.I. Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Heredity 2007, 144, 244–253. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Oddou-Muratorio, S.; Vendramin, G.G.; Buiteveld, J.; Fady, B. Population estimators or progeny tests: What is the best method to assess null allele frequencies at SSR loci? Conserv. Genet. 2009, 10, 1343. [Google Scholar] [CrossRef]
- Ellis, J.R.; Burke, J.M. EST-SSRs as a resource for population genetic analyses. Heredity 2007, 99, 125–132. [Google Scholar] [CrossRef]

| Accession | Origin | Source | Race 1 | Type |
|---|---|---|---|---|
| Walter Hole | California, USA | CATAS, Hainan, China | M | C |
| Duke 7 | California, USA | CATAS-SSCRI, Guangdong, China | M | RS |
| Nabal | Antigua, Guatemala | CATAS, Hainan, China | G | C |
| Reed | California, USA | CATAS-SSCRI, Guangdong, China | G | C |
| Pollock | Florida, US | CATAS, Hainan, China | WI | C |
| Donnie | Florida, USA | CATAS, Hainan, China | WI | C |
| Simmonds | Florida, USA | CATAS-SSCRI, Guangdong, China | WI | C |
| Bacon | California, USA | CATAS, Hainan, China | G × M | C |
| Hass | California, USA | CATAS, Hainan, China | G × M | C |
| Pinkerton | California, USA | CATAS, Hainan, China | G × M | C |
| Zutano | California, USA | CATAS, Hainan, China | G × M | C |
| Ettinger | Kefar Malal, Israel | CATAS, Hainan, China | G × M | C |
| Fuerte | Puebla, Mexico | CATAS-SSCRI, Guangdong, China | G × M | C |
| Dusa | Westfalia Estate, South Africa | CATAS, Hainan, China | G × M | RS |
| Miguel | Florida, USA | CATAS, Hainan, China | G × WI | C |
| Loretta | Florida, USA | CATAS, Hainan, China | G × WI | C |
| Beta | Florida, USA | CATAS-SSCRI, Guangdong, China | G × WI | C |
| Choquette | Florida, USA | CATAS, Hainan, China | G × WI | C |
| Lula | Florida, USA | CATAS, Hainan, China | G × WI | C |
| Tonnage | Florida, USA | CATAS-SSCRI, Guangdong, China | G × WI | C |
| Guikenda No. 2 | Guangxi, China | CATAS, Hainan, China | G × WI | C |
| Guikenda No. 3 | Guangxi, China | CATAS, Hainan, China | G × WI | LS |
| Guikenda No. 4 | Guangxi, China | CATAS, Hainan, China | G × WI | LS |
| Guiyan No. 8 | Guangxi, China | CATAS, Hainan, China | G × WI | LS |
| Guiyan No. 10 | Guangxi, China | CATAS, Hainan, China | G × WI | LS |
| Daling No. 2 | Guangxi, China | CATAS, Hainan, China | G × WI | LS |
| Daling No. 4 | Guangxi, China | CATAS, Hainan, China | G × WI | LS |
| Mian No. 1 | Guangxi, China | CATAS, Hainan, China | G × WI | LS |
| Category | Transcripts | Unigenes |
|---|---|---|
| Min length | 201 | 201 |
| Mean length | 731 | 922 |
| Median length | 382 | 627 |
| Max length | 17,239 | 17,230 |
| N50 | 1271 | 1283 |
| N90 | 269 | 432 |
| Total nucleotides | 268,157,161 | 142,337,653 |
| Sample Name | No. of DEGs | No. of Up-Regulated DEGs | No. of Down-Regulated DEGs |
|---|---|---|---|
| Duke 7 versus Reed | 2519 | 1376 | 1143 |
| Duke 7 versus Simmonds | 3559 | 1792 | 1767 |
| Duke 7 versus Fuerte | 1808 | 954 | 854 |
| Duke 7 versus Beta | 2115 | 1051 | 1064 |
| Duke 7 versus Tonnage | 2978 | 1510 | 1468 |
| Reed versus Simmonds | 2184 | 1096 | 1088 |
| Reed versus Fuerte | 1265 | 579 | 686 |
| Reed versus Beta | 2038 | 870 | 1168 |
| Reed versus Tonnage | 1313 | 636 | 677 |
| Simmonds versus Fuerte | 2467 | 1213 | 1254 |
| Simmonds versus Beta | 1112 | 406 | 706 |
| Simmonds versus Tonnage | 1048 | 472 | 576 |
| Fuerte versus Beta | 2467 | 1110 | 1313 |
| Fuerte versus Tonnage | 1943 | 1010 | 933 |
| Beta versus Tonnage | 1456 | 847 | 609 |
| Source | Number |
|---|---|
| Total number of sequences examined | 154,310 |
| Total size of examined sequences (bp) | 142,337,653 |
| Total number of identified SSRs | 55,558 |
| Number of SSR containing sequences | 43,270 |
| Number of sequences containing more than 1 SSR | 9789 |
| Number of SSRs present in compound formation | 2661 |
| SSR Motif Length | Repeat Unit Number | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | 10 | >10 | Total | % | |
| Mono- | - | - | - | - | - | 10,465 | 23,639 | 34,104 | 61.38 |
| Di- | - | 3628 | 2638 | 2856 | 3051 | 1354 | 193 | 13,720 | 24.69 |
| Tri- | 4461 | 1992 | 679 | 25 | 4 | - | - | 7161 | 12.89 |
| Tetra- | 473 | 35 | 0 | 2 | 0 | 1 | - | 511 | 0.92 |
| Penta- | 25 | 7 | 1 | 1 | - | - | - | 34 | 0.06 |
| Hexa- | 17 | 8 | 0 | 1 | 2 | - | - | 28 | 0.05 |
| Total | 4976 | 5670 | 3318 | 2885 | 3057 | 11,820 | 23,832 | 55,558 | |
| % | 8.96 | 10.21 | 5.97 | 5.19 | 5.50 | 2.13 | 42.90 | ||
| Marker Name | Unigene ID | Na 1 | Ne 2 | Ho 3 | He 4 | PIC 5 |
|---|---|---|---|---|---|---|
| Pa-eSSR-1 | c20669_g0_i0 | 10 | 4.31 | 0.39 | 0.78 | 0.74 |
| Pa-eSSR-2 | c92252_g2_i0 | 5 | 3.53 | 0.00 | 0.72 | 0.67 |
| Pa-eSSR-3 | c77919_g2_i1 | 4 | 2.23 | 0.00 | 0.55 | 0.46 |
| Pa-eSSR-4 | c63541_g0_i1 | 5 | 2.48 | 0.14 | 0.60 | 0.54 |
| Pa-eSSR-5 | c40793_g0_i0 | 9 | 4.98 | 0.11 | 0.80 | 0.77 |
| Pa-eSSR-6 | c40000_g0_i0 | 6 | 2.91 | 0.00 | 0.66 | 0.60 |
| Pa-eSSR-7 | c22125_g0_i0 | 4 | 3.09 | 0.00 | 0.68 | 0.62 |
| Pa-eSSR-8 | c76175_g0_i2 | 8 | 4.78 | 0.00 | 0.79 | 0.77 |
| Pa-eSSR-9 | c90335_g0_i0 | 12 | 6.75 | 0.15 | 0.85 | 0.84 |
| Pa-eSSR-10 | c83091_g0_i0 | 3 | 1.56 | 0.00 | 0.36 | 0.33 |
| Pa-eSSR-11 | c64286_g0_i0 | 4 | 2.65 | 0.00 | 0.62 | 0.55 |
| Pa-eSSR-12 | c92291_g1_i0 | 10 | 7.04 | 0.19 | 0.86 | 0.84 |
| Pa-eSSR-13 | c79749_g0_i0 | 5 | 2.61 | 0.04 | 0.62 | 0.57 |
| Pa-eSSR-14 | c80732_g1_i1 | 4 | 2.51 | 0.00 | 0.60 | 0.54 |
| Pa-eSSR-15 | c115429_g2_i1 | 6 | 3.56 | 0.00 | 0.72 | 0.68 |
| Pa-eSSR-16 | c85149_g0_i0 | 3 | 1.44 | 0.00 | 0.30 | 0.27 |
| Total | 98 | |||||
| Mean | 6.13 | 3.53 | 0.06 | 0.66 | 0.61 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Y.; Tan, L.; Wu, B.; Wang, T.; Zhang, T.; Chen, H.; Zou, M.; Ma, F.; Xu, Z.; Zhan, R. Transcriptome Sequencing of Different Avocado Ecotypes: de novo Transcriptome Assembly, Annotation, Identification and Validation of EST-SSR Markers. Forests 2019, 10, 411. https://doi.org/10.3390/f10050411
Ge Y, Tan L, Wu B, Wang T, Zhang T, Chen H, Zou M, Ma F, Xu Z, Zhan R. Transcriptome Sequencing of Different Avocado Ecotypes: de novo Transcriptome Assembly, Annotation, Identification and Validation of EST-SSR Markers. Forests. 2019; 10(5):411. https://doi.org/10.3390/f10050411
Chicago/Turabian StyleGe, Yu, Lin Tan, Bin Wu, Tao Wang, Teng Zhang, Haihong Chen, Minghong Zou, Funing Ma, Zining Xu, and Rulin Zhan. 2019. "Transcriptome Sequencing of Different Avocado Ecotypes: de novo Transcriptome Assembly, Annotation, Identification and Validation of EST-SSR Markers" Forests 10, no. 5: 411. https://doi.org/10.3390/f10050411
APA StyleGe, Y., Tan, L., Wu, B., Wang, T., Zhang, T., Chen, H., Zou, M., Ma, F., Xu, Z., & Zhan, R. (2019). Transcriptome Sequencing of Different Avocado Ecotypes: de novo Transcriptome Assembly, Annotation, Identification and Validation of EST-SSR Markers. Forests, 10(5), 411. https://doi.org/10.3390/f10050411





