De novo Characterization of the Platycladus orientalis Transcriptome and Analysis of Photosynthesis-Related Genes during Aging
Abstract
:1. Introduction
2. Materials and Methods
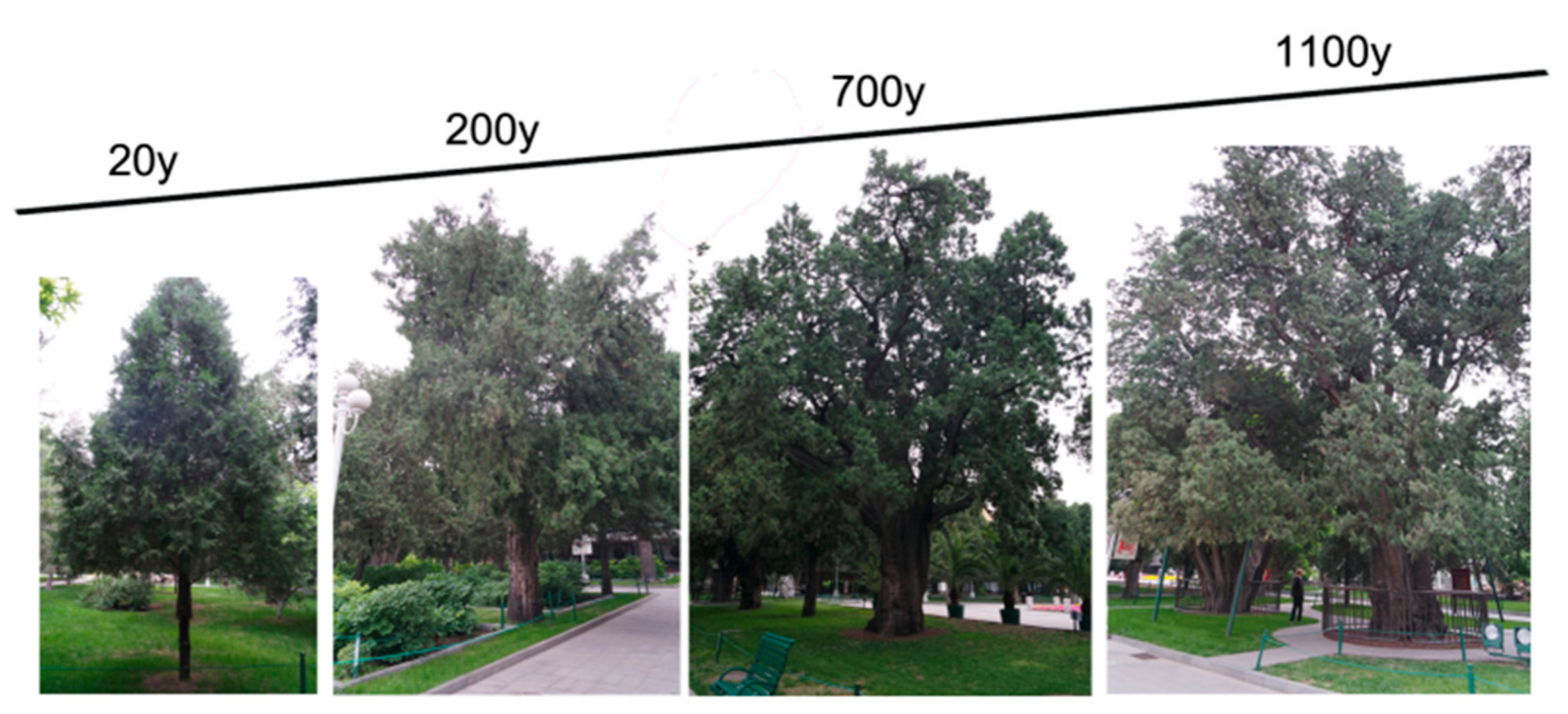
2.1. Plant Materials
2.2. Physiological Index Determination
2.3. Total RNA Extraction
2.4. Construction and Sequencing of cDNA Libraries
2.5. Transcriptome Assembly and Functional Annotation
2.6. Quantification of Gene Expression Levels and Analyses of Differentially Expressed Genes
2.7. Quantitative Real-Time PCR
3. Results
3.1. Biochemical Changes during Senescence
3.2. De novo Assembly of the P. orientalis Transcriptome
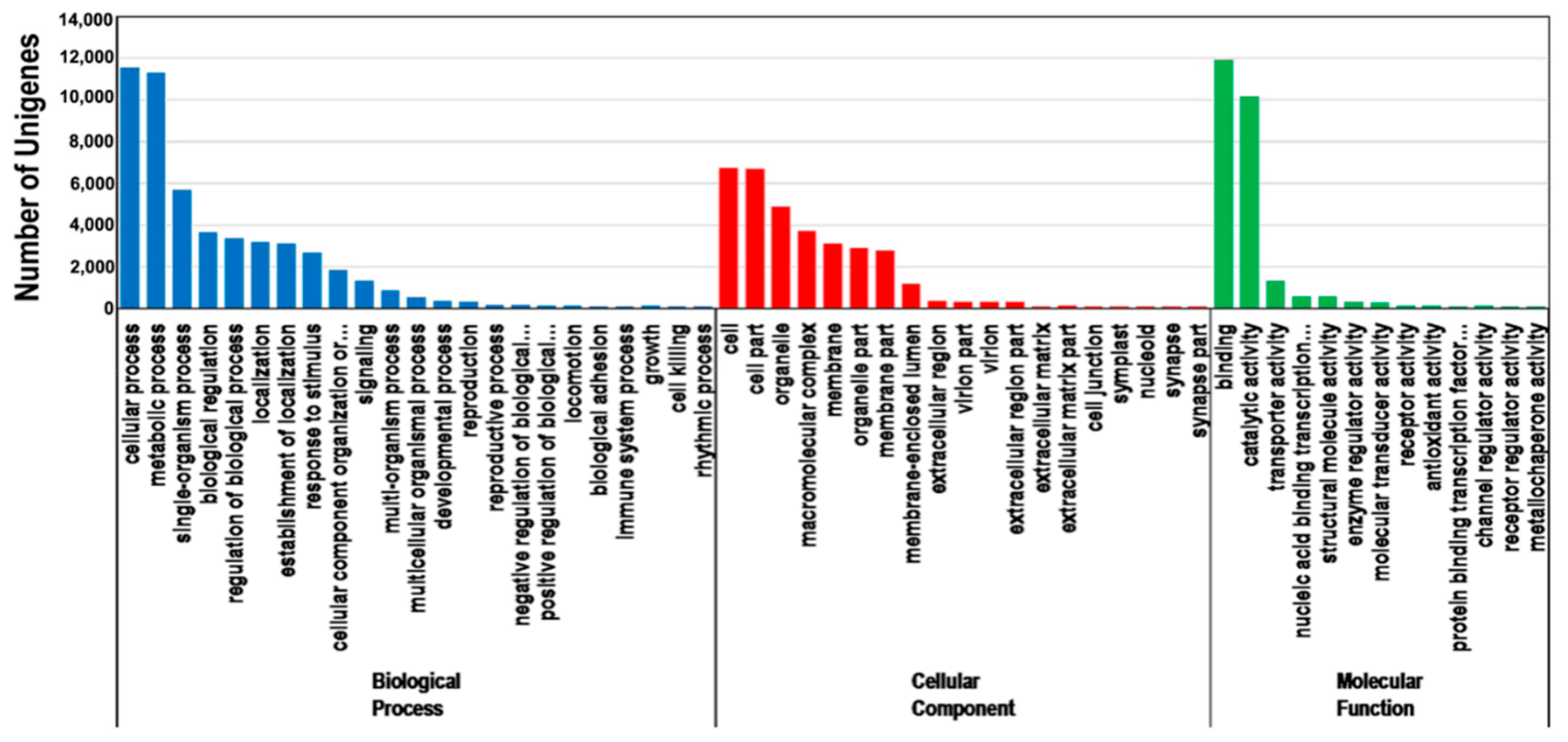
3.3. Functional Classification of P. orientalis Unigenes
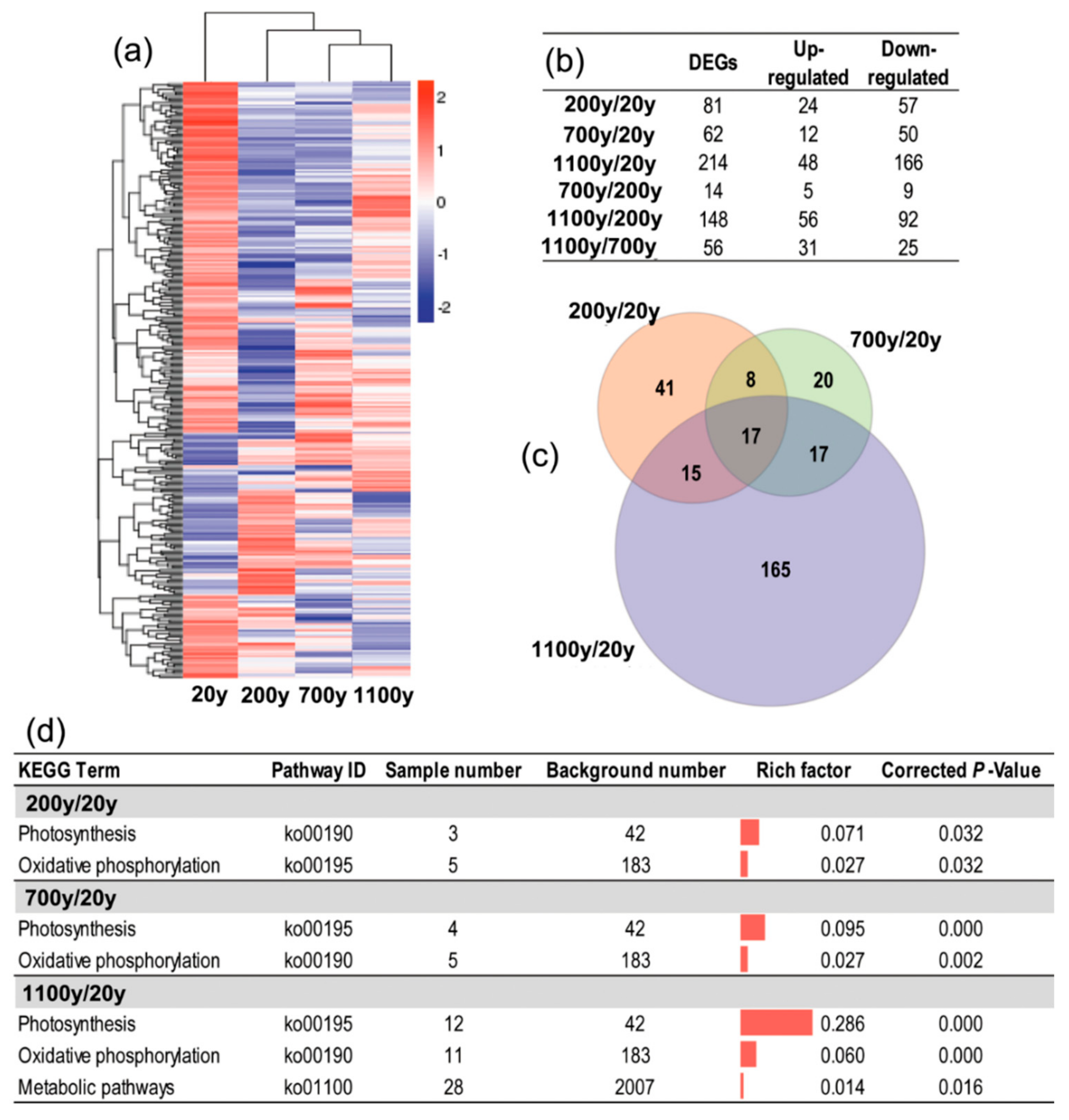
3.4. Identification of DEGs during Senescence
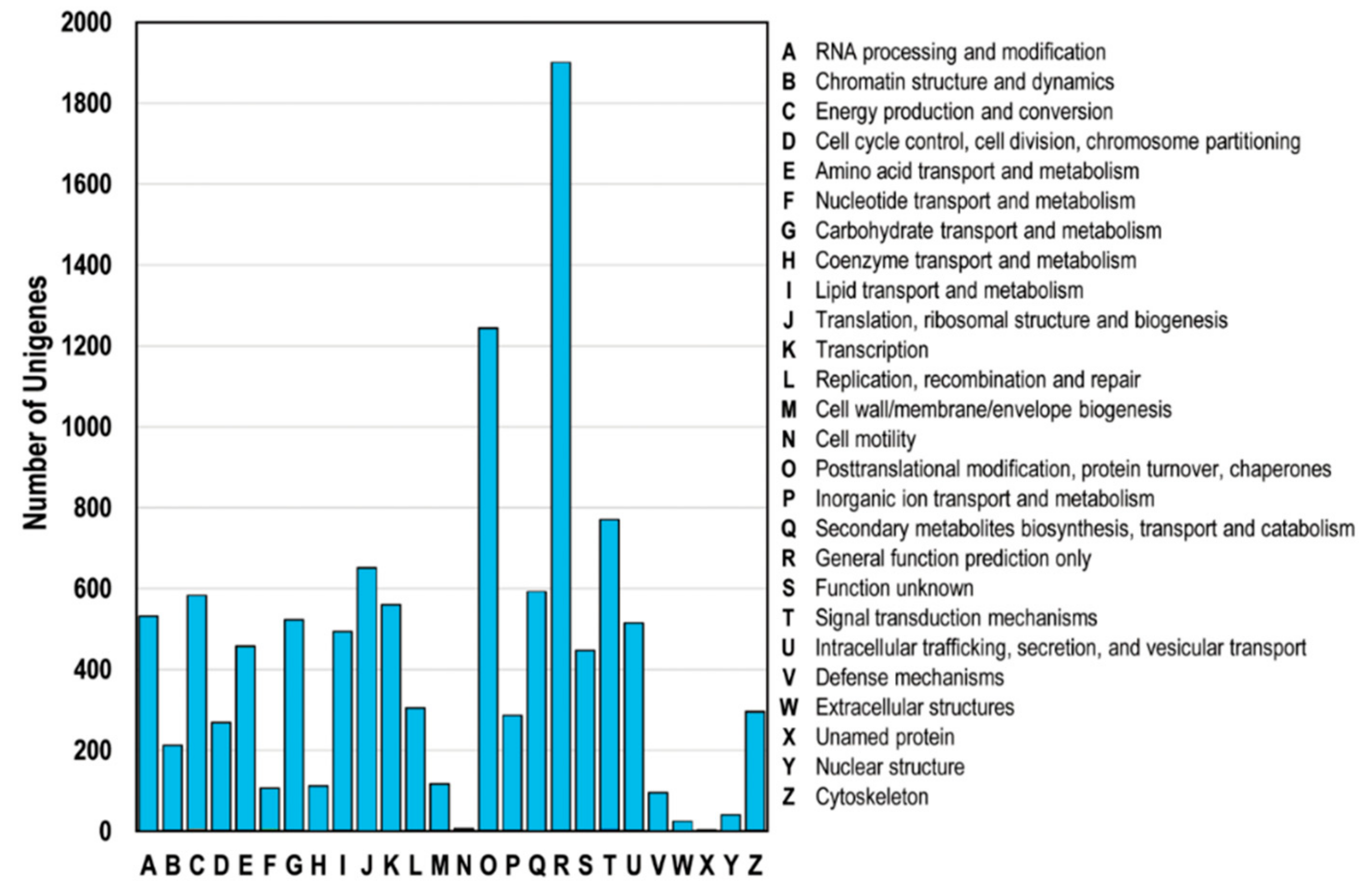
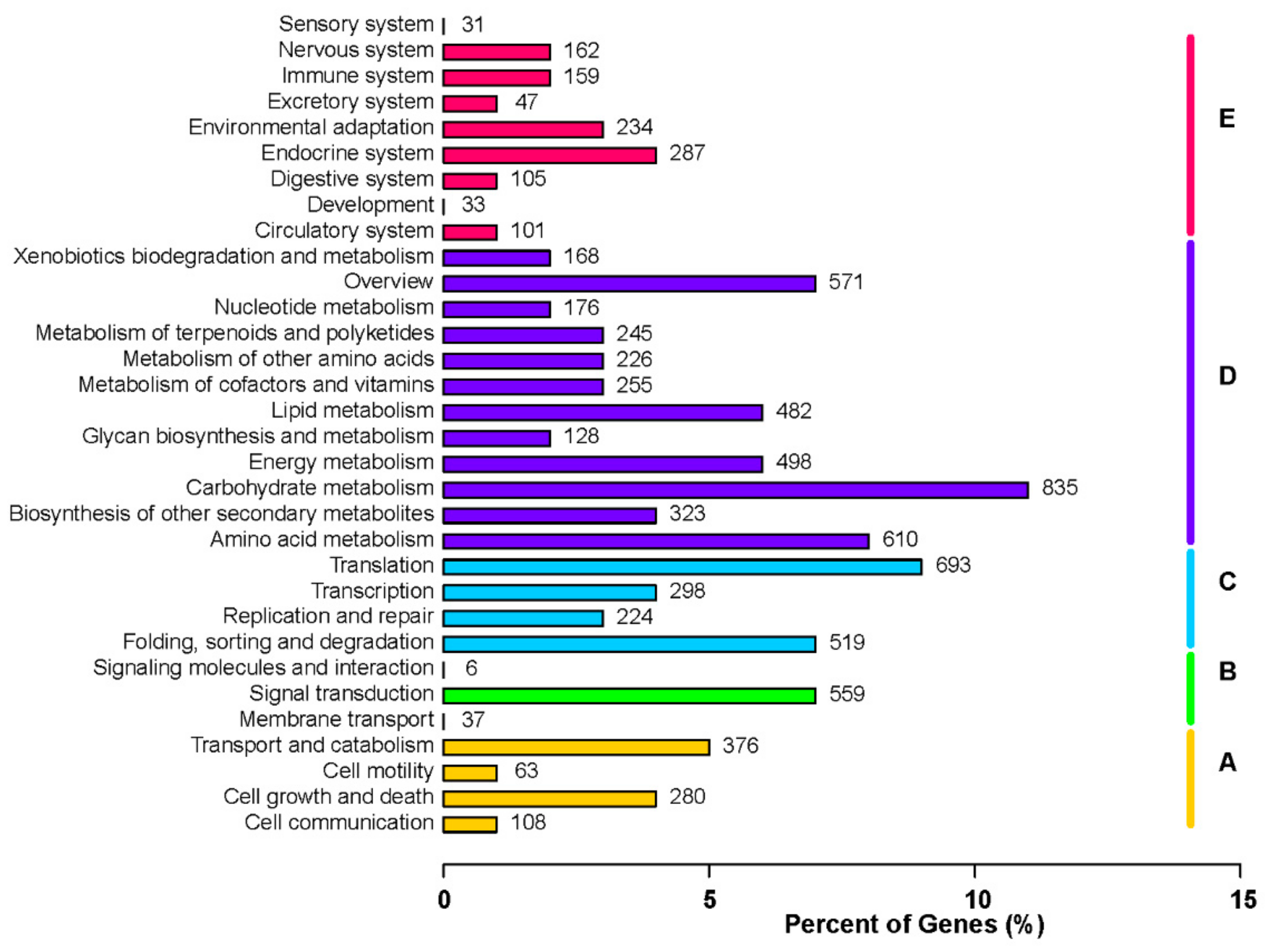
3.5. Functional Classification of DEGs
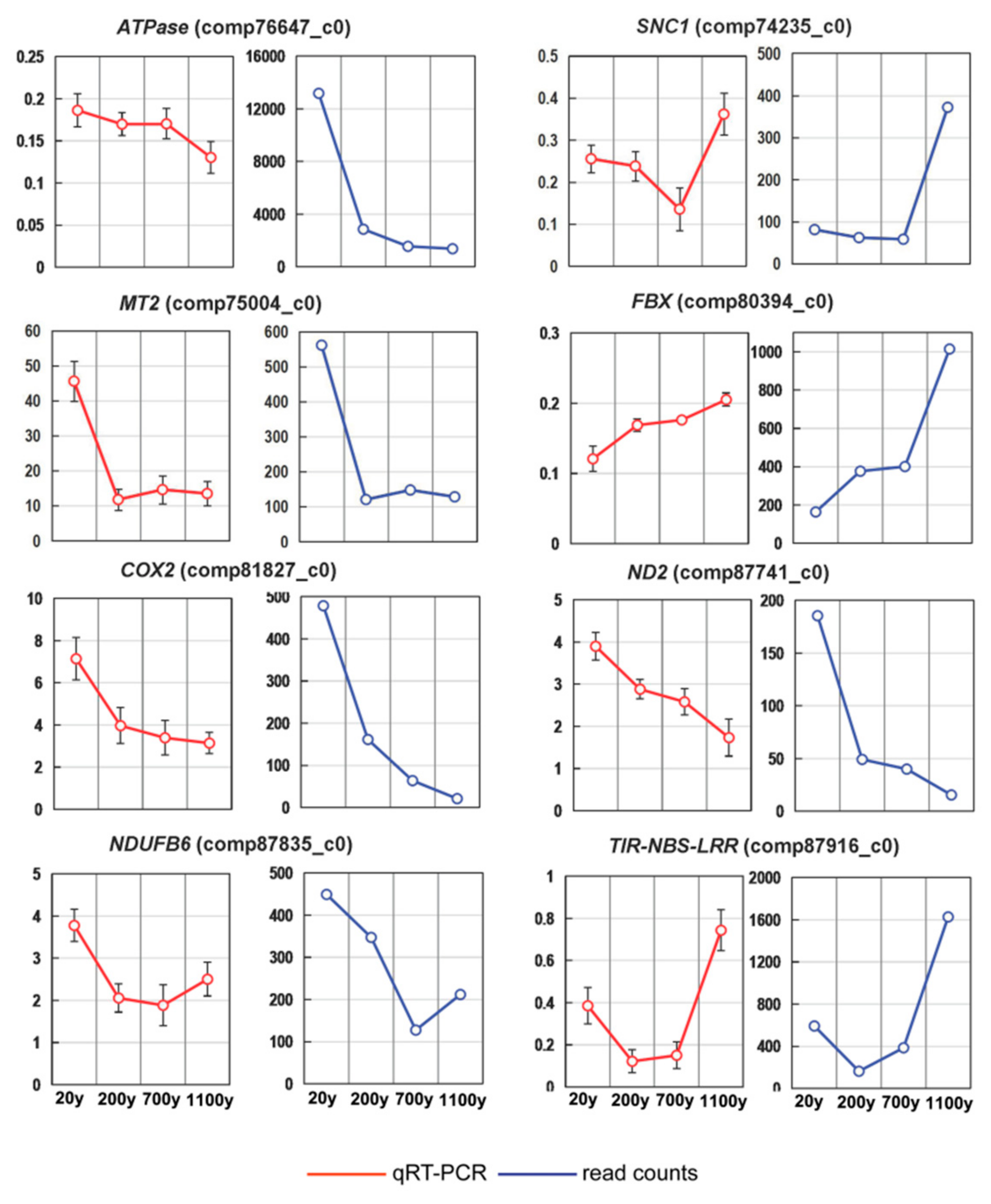
3.6. Expression Profiling of Differentially Regulated Genes by RT-qPCR
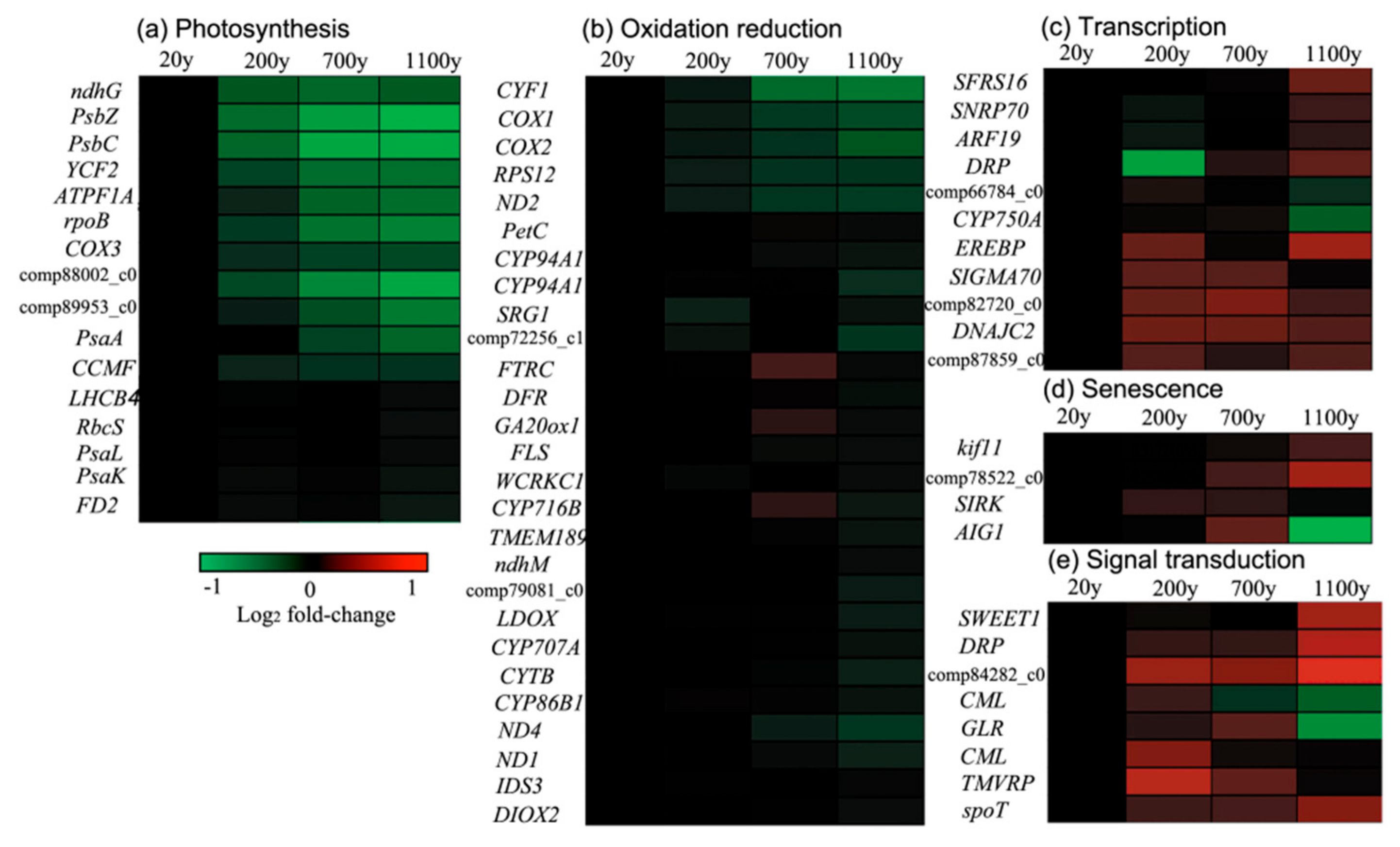
3.7. Down-Regulated Gene Clusters Resulted in Changes to Photosynthesis and the Redox Reaction
3.8. Up-Regulated Gene Clusters Resulted in Changes to Transcription, Signal Transduction, and Senescence
3.9. Validation of the Expression Kinetics of the Photosynthesis-Related Genes during Senescence
3.10. Age-Related Chlorophyll Level Changes in P. orientalis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Munné-Bosch, S. Aging in perennials. Crit. Rev. Plant Sci. 2007, 26, 123–138. [Google Scholar]
- Chang, E.; Jin, Z.; Nan, D.; Yao, X.; Liu, J.; Zhao, X.; Jiang, Z.; Shi, S. Transcriptome differences between 20- and 3000-year-old Platycladus orientalis reveals ROS involved in senescence regulation. Electron. J. Biotechn. 2017, 29, 68–77. [Google Scholar]
- Zhang, S.; Zhang, L.; Chai, Y.; Wang, F.; Li, Y.; Su, L.; Zhao, Z. Physiology and proteomics research on the leaves of ancient Platycladus orientalis (L.) during winter. J. Proteomics 2015, 126, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, S.; Finkel, T. Ageing and the mystery at Arles. Nature 2004, 429, 149–152. [Google Scholar] [CrossRef]
- Klimešová, J.; Nobis, M.P.; Herben, T. Senescence, ageing and death of the whole plant: Morphological prerequisites and constraints of plant immortality. New Phytol. 2015, 206, 14–18. [Google Scholar] [CrossRef]
- Sillett, S.C.; Van Pelt, R.; Carroll, A.L.; Kramer, R.D.; Ambrose, A.R.; Trask, D. How do tree structure and old age affect growth potential of California redwoods? Ecol. Monogr. 2015, 85, 181–212. [Google Scholar] [CrossRef]
- Lanner, R.M.; Connor, K.F. Does bristlecone pine senesce? Exp. Gerontol. 2001, 36, 675–685. [Google Scholar] [CrossRef]
- Flanary, B.E.; Kletetschka, G. Analysis of telomere length and telomerase activity in tree species of various life-spans, and with age in the bristlecone pine Pinus longaeva. Biogerontology 2005, 6, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Munné-Bosch, S. Do perennials really senesce? Trends Plant Sci. 2008, 13, 216–220. [Google Scholar] [CrossRef]
- Martínez, D.E.; Guiamet, J.J. Senescence-related changes in the leaf apoplast. J. Plant Growth Regul. 2014, 33, 44–55. [Google Scholar] [CrossRef]
- Peñuelas, J. Plant physiology: A big issue for trees. Nature 2005, 437, 965–966. [Google Scholar] [CrossRef] [PubMed]
- Issartel, J.; Coiffard, C. Extreme longevity in trees: Live slow, die old? Oecologia 2011, 165, 1–5. [Google Scholar] [CrossRef]
- Gupta, D.K.; Pena, L.B.; Romero Puertas, M.C.; Hernández, A.; Inouhe, M.; Sandalio, L.M. NADPH oxidases differentially regulate ROS metabolism and nutrient uptake under cadmium toxicity. Plant Cell Environ. 2016, 40, 509–526. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Palma, J.M.; Río, L.A.D. Antioxidative enzymes from chloroplasts, mitochondria, and peroxisomes during leaf senescence of nodulated pea plants. J. Exp. Botany 2006, 57, 1747–1758. [Google Scholar] [CrossRef]
- Keskitalo, J.; Bergquist, G.; Gardeström, P.; Jansson, S. A cellular timetable of autumn senescence. Plant Physiol. 2005, 139, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Munné-Bosch, S.; Alegre, L. Plant aging increases oxidative stress in chloroplasts. Planta 2002, 214, 608–615. [Google Scholar]
- Juvany, M.; Müller, M.; Munné-Bosch, S. Plant age-related changes in cytokinins, leaf growth and pigment accumulation in juvenile mastic trees. Environ. Exp. Botany 2013, 87, 10–18. [Google Scholar] [CrossRef]
- Luo, Z.; Guan, H.; Zhang, X.; Liu, N. Photosynthetic capacity of senescent leaves for a subtropical broad leaf deciduous tree species Liquidambar formosana Hance. Sci. Rep. 2017, 7, 6323. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, G.; Gao, H.; Zhang, L.; Yang, C.; Liu, P.; Meng, Q. Characterization of photosynthetic performance during senescence in stay-green and quick-leaf-senescence Zea mays L. Inbred Lines. PLoS ONE 2012, 7, e42936. [Google Scholar] [CrossRef]
- Gonzales, D.H.; Neupert, W. Biogenesis of mitochondrialc-type cytochromes. J. Bioenerg. Biomembr. 1990, 22, 753–768. [Google Scholar]
- Reich, P.B.; Tjoelker, M.G.; Machado, J.; Oleksyn, J. Universal scaling of respiratory metabolism, size and nitrogen in plants. Nature 2006, 439, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, C.; Yang, T.J.; Stead, A.D.; Buchanan Wollaston, V.; Roberts, J.A. A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J. 2009, 57, 690–705. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cai, Z.; Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004, 27, 521–549. [Google Scholar] [CrossRef]
- Wang, B.; Du, Q.; Yang, X.; Zhang, D. Identification and characterization of nuclear genes involved in photosynthesis in Populus. BMC Plant Biol. 2014, 14, 81. [Google Scholar] [CrossRef]
- Bhalerao, R.; Keskitalo, J.; Sterky, F.; Erlandsson, R.; Björkbacka, H.; Birve, S.J.; Karlsson, J.; Gardeström, P.; Gustafsson, P.; Lundeberg, J. Gene expression in autumn leaves. Plant Physiol. 2003, 131, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Bi, W.; Hao, X.; Xu, Y.; Li, P.; Walker, M.A.; Wang, Q. Responses of in vitro-grown plantlets (Vitis vinifera) to grapevine leaf roll-associated virus-3 and PEG-induced drought stress. Front. Physiol. 2016, 7, 203. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Plumbdhindsa, P.; Thorpe, T.A. Leaf senescence: Correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Botany 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.H.; Hughes, L.; Hickman, H.; Hill, C.; Kiddle, S.; Kim, Y.; Penfold, C.A.; Jenkins, D. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–94. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Moran, Y.; Levin, J.Z.; Thompson, D.A.; Ido, A.; Xian, A.; Lin, F.; Raktima, R.; Qiandong, Z. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Conesa, A.; Gã Tz, S.; Garcã A-Gã Mez, J.M.; Terol, J.; Talã N, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Tang, Q.; Ma, X.; Mo, C.; Wilson, I.W.; Song, C.; Zhao, H.; Yang, Y.; Fu, W.; Qiu, D. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genom. 2011, 12, 343. [Google Scholar] [CrossRef]
- Bo, L.; Dewey, C.N.; Li, B. Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar]
- Springer, A.; Acker, G.; Bartsch, S.; Bauerschmitt, H.; Reinbothe, S.; Reinbothe, C. Differences in gene expression between natural and artificially induced leaf senescence in barley. J. Plant Physiol. 2015, 176, 180–191. [Google Scholar] [CrossRef]
- Deng, N.; Chang, E.; Li, M.; Ji, J.; Yao, X.; Bartish, I.V.; Liu, J.; Ma, J.; Chen, L.; Jiang, Z. Transcriptome characterization of Gnetum parvifolium reveals candidate genes involved in important secondary metabolic pathways of flavonoids and stilbenoids. Front. Plant Sci. 2016, 7, 174. [Google Scholar] [CrossRef]
- Andersson, A.; Keskitalo, J.; Sjödin, A.; Bhalerao, R.; Sterky, F.; Wissel, K.; Tandre, K.; Aspeborg, H.; Moyle, R.; Ohmiya, Y. A transcriptional timetable of autumn senescence. Genome Biol. 2004, 5, 1–13. [Google Scholar] [CrossRef]
- Wang, P.; Sun, X.; Li, C.; Wei, Z.; Liang, D.; Ma, F. Long-term exogenous application of melatonin delays drought-induced leaf senescence in apple. J. Pineal Res. 2013, 54, 292–302. [Google Scholar] [CrossRef]
- Amunts, A.; Toporik, H.; Borovikova, A.; Nelson, N. Structure determination and improved model of plant photosystem I. J. Biol. Chem. 2010, 285, 3478–3486. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, R.M.; Ryan, M.V.; Sperry, J.S. Stomatal conductance and photosynthesis vary linearly with plant hydraulic conductance in ponderosa pine. Tree Physiol. 2010, 24, 113–121. [Google Scholar] [CrossRef]
- Blokhina, O.; Fagerstedt, K.V. Reactive oxygen species and nitric oxide in plant mitochondria: Origin and redundant regulatory systems. Physiol. Plantarum 2010, 138, 447–462. [Google Scholar] [CrossRef]
- Rivas, F.; Fornes, F.; Rodrigo, M.J.; Zacarías, L.; Agustí, M. Changes in carotenoids and ABA content in citrus leaves in response to girdling. Sci. Horticult. 2011, 127, 482–487. [Google Scholar] [CrossRef]
- Małgorzata, G.; Piotr, L.; Krystyna, R. Two-dimensional zymography in detection of proteolytic enzymes in wheat leaves. Acta Physiol. Plant. 2013, 35, 3477–3482. [Google Scholar]
- Yutaka, S.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2008, 57, 120–131. [Google Scholar]
- Locke, A.M.; Ort, D.R. Leaf hydraulic conductance declines in coordination with photosynthesis, transpiration and leaf water status as soybean leaves age regardless of soil moisture. J. Exp. Bot. 2014, 65, 6617–6627. [Google Scholar] [CrossRef]
- Liu, A.; Chen, S.; Wang, M.; Wang, Z.; Zheng, C.; Zhao, P.; Guo, D.; Ahammed, G.J. Silencing of mitochondrial uncoupling protein gene aggravates chilling stress by altering mitochondrial respiration and electron transport in tomato. Acta Physiol. Plant. 2015, 37, 1–9. [Google Scholar] [CrossRef]
- Ott, M.; Gogvadze, V.; Orrenius, S.; Zhivotovsky, B. Mitochondria, oxidative stress and cell death. Apoptosis 2007, 12, 913–922. [Google Scholar] [CrossRef]
- Wang, C.; Yang, Y.; Wang, H.; Ran, X.; Li, B.; Zhang, J.; Zhang, H. Ectopic expression of a cytochrome P450 monooxygenase gene PtCYP714A3 from Populus trichocarpa reduces shoot growth and improves tolerance to salt stress in transgenic rice. Plant Biotechnol. J. 2016, 14, 1838–1851. [Google Scholar] [CrossRef] [PubMed]
- Pinot, F.; Skrabs, M.; Compagnon, V.; Salaün, J.P.; Benveniste, I.; Schreiber, L.; Durst, F. Omega-Hydroxylation of epoxy- and hydroxy-fatty acids by CYP94A1: Possible involvement in plant defence. Biochem. Soc. Trans. 2000, 28, 867–870. [Google Scholar] [CrossRef]




| Annotated Databases | Number of Unigenes | Percentage (%) |
|---|---|---|
| Annotated in NR | 24,080 | 50.12 |
| Annotated in NT | 8380 | 17.44 |
| Annotated in KO | 7783 | 16.19 |
| Annotated in Swiss-Prot | 18,483 | 38.47 |
| Annotated in PFAM | 18,546 | 38.6 |
| Annotated in GO | 20,029 | 41.68 |
| Annotated in KOG | 9877 | 20.55 |
| Annotated in all databases | 3105 | 6.46 |
| Annotated in at least one database | 26,226 | 54.58 |
| Total Unigenes | 48,044 | 100 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, E.; Zhang, J.; Yao, X.; Tang, S.; Zhao, X.; Deng, N.; Shi, S.; Liu, J.; Jiang, Z. De novo Characterization of the Platycladus orientalis Transcriptome and Analysis of Photosynthesis-Related Genes during Aging. Forests 2019, 10, 393. https://doi.org/10.3390/f10050393
Chang E, Zhang J, Yao X, Tang S, Zhao X, Deng N, Shi S, Liu J, Jiang Z. De novo Characterization of the Platycladus orientalis Transcriptome and Analysis of Photosynthesis-Related Genes during Aging. Forests. 2019; 10(5):393. https://doi.org/10.3390/f10050393
Chicago/Turabian StyleChang, Ermei, Jin Zhang, Xiamei Yao, Shuo Tang, Xiulian Zhao, Nan Deng, Shengqing Shi, Jianfeng Liu, and Zeping Jiang. 2019. "De novo Characterization of the Platycladus orientalis Transcriptome and Analysis of Photosynthesis-Related Genes during Aging" Forests 10, no. 5: 393. https://doi.org/10.3390/f10050393
APA StyleChang, E., Zhang, J., Yao, X., Tang, S., Zhao, X., Deng, N., Shi, S., Liu, J., & Jiang, Z. (2019). De novo Characterization of the Platycladus orientalis Transcriptome and Analysis of Photosynthesis-Related Genes during Aging. Forests, 10(5), 393. https://doi.org/10.3390/f10050393





