Leaf-Associated Shifts in Bacterial and Fungal Communities in Response to Chicken Rearing Under Moso Bamboo Forests in Subtropical China
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Design
2.2. Sampling
2.3. Illumina High-Throughput Sequencing
2.4. Data Analyses
3. Results
3.1. Compositions of Bacterial and Fungal Communities
3.2. Bacterial and Fungal Community Diversities
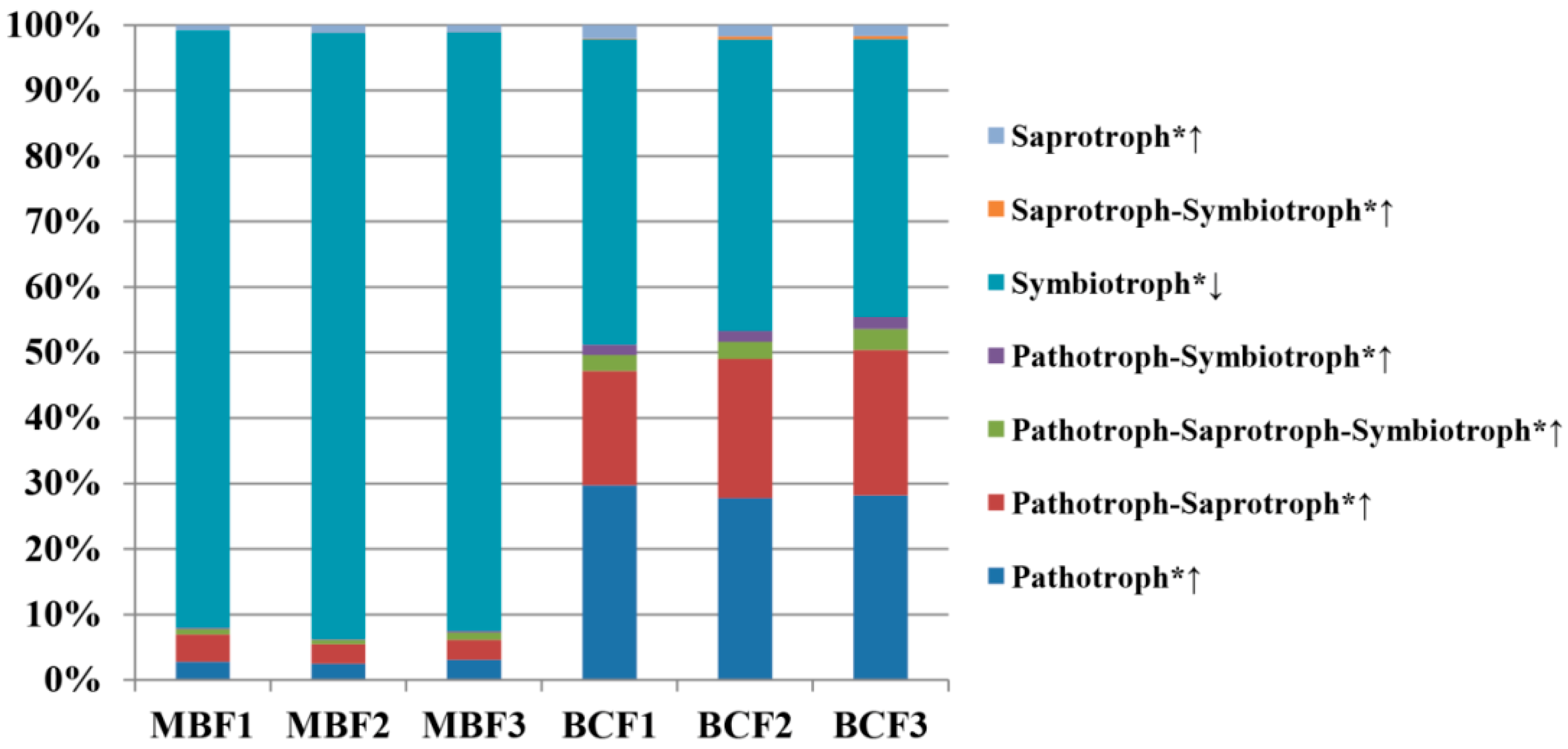
3.3. Ecological Function Evaluation of the Niche Shift of Leaf-Associated Microbiome
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- FAO. Global Forest Resources Assessment; FAO Forestry Paper: Rome, Italy, 2010. [Google Scholar]
- Xie, L.; Li, X.; Hou, D.; Cheng, Z.; Liu, J.; Li, J.; Mu, S.; Gao, J. Genome-Wide Analysis and Expression Profiling of the Heat Shock Factor Gene Family in Phyllostachys edulis during Development and in Response to Abiotic Stresses. Forests 2019, 10, 100. [Google Scholar] [CrossRef]
- Li, L.; Cheng, Z.; Ma, Y.; Bai, Q.; Li, X.; Cao, Z.; Wu, Z.; Gao, J. The association of hormone signaling genes, transcription, and changes in shoot anatomy during moso bamboo growth. Plant Biotechnol. J. 2017, 16, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Shi, Y.; Zhou, G.; Xu, X.; Liu, E.; Zhou, Y.; Li, C.; Fang, H.; Deng, X. Temporal Change in Aboveground Culms Carbon Stocks in the Moso Bamboo Forests and Its Driving Factors in Zhejiang Province, China. Forests 2017, 8, 371. [Google Scholar] [CrossRef]
- Gu, L.; Zhou, Y.; Mei, T.; Zhou, G.; Xu, L. Carbon Footprint Analysis of Bamboo Scrimber Flooring—Implications for Carbon Sequestration of Bamboo Forests and Its Products. Forests 2019, 10, 51. [Google Scholar] [CrossRef]
- Flynn, A.; Chan, K.W.; Zhu, Z.H.; Yu, L. Sustainability, space and supply chains: The role of bamboo in Anji County, China. J. Rural. Stud. 2017, 49, 128–139. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, H.; Cai, Z.; Zhou, X.; Peng, C. The response of the net primary production of Moso bamboo forest to the On and Off-year management: A case study in Anji County, Zhejiang, China. For. Ecol. Manag. 2018, 409, 1–7. [Google Scholar] [CrossRef]
- Song, X.; Peng, C.; Zhou, G.; Gu, H.; Li, Q.; Zhang, C. Dynamic allocation and transfer of non-structural carbohydrates, a possible mechanism for the explosive growth of Moso bamboo (Phyllostachys heterocycla). Sci. Rep. 2016, 6, 25908. [Google Scholar] [CrossRef] [PubMed]
- FAO. FAO Projects. 2015. Available online: http://www.fao.org/forestry/agroforestry/90030/en/ (accessed on 27 March 2017).
- Kittur, B.; Sudhakara, K.; Kumar, B.M.; Kunhamu, T.; Sureshkumar, P. Bamboo based agroforestry systems in Kerala, India: Performance of turmeric (Curcuma longa L.) in the subcanopy of differentially spaced seven year-old bamboo stand. Agroforest. Syst. 2016, 90, 237–250. [Google Scholar] [CrossRef]
- Zhang, X.; Wen, T.; Zhang, Z.; Huang, S.; Huo, D.; Jiang, X.; Yuan, K.; Shen, X.; Huang, X. A Study of Vegetation on Biomass Changes and Black-Bone Chicken Breeding Technologies under Phyllostachys Pubesebs Forest in Chishui River Basin. World Bamboo Rattan 2015, 13, 1–7. [Google Scholar]
- Zhu, C.; Yang, C.; Shen, X.; Wang, B. Effects of Raising Chicken on Soil Quality and Bamboo Growth in Phyllostachys edulis Forest. J. Bamboo Res. 2018, 37, 49–53. [Google Scholar]
- Levy, A.; Conway, J.M.; Dangl, J.L.; Woyke, T. Elucidating Bacterial Gene Functions in the Plant Microbiome. Cell Host Microbe 2018, 24, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Rothballer, M.; Schmid, M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 2008, 312, 7–14. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bulgarelli, D. The plant microbiome at work. Mol. Plant-Microbe Interact. 2015, 28, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Liu, F.; Zhang, G. Isolation of culturable endophytic bacteria from Moso bamboo (Phyllostachys edulis) and 16S rDNA diversity analysis. Arch. Biol. Sci. 2015, 67, 1001–1008. [Google Scholar] [CrossRef]
- Shen, X.-Y.; Cheng, Y.-L.; Cai, C.-J.; Fan, L.; Gao, J.; Hou, C.-L. Diversity and antimicrobial activity of culturable endophytic fungi isolated from moso bamboo seeds. PLoS ONE 2014, 9, e95838. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yuan, Z.; Zhang, X.; Zhang, G.; Xie, B. Characteristics and diversity of endophytic bacteria in moso bamboo (Phyllostachys edulis) based on 16S rDNA sequencing. Arch. Microbiol. 2017, 199, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; O’Connor, T.K.; Arnold, H.K.; Hubbell, S.P.; Wright, S.J.; Green, J.L. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. USA 2014, 111, 13715–13720. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Coaker, G.L.; Leveau, J.H. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 2013, 348, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, C.; Liese, W. Diseases of bamboos. Int. J. Trop. Plant Dis. 1990, 8, 1–20. [Google Scholar]
- Yang, C.; Zhong, Z.; Zhang, X.; Bian, F.; Du, X. Responses of Soil Organic Carbon Sequestration Potential and Bacterial Community Structure in Moso Bamboo Plantations to Different Management Strategies in Subtropical China. Forests 2018, 9, 657. [Google Scholar] [CrossRef]
- Chen, C.R.; Xu, Z.H.; Mathers, N.J. Soil Carbon Pools in Adjacent Natural and Plantation Forests of Subtropical Australia. Soil Sci. Soc. Am. J. 2004, 68, 282–291. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Tedersoo, L.; Ryberg, M.; Kristiansson, E.; Hartmann, M.; Unterseher, M.; Porter, T.M.; Bengtsson-Palme, J.; Walker, D.M.; De Sousa, F. A comprehensive, automatically updated fungal ITS sequence dataset for reference-based chimera control in environmental sequencing efforts. Microbes Environ. 2015, 30, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Henrik Nilsson, R.; Larsson, K.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T. The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Arndt, D.; Xia, J.; Liu, Y.; Zhou, Y.; Guo, A.C.; Cruz, J.A.; Sinelnikov, I.; Budwill, K.; Nesbø, C.L.; Wishart, D.S. METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012, 40, W88–W95. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Yen, T.-M.; Lee, J.-S. Comparing aboveground carbon sequestration between moso bamboo (Phyllostachys heterocycla) and China fir (Cunninghamia lanceolata) forests based on the allometric model. For. Ecol. Manag. 2011, 261, 995–1002. [Google Scholar] [CrossRef]
- Christanty, L.; Mailly, D.; Kimmins, J. “Without bamboo, the land dies”: Biomass, litterfall, and soil organic matter dynamics of a Javanese bamboo talun-kebun system. For. Ecol. Manag. 1996, 87, 75–88. [Google Scholar] [CrossRef]
- Lobovikov, M.; Schoene, D.; Yping, L. Bamboo in climate change and rural livelihoods. Mitig. Adapt. Strateg. Glob. Chang. 2012, 17, 261–276. [Google Scholar] [CrossRef]
- Rigonato, J.; Gonçalves, N.; Andreote, A.P.D.; Lambais, M.R.; Fiore, M.F. Estimating genetic structure and diversity of cyanobacterial communities in Atlantic forest phyllosphere. Can. J. Microbiol. 2016, 62, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Klawonn, I.; Nahar, N.; Walve, J.; Andersson, B.; Olofsson, M.; Svedén, J.; Littmann, S.; Whitehouse, M.J.; Kuypers, M.; Ploug, H. Cell-specific nitrogen-and carbon-fixation of cyanobacteria in a temperate marine system (Baltic Sea). Environ. Microbiol. 2016, 18, 4596–4609. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, M.; Egardt, J.; Singh, A.; Ploug, H. Inorganic phosphorus enrichments in Baltic Sea water have large effects on growth, carbon fixation, and N2 fixation by Nodularia spumigena. Aquat. Microb. Ecol. 2016, 77, 111–123. [Google Scholar] [CrossRef]
- Chaverri, P.; Vilchez, B. Hypocrealean (Hypocreales, Ascomycota) Fungal Diversity in Different Stages of Tropical Forest Succession in Costa Rica. Biotropica 2006, 38, 531–543. [Google Scholar] [CrossRef]
- Trappe, J.M.; Castellano, M.A. New sequestrate Ascomycota and Basidiomycota covered by the northwest forest plan. Mycotaxon 2000, 75, 153–180. [Google Scholar]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Dimitrijević, S.; Pavlović, M.; Maksimović, S.; Ristić, M.; Filipović, V.; Antonović, D.; Dimitrijević-Branković, S. Plant growth-promoting bacteria elevate the nutritional and functional properties of black cumin and flaxseed fixed oil. J. Sci. Food Agric. 2018, 98, 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.P.; Sobral, B.W.; Dickerman, A.W. A robust species tree for the alphaproteobacteria. J. Bacteriol. 2007, 189, 4578–4586. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xing, K.; Jiang, J.-H.; Xu, L.-H.; Li, W.-J. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 2011, 89, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Aviles-Garcia, M.; Flores-Cortez, I.; Hernández-Soberano, C.; Santoyo, G.; Valencia-Cantero, E. The plant growth-promoting rhizobacterium Arthrobacter agilis UMCV2 endophytically colonizes Medicago truncatula. Rev. Argent. Microbiol. 2016, 48, 342–346. [Google Scholar] [PubMed]
- Chen, Z.; Li, L.; Shan, Z.; Huang, H.; Chen, H.; Ding, X.; Guo, J.; Liu, L. Transcriptome sequencing analysis of novel sRNAs of Kineococcus radiotolerans in response to ionizing radiation. Microbiol. Res. 2016, 192, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Dourado, M.N.; Bogas, A.C.; Pomini, A.M.; Andreote, F.D.; Quecine, M.C.; Marsaioli, A.J.; Araújo, W.L. Methylobacterium-plant interaction genes regulated by plant exudate and quorum sensing molecules. Braz. J. Microbiol. 2013, 44, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Fries, J.; Pfeiffer, S.; Kuffner, M.; Sessitsch, A. Spirosomaendophyticum sp. nov.; isolated from Zn-and Cd-accumulating Salix caprea. Int. J. Syst. Evol. Microbiol. 2013, 63, 4586–4590. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-J.; Srinivasan, S.; Lim, S.; Joe, M.; Im, S.; Bae, S.I.; Park, K.R.; Han, J.-H.; Park, S.-H.; Joo, B.-m. Spirosoma radiotolerans sp. nov.; a gamma-radiation-resistant bacterium isolated from gamma ray-irradiated soil. Curr. Microbiol. 2014, 69, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, S.; Islam, E.; Chen, J.-r.; Wu, J.-s.; Ye, Z.-q.; Peng, D.-l.; Yan, W.-b.; Lu, K.-p. Lead accumulation and tolerance of Moso bamboo (Phyllostachys pubescens) seedlings: Applications of phytoremediation. J. Zhejiang Univ. Sci. B 2015, 16, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Bian, F.; Zhong, Z.; Zhang, X.; Yang, C. Phytoremediation potential of moso bamboo (Phyllostachys pubescens) intercropped with Sedum plumbizincicola in metal-contaminated soil. Environ. Sci. Pollut. 2017, 24, 27244–27253. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Amna, A.; Opiyo, S.O. The culturable endophytic fungal communities of switchgrass grown on a coal-mining site and their effects on plant growth. PLoS ONE 2018, 13, e0198994. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Gond, S.K.; Mishra, A.; Sharma, V.K.; Kumar, J.; Singh, D.K.; Kumar, A.; Kharwar, R.N. Fungal Endophytes Representing Diverse Habitats and Their Role in Plant Protection. In Developments in Fungal Biology and Applied Mycology; Springer: Singapore, 2017; pp. 135–157. [Google Scholar]
- Rodriguez, R.J.; Henson, J.; Van Volkenburgh, E.; Hoy, M.; Wright, L.; Beckwith, F.; Kim, Y.-O.; Redman, R.S. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008, 2, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Gundel, P.; Helander, M.; Garibaldi, L.; Vázquez-de-Aldana, B.; Zabalgogeazcoa, I.; Saikkonen, K. Direct and indirect effects of the fungal endophyte Epichloë uncinatum on litter decomposition of the host grass, Schedonorus pratensis. Plant Ecol. 2017, 218, 1107–1115. [Google Scholar] [CrossRef]
- Rodriguez, R.; Redman, R. More than 400 million years of evolution and some plants still can’t make it on their own: Plant stress tolerance via fungal symbiosis. J. Exp. Bot. 2008, 59, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Gafni, A.; Calderon, C.E.; Harris, R.; Buxdorf, K.; Dafa-Berger, A.; Zeilinger-Reichert, E.; Levy, M. Biological control of the cucurbit powdery mildew pathogen Podosphaera xanthii by means of the epiphytic fungus Pseudozyma aphidis and parasitism as a mode of action. Front. Plant. Sci. 2015, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Barda, O.; Shalev, O.; Alster, S.; Buxdorf, K.; Gafni, A.; Levy, M. Pseudozyma aphidis induces salicylic-acid-independent resistance to Clavibacter michiganensis in tomato plants. Plant Dis. 2015, 99, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Osono, T.; Bhatta, B.K.; Takeda, H. Phyllosphere fungi on living and decomposing leaves of giant dogwood. Mycoscience 2004, 45, 35–41. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Baldauf, S.L.; Leyval, C.; Straczek, J.; Young, J.P.W. Extensive fungal diversity in plant roots. Science 2002, 295, 2051. [Google Scholar] [CrossRef] [PubMed]
- Bastian, F.; Bouziri, L.; Nicolardot, B.; Ranjard, L. Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol. Biochem. 2009, 41, 262–275. [Google Scholar] [CrossRef]
- Osono, T.; Takeda, H. Fungal decomposition of Abies needle and Betula leaf litter. Mycologia 2006, 98, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Yelle, D.J.; Ralph, J.; Lu, F.; Hammel, K.E. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ. Microbiol. 2008, 10, 1844–1849. [Google Scholar] [CrossRef] [PubMed]
- Lundell, T.K.; Mäkelä, M.R.; Hildén, K. Lignin-modifying enzymes in filamentous basidiomycetes–ecological, functional and phylogenetic review. J. Basic Microbiol. 2010, 50, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Bardgett, R.; Leemans, D.; Cook, R.; Hobbs, P. Seasonality of the soil biota of grazed and ungrazed hill grasslands. Soil Biol. Biochem. 1997, 29, 1285–1294. [Google Scholar] [CrossRef]
- Kohler, F.; Hamelin, J.; Gillet, F.; Gobat, J.-M.; Buttler, A. Soil microbial community changes in wooded mountain pastures due to simulated effects of cattle grazing. Plant Soil 2005, 278, 327–340. [Google Scholar] [CrossRef]
- Kauffman, J.B.; Thorpe, A.S.; Brookshire, E.J. Livestock exclusion and belowground ecosystem responses in riparian meadows of eastern Oregon. Ecol. Appl. 2004, 14, 1671–1679. [Google Scholar] [CrossRef]
- Kobayashi, T.; Hori, Y.; Nomoto, N. Effects of trampling and vegetation removal on species diversity and micro-environment under different shade conditions. J. Veg. Sci. 1997, 8, 873–880. [Google Scholar] [CrossRef]
- Skraban, J.; Dzeroski, S.; Zenko, B.; Tusar, L.; Rupnik, M. Changes of poultry faecal microbiota associated with Clostridium difficile colonisation. Vet. Microbiol. 2013, 165, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Hughes, R.J.; Moore, R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014, 98, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Maciorowski, K.G.; Herrera, P.; Jones, F.T.; Pillai, S.D.; Ricke, S.C. Effects on poultry and livestock of feed contamination with bacteria and fungi. Anim. Feed Sci. Technol. 2007, 133, 109–136. [Google Scholar] [CrossRef]
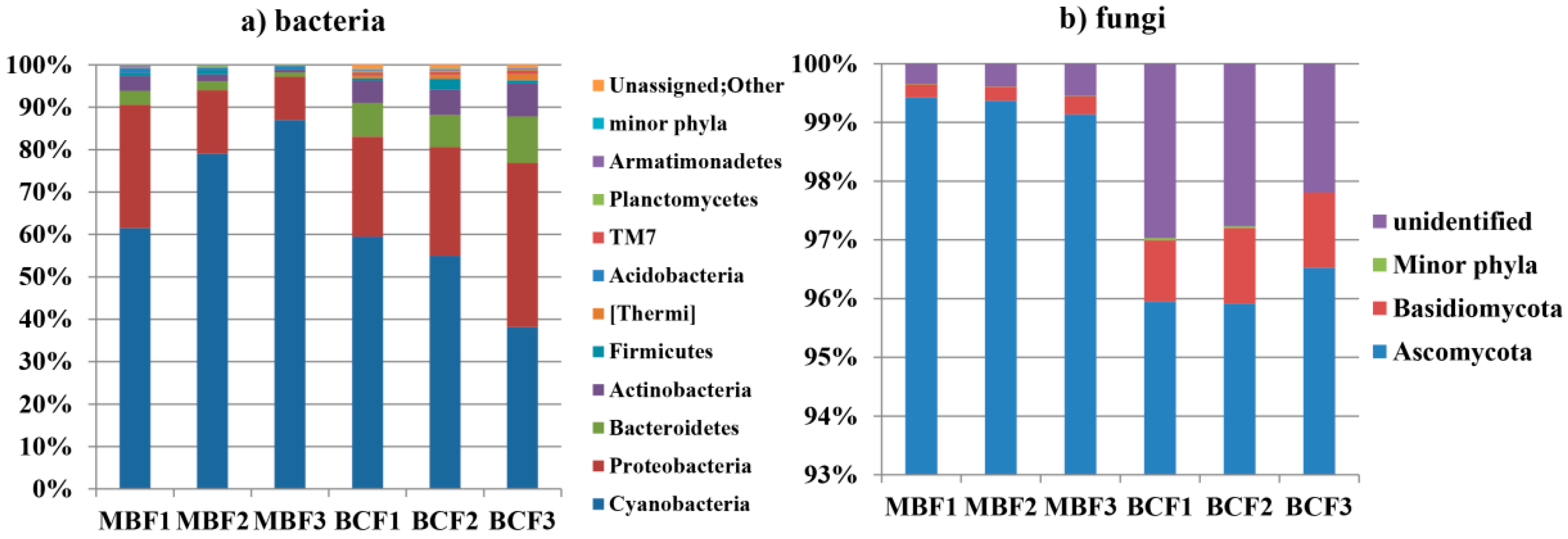
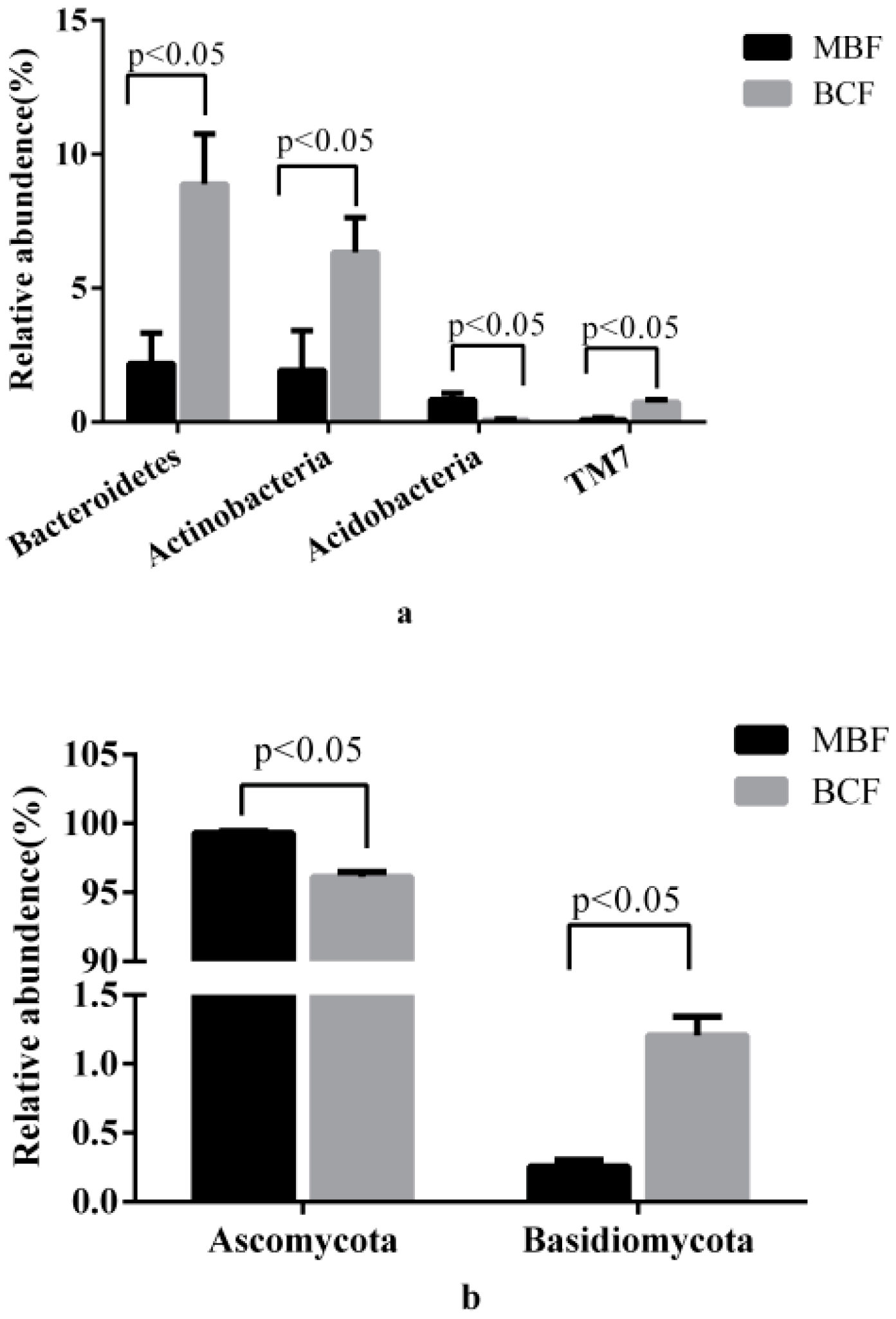
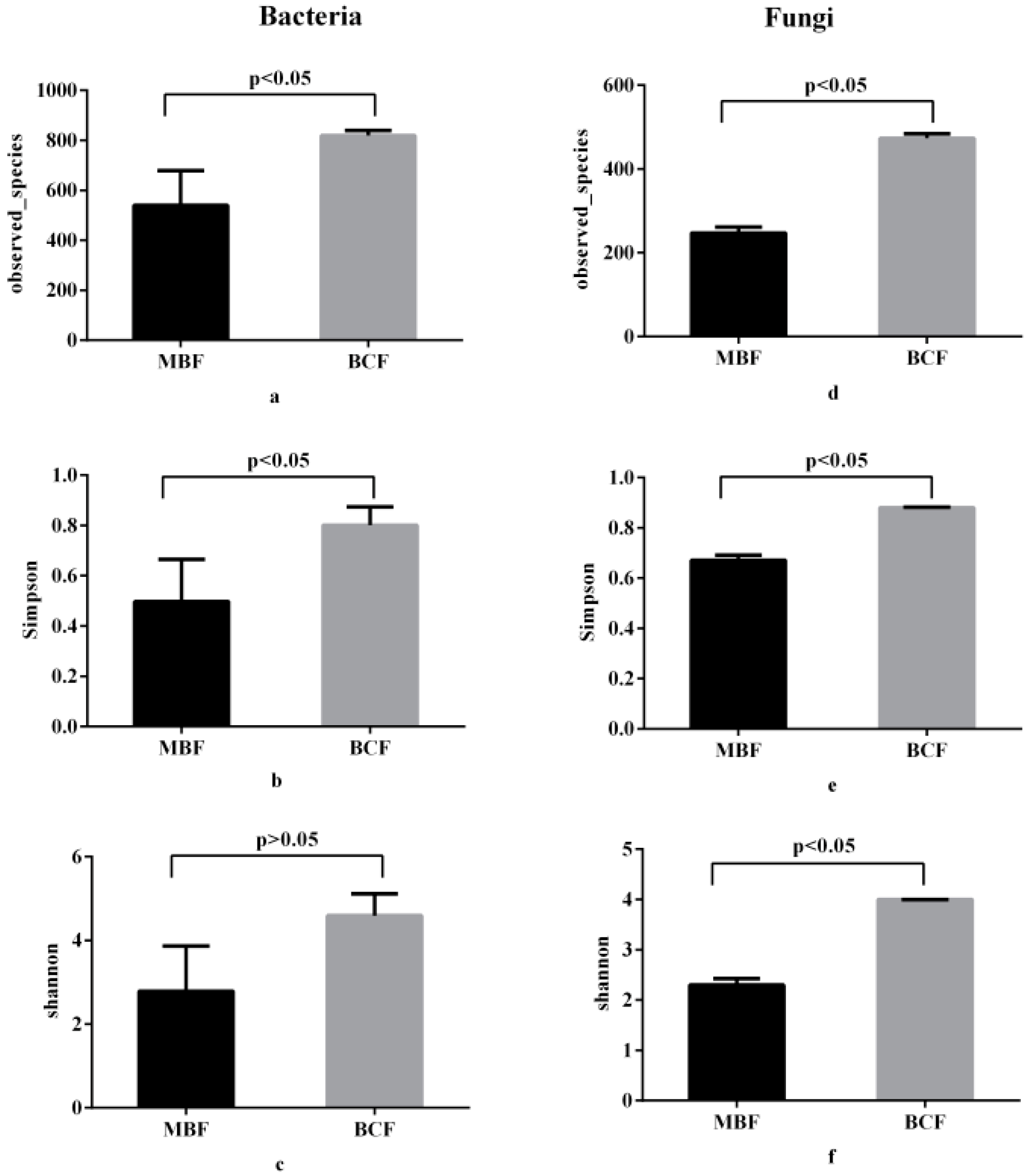

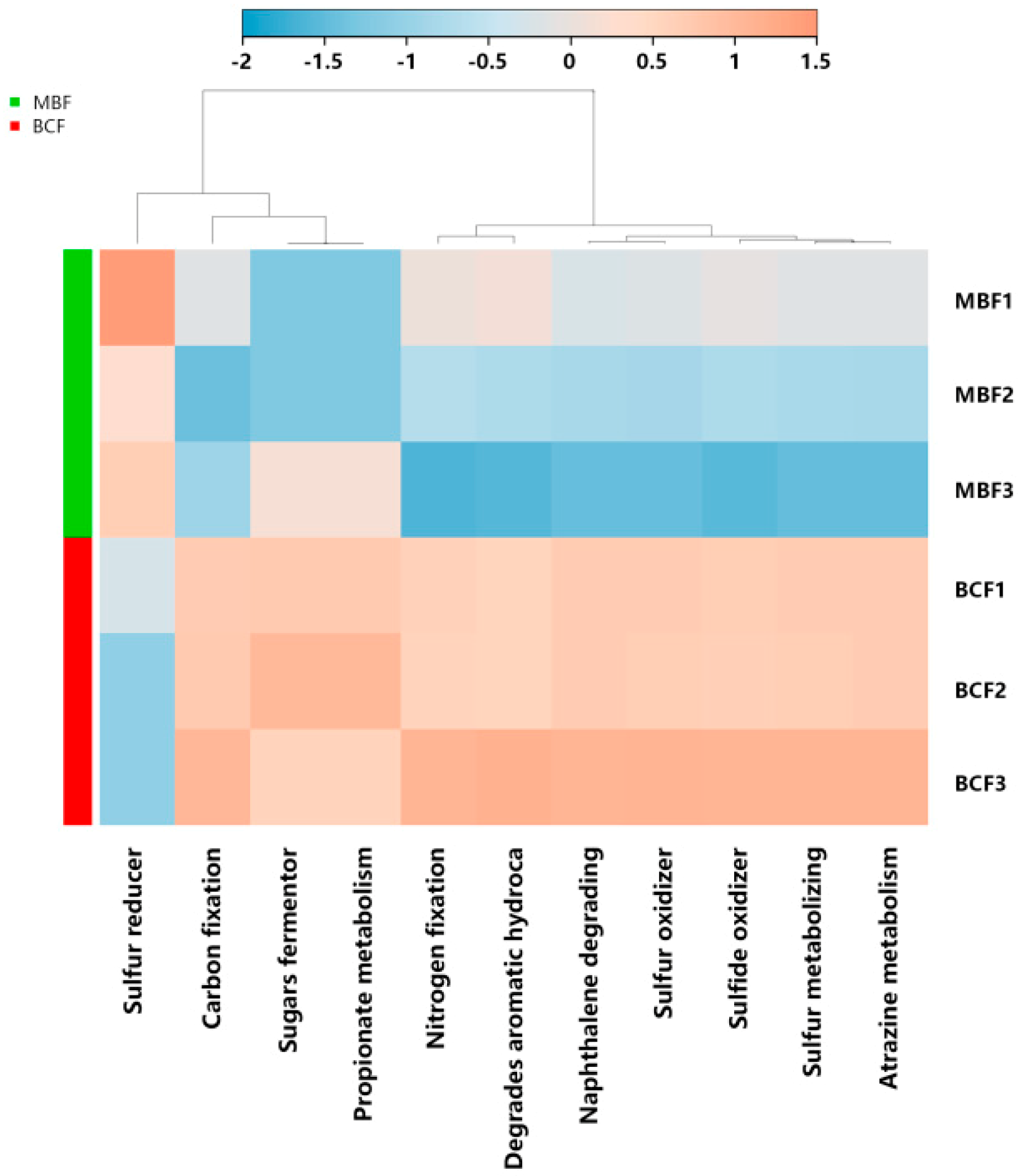
| Phylum | Genus | MBF | BCF | p Value | |
|---|---|---|---|---|---|
| Bacteria | Proteobacteria | Methylobacterium | 2.05% ± 1.46% | 13.40% ± 3.96% | 0.010 |
| Bacteroidetes | Hymenobacter | 1.84% ± 0.88% | 7.64% ± 2.14% | 0.012 | |
| Proteobacteria | Sphingomonas | 3.57% ± 2.41% | 5.84% ± 2.20% | 0.297 | |
| [Thermi] | Deinococcus | 0.00% ± 0.01% | 1.02% ± 0.42% | 0.052 | |
| Actinobacteria | Kineococcus | 0.08% ± 0.05% | 0.88% ± 0.12% | 0.003 | |
| Bacteroidetes | Spirosoma | 0.02% ± 0.01% | 0.79% ± 0.23% | 0.027 | |
| Proteobacteria | Beijerinckia | 0.58% ± 0.46% | 0.11% ± 0.07% | 0.158 | |
| Actinobacteria | Curtobacterium | 0.11% ± 0.09% | 0.26% ± 0.15% | 0.203 | |
| Actinobacteria | Friedmanniella | 0.00% ± 0.00% | 0.33% ± 0.06% | 0.001 | |
| Actinobacteria | Microbacterium | 0.22% ± 0.22% | 0.10% ± 0.04% | 0.446 | |
| Proteobacteria | Ralstonia | 0.18% ± 0.20% | 0.14% ± 0.11% | 0.790 | |
| Proteobacteria | Burkholderia | 0.25% ± 0.20% | 0.03% ± 0.01% | 0.197 | |
| Proteobacteria | Bdellovibrio | 0.08% ± 0.05% | 0.17% ± 0.03% | 0.039 | |
| Acidobacteria | Terriglobus | 0.20% ± 0.07% | 0.04% ± 0.02% | 0.019 | |
| Actinobacteria | Arthrobacter | 0.01% ± 0.01% | 0.22% ± 0.08% | 0.012 | |
| Fungi | Ascomycota | Alatosessilispora | 0.02% ± 0.02% | 25.30% ± 2.14% | 0.002 |
| Ascomycota | Strelitziana | 0.03% ± 0.02% | 24.17% ± 0.84% | 0.000 | |
| Ascomycota | Shiraia | 3.11% ± 0.69% | 0.40% ± 0.10% | 0.003 | |
| Ascomycota | Cladosporium | 0.11% ± 0.05% | 1.91% ± 0.35% | 0.001 | |
| Ascomycota | Camptophora | 0.05% ± 0.01% | 1.46% ± 0.13% | 0.000 | |
| Ascomycota | Geastrumia | 0.76% ± 0.30% | 0.02% ± 0.02% | 0.013 | |
| Ascomycota | Mycosphaerella | 0.54% ± 0.04% | 0.18% ± 0.03% | 0.000 | |
| Ascomycota | Ramularia | 0.01% ± 0.01% | 0.70% ± 0.15% | 0.001 | |
| Ascomycota | Bacidina | 0.69% ± 0.18% | 0.01% ± 0.01% | 0.003 | |
| Ascomycota | Trichomerium | 0.00% ± 0.00% | 0.53% ± 0.05% | 0.003 | |
| Ascomycota | Hortaea | 0.40% ± 0.09% | 0.00% ± 0.00% | 0.015 | |
| Basidiomycota | Hygrocybe | 0.00% ± 0.00% | 0.29% ± 0.19% | 0.118 | |
| Ascomycota | Arthrinium | 0.07% ± 0.03% | 0.20% ± 0.07% | 0.043 | |
| Ascomycota | Didymella | 0.06% ± 0.01% | 0.14% ± 0.02% | 0.006 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhong, Z.; Gai, X.; Ying, J.; Li, W.; Du, X.; Bian, F.; Yang, C. Leaf-Associated Shifts in Bacterial and Fungal Communities in Response to Chicken Rearing Under Moso Bamboo Forests in Subtropical China. Forests 2019, 10, 216. https://doi.org/10.3390/f10030216
Zhang X, Zhong Z, Gai X, Ying J, Li W, Du X, Bian F, Yang C. Leaf-Associated Shifts in Bacterial and Fungal Communities in Response to Chicken Rearing Under Moso Bamboo Forests in Subtropical China. Forests. 2019; 10(3):216. https://doi.org/10.3390/f10030216
Chicago/Turabian StyleZhang, Xiaoping, Zheke Zhong, Xu Gai, Jiafu Ying, Weifen Li, Xuhua Du, Fangyuan Bian, and Chuanbao Yang. 2019. "Leaf-Associated Shifts in Bacterial and Fungal Communities in Response to Chicken Rearing Under Moso Bamboo Forests in Subtropical China" Forests 10, no. 3: 216. https://doi.org/10.3390/f10030216
APA StyleZhang, X., Zhong, Z., Gai, X., Ying, J., Li, W., Du, X., Bian, F., & Yang, C. (2019). Leaf-Associated Shifts in Bacterial and Fungal Communities in Response to Chicken Rearing Under Moso Bamboo Forests in Subtropical China. Forests, 10(3), 216. https://doi.org/10.3390/f10030216




