Effect of Microenvironment on Species Distribution Patterns in the Regeneration Layer of Forest Gaps and Non-Gaps in a Subtropical Natural Forest, China
Abstract
1. Introduction
2. Materials and Methods
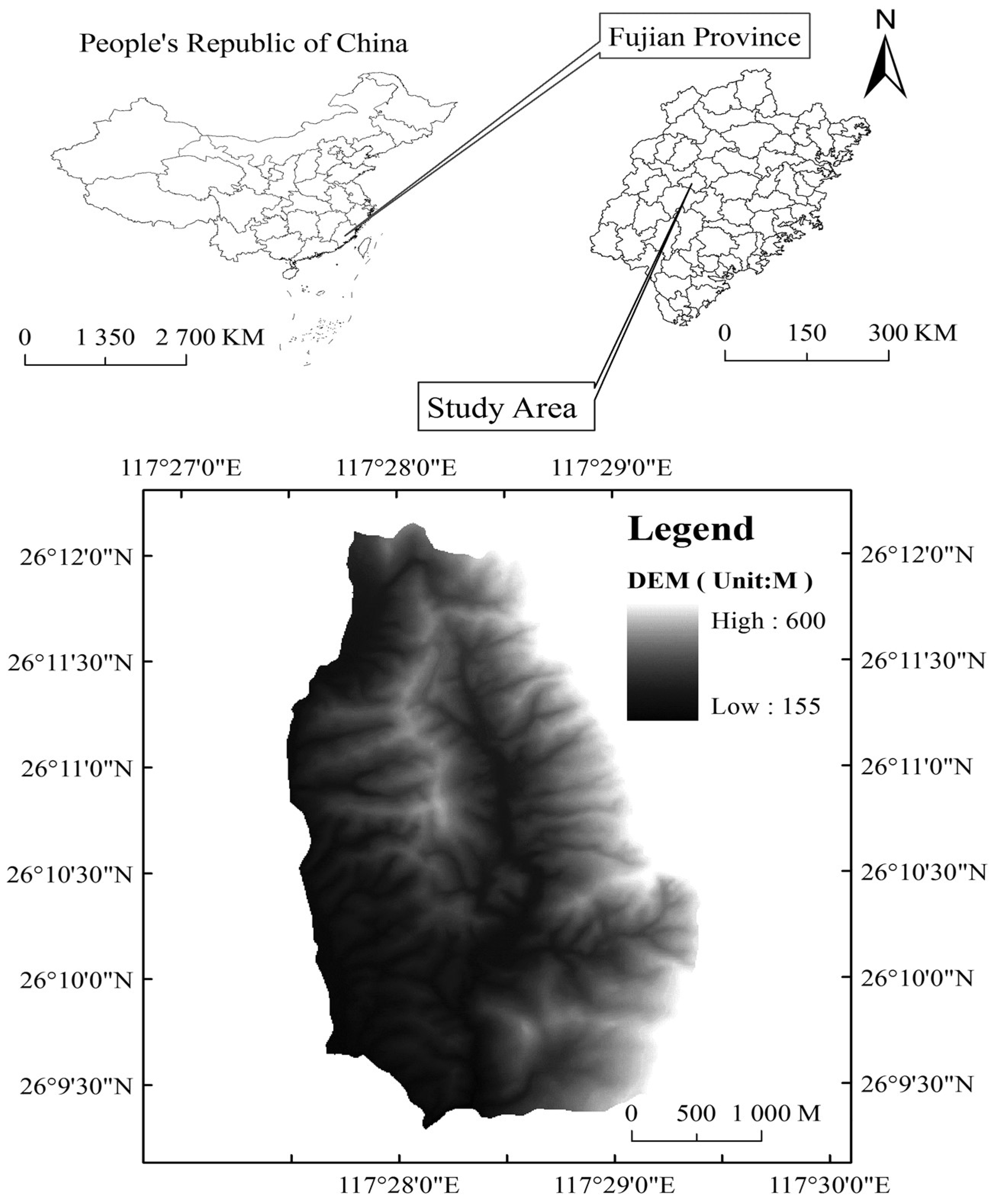
2.1. Study Site and Stand History
2.2. Community Survey in the Regeneration Layer
2.3. Study of Microenvironments in the Forest Gaps and Non-Gaps
2.4. Coupling Analysis Method
3. Results
3.1. Community Characteristics of the Regeneration Layer in Different Habitats
3.2. Screening of Environmental Factors in Different Habitats
3.3. The Coupling Relationship Between Plant Species and the Microenvironment
4. Discussion
4.1. The Dominant Species in the Regeneration Layer of the Forest Gaps and Non-Gaps
4.2. The Impact of Environmental Factors to the Species Coexistence in the Forest Gaps and Non-Gaps
4.3. The Relationships Between Species Distribution and Micro-Environment in the Forest Gaps and Non-Gaps
4.4. Implications for the Regeneration of the C. kawakamii Population
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muscolo, A.; Bagnato, S.; Sidari, M.; Mercurio, R. A review of the roles of forest canopy gaps. J. For. Res. 2014, 25, 725–736. [Google Scholar] [CrossRef]
- Zang, R.G.; Liu, G.Y. Gap dynamic and forest biodiversity; China Forestry Publish Press: Beijing, China, 1999. [Google Scholar]
- He, Z.S.; Liu, J.F.; Wu, C.T.; Zheng, S.Q.; Hong, W.; Su, S.J.; Wu, C.Z. Effects of forest gaps on some microclimate variables in Castanopsis kawakamii natural forest. J. Mt. Sci. 2012, 9, 706–714. [Google Scholar] [CrossRef]
- He, Z.S.; Liu, J.F.; Su, S.J.; Zheng, S.Q.; Xu, D.W.; Wu, Z.Y.; Hong, W.; Wang, J.L. Effects of forest gaps on soil properties in Castanopsis kawakamii nature forest. PLoS ONE 2015, 10, e0141203. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Osumi, K.; Takahashi, K.; Hoshizaki, K.; Matsune, K.; Suzuki, W. Effects of microenvironmental heterogeneity on the seed-to-seedling process and tree coexistence in a riparian forest. Ecol. Res. 2007, 22, 724–734. [Google Scholar] [CrossRef]
- Schliemann, S.A.; Bockheim, J.G. Methods for studying treefall gaps: A review. For. Ecol. Manag. 2011, 261, 1143–1151. [Google Scholar] [CrossRef]
- Kirchner, K.; Kammermeier, S.; Bruelheide, H. The response of the pseudoannual species Trientalis europaea L. to forest gap dynamics in a near-natural spruce forest. For. Ecol. Manag. 2008, 257, 1070–1077. [Google Scholar] [CrossRef]
- Liao, J.B.; Bogaert, J.; Nijs, I. Species interactions determine the spatial mortality patterns emerging in plant communities after extreme events. Sci. Rep. 2015, 5, 11229. [Google Scholar] [CrossRef]
- Chen, L.; Liu, G.H.; Liu, D. How forest gap and elevation shaped Abies faxoniana Rehd. et Wils. regeneration in a Subalpine coniferous forest, Southwestern China. Forests 2018, 9, 271. [Google Scholar] [CrossRef]
- Thorson, J.T.; Ianelli, J.N.; Larsen, E.A.; Ries, L.; Scheuerell, M.D.; Szuwalski, C.; Zipkin, E.F. Joint dynamic species distribution models: A tool for community ordination and spatio-temporal monitoring. Glob. Ecol. Biogeogr. 2016, 25, 1144–1158. [Google Scholar] [CrossRef]
- Kent, M. Vegetation Description and Data Analysis: A Practical Approach; John Wiley and Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Veneklaas, E.J.; Fajardo, A.; Lozano, J. Gallery forest types and their environmental correlates in a Colombian savanna landscape. Ecography 2005, 28, 236–252. [Google Scholar] [CrossRef]
- Kargar-Chigani, H.; Javadi, S.; Zahedi-Amiri, G.; Khajeddin, S.; Jafari, M. Vegetation composition differentiation and species-environment relationships in the northern part of Isfahan Province, Iran. J. Arid Land 2017, 9, 161–175. [Google Scholar] [CrossRef]
- Sarvade, S.; Gupta, B.; Singh, M. Composition, diversity and distribution of tree species in response to changing soil properties with increasing distance from water source—A case study of Gobind Sagar reservoir in India. J. Mt. Sci. 2016, 13, 522–533. [Google Scholar] [CrossRef]
- Du, H.; Peng, W.X.; Song, T.Q.; Zeng, F.P.; Wang, K.L.; Song, M.; Zhang, H. Spatial pattern of woody plants and their environmental interpretation in the karst forest of southwest China. Plant Biosyst. 2013, 149, 1–10. [Google Scholar] [CrossRef]
- Liu, J.; Hong, W.; Li, J.; Yang, W. Gap natural disturbance regime in the Castanopsis kawakamii forest. Acta Ecol. Sinica 2003, 23, 1991–1999. [Google Scholar]
- Liu, J.F.; He, Z.S.; Hong, W.; Zheng, S.Q.; Wang, Z.J. Conservation ecology of endangered plant Castanopsis kawakamii. J. Beijing Univ. 2011, 33, 136–143. [Google Scholar]
- He, Z.S.; Liu, J.F.; Zheng, S.Q.; Hong, W.; Wu, C.Z.; Wu, Z.Y.; Niu, J.; Lin, Y.J. Studies on the seeds dispersal and seedlings regeneration in gaps and understory of Castanopsis kawakamii natural forest. J. Trop. Subtrop. Bot. 2011, 19, 230–236. [Google Scholar] [CrossRef]
- Buajan, S.; Liu, J.F.; He, Z.S.; Feng, X.P. Effect of gap sizes on specific leaf area and chlorophyll contents at the Castanopsis kawakamii natural reserve forest, China. Forests 2018, 9, 682. [Google Scholar] [CrossRef]
- Hilborn, R.; Mangel, M. The Ecological Detective: Confronting Models with Data; Princeton University Press: Princeton, NJ, USA, 1997. [Google Scholar]
- Zhang, W.R.; Yang, G.Y.; Tu, X.N. Forest Soil Analysis Method; China Forestry Publish Press: Beijing, China, 1999. [Google Scholar]
- Braak, C.J.F. Canonical correspondence-analysis—A new eigenvector technique for multivariate direct gradient analysis. Ecology 1986, 67, 1167–1179. [Google Scholar] [CrossRef]
- Braak, C.J.F.; Barendregt, L.G. Weighted averaging of species indicator values—Its efficiency in environmental calibration. Math. Biosci. 1986, 78, 57–72. [Google Scholar] [CrossRef]
- Šmilauer, P.; Lepš, J. Multivariate Analysis of Ecological Data Using CANOCO 5; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Braak, C.J.F.; Smilauer, P. CANOCO Reference Manual and User’s Guide: Software for Ordination (Version 5.0); Microcomputer Power: Ithaca, NY, USA, 2012. [Google Scholar]
- Tesfaye, G.; Teketay, D.; Fetene, M.; Beck, E. Regeneration of seven indigenous tree species in a dry Afromontane forest, Southern Ethiopia. Flora-Morphol. Distrib. Funct. Ecol. Plants 2010, 205, 135–143. [Google Scholar] [CrossRef]
- Lu, D.L.; Wang, G.G.; Yan, Q.L.; Gao, T.; Zhu, J.J. Effects of gap size and within-gap position on seedling growth and biomass allocation: Is the gap partitioning hypothesis applicable to the temperate secondary forest ecosystems in Northeast China? For. Ecol. Manag. 2018, 429, 351–362. [Google Scholar] [CrossRef]
- Donoso, P.J. Crown Index: A canopy balance indicator to assess growth and regeneration in uneven-aged forest stands of the coastal range of Chile. Forestry 2005, 78, 337–351. [Google Scholar] [CrossRef]
- Zhang, T.; Yan, Q.L.; Wang, J.; Zhu, J.J. Restoring temperate secondary forests by promoting sprout regeneration: Effects of gap size and within-gap position on the photosynthesis and growth of stump sprouts with contrasting shade tolerance. For. Ecol. Manag. 2018, 429, 267–277. [Google Scholar] [CrossRef]
- Madsen, P.; Hahn, K. Natural regeneration in a beech-dominated forest managed by close-to-nature principles-a gap cutting based experiment. Can. J. For. Res. 2008, 38, 1716–1729. [Google Scholar] [CrossRef]
- Zhu, J.J.; Lu, D.L.; Zhang, W.D. Effects of gaps on regeneration of woody plants: A meta-analysis. J. For. Res. 2014, 25, 501–510. [Google Scholar] [CrossRef]
- Bottero, A.; Garbarino, M.; Dukic, V.; Govedar, Z.; Lingua, E.; Nagel T, A.; Motta, R. Gap-phase dynamics in the old-growth forest of Lom, Bosnia and Herzegovina. Silva Fennica 2011, 45, 875–887. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Yang, W.Q.; Wu, F.Z.; Xu, Z.F.; Tan, B.; Zhang, L.; He, X.H.; Guo, L. Canopy gaps accelerate soil organic carbon retention by soil microbial biomass in the organic horizon in a subalpine fir forest. Appl. Soil Ecol. 2018, 125, 169–176. [Google Scholar] [CrossRef]
- Haghverdi, K.; Kiadaliri, H.; Sagheb-Talebi K, Y.; Kooch, Y. Variability of plant diversity and soil features following gap creation in Caspian beech forests of Iran. Ann. Biol. Res. 2012, 3, 4622–4635. [Google Scholar]
- Brown, C.; Burslem, D.F.; Illian, J.B.; Bao, L.; Brockelman, W.; Cao, M.; Chang, L.W.; Dattaraja, H.S.; Davies, S.; Gunatilleke, C.V.; et al. Multispecies coexistence of trees in tropical forests: Spatial signals of topographic niche differentiation increase with environmental heterogeneity. Proc. Biol. Sci. 2013, 280, 20130502. [Google Scholar] [CrossRef]
- Edwards, R.D.; Crisp, M.D.; Cook, L.G. Niche differentiation and spatial partitioning in the evolution of two Australian monsoon tropical tree species. J. Biogeogr. 2013, 40, 559–569. [Google Scholar] [CrossRef]
- Zhang, Q.; Zak, J.C. Effects of gap size on litter decomposition and microbial activity in a subtropical forest. Ecology 1995, 76, 2196–2204. [Google Scholar] [CrossRef]


| GAP1 | GAP2 | GAP3 | GAP4 | GAP5 | GAP6 | GAP7 | GAP8 | GAP9 | GAP10 | GAP11 | GAP12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aja | 0.015 | 0.007 | 0.022 | 0 | 0.015 | 0.046 | 0.046 | 0.031 | 0.016 | 0.015 | 0.157 | 0 |
| Apu | 0.012 | 0.194 | 0.088 | 0.101 | 0.012 | 0.032 | 0.010 | 0.071 | 0.053 | 0.020 | 0.074 | 0.007 |
| Cfa | 0.012 | 0 | 0.048 | 0.018 | 0.012 | 0.024 | 0.022 | 0 | 0.009 | 0.005 | 0.028 | 0 |
| Cka | 0.041 | 0.065 | 0.065 | 0.088 | 0.018 | 0.024 | 0.058 | 0.053 | 0.006 | 0.005 | 0.035 | 0.038 |
| Coc | 0.027 | 0.015 | 0.006 | 0.005 | 0.027 | 0.028 | 0.078 | 0 | 0.026 | 0.014 | 0.003 | 0 |
| Cth | 0.375 | 0 | 0 | 0 | 0.375 | 0.127 | 0.092 | 0 | 0.047 | 0.040 | 0.010 | 0 |
| Ddu | 0.102 | 0.023 | 0.020 | 0.005 | 0.037 | 0.020 | 0.006 | 0.140 | 0.043 | 0.043 | 0.022 | 0.067 |
| Fol | 0 | 0 | 0 | 0 | 0.015 | 0 | 0 | 0 | 0.013 | 0.246 | 0.039 | 0.003 |
| Ich | 0 | 0.071 | 0.020 | 0.010 | 0 | 0 | 0 | 0.081 | 0.048 | 0.009 | 0.015 | 0 |
| Lsu | 0.082 | 0.121 | 0.127 | 0.088 | 0.148 | 0.094 | 0.086 | 0.050 | 0.300 | 0.116 | 0.127 | 0.117 |
| Mgr | 0.011 | 0.056 | 0.064 | 0.021 | 0.011 | 0.010 | 0.024 | 0.086 | 0.002 | 0.011 | 0.051 | 0.015 |
| Phi | 0 | 0.147 | 0 | 0 | 0 | 0.005 | 0 | 0.024 | 0.002 | 0 | 0 | 0 |
| Rco | 0 | 0.023 | 0.019 | 0 | 0 | 0.029 | 0.005 | 0.033 | 0.008 | 0.035 | 0.063 | 0 |
| Sbu | 0 | 0 | 0.028 | 0.067 | 0.020 | 0.053 | 0.169 | 0.014 | 0.033 | 0.078 | 0.020 | 0.063 |
| Sgr | 0 | 0.031 | 0.006 | 0.005 | 0 | 0 | 0.025 | 0.046 | 0.006 | 0.003 | 0.004 | 0 |
| Sla | 0.104 | 0.051 | 0.029 | 0.013 | 0.016 | 0.005 | 0.007 | 0.035 | 0.029 | 0.069 | 0.043 | 0.026 |
| Ssu | 0 | 0.050 | 0.056 | 0.024 | 0 | 0.009 | 0.034 | 0.008 | 0.028 | 0.003 | 0.003 | 0 |
| NG1 | NG2 | NG3 | NG4 | NG5 | NG6 | NG7 | NG8 | NG9 | NG10 | NG11 | NG12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aja | 0.030 | 0.050 | 0.004 | 0.005 | 0.017 | 0.041 | 0.075 | 0.010 | 0.012 | 0.020 | 0.026 | 0.034 |
| Apu | 0.216 | 0.246 | 0.096 | 0.035 | 0.006 | 0.062 | 0.014 | 0.153 | 0.106 | 0.077 | 0.040 | 0.111 |
| Cfa | 0 | 0.014 | 0.021 | 0.028 | 0.006 | 0.005 | 0.005 | 0 | 0.008 | 0.039 | 0.059 | 0.007 |
| Cka | 0.060 | 0.049 | 0.009 | 0.030 | 0.003 | 0.031 | 0.009 | 0.019 | 0.014 | 0.047 | 0.088 | 0.021 |
| Coc | 0 | 0 | 0.015 | 0.046 | 0.012 | 0.036 | 0.139 | 0 | 0 | 0 | 0.103 | 0.008 |
| Cth | 0 | 0 | 0 | 0.041 | 0.329 | 0.108 | 0.168 | 0.004 | 0.042 | 0.015 | 0 | 0.131 |
| Ddu | 0.038 | 0.035 | 0.013 | 0.116 | 0.003 | 0.019 | 0.009 | 0.114 | 0.031 | 0.130 | 0.016 | 0.016 |
| Ich | 0.029 | 0.035 | 0.072 | 0 | 0 | 0 | 0 | 0.019 | 0.173 | 0 | 0.082 | 0.081 |
| Lsu | 0.140 | 0.081 | 0.082 | 0.024 | 0.258 | 0.102 | 0.091 | 0.037 | 0.252 | 0.113 | 0.084 | 0.090 |
| Mgr | 0.119 | 0.010 | 0.081 | 0.012 | 0 | 0.013 | 0.033 | 0.044 | 0.021 | 0.064 | 0.035 | 0.020 |
| Mja | 0 | 0 | 0 | 0.008 | 0.082 | 0.138 | 0.074 | 0 | 0.006 | 0 | 0.112 | 0.128 |
| Nca | 0.005 | 0 | 0 | 0 | 0 | 0.008 | 0.023 | 0 | 0.069 | 0.045 | 0 | 0.030 |
| Sbu | 0 | 0 | 0.008 | 0.083 | 0.006 | 0.033 | 0.060 | 0.004 | 0.007 | 0.177 | 0.036 | 0.071 |
| Sgl | 0.143 | 0.016 | 0.021 | 0.004 | 0.018 | 0.030 | 0.014 | 0.068 | 0.034 | 0.031 | 0.008 | 0.014 |
| Sgr | 0.006 | 0.058 | 0.026 | 0 | 0.003 | 0.010 | 0.007 | 0.018 | 0.024 | 0.007 | 0 | 0 |
| Sla | 0.026 | 0 | 0.021 | 0.024 | 0.009 | 0.018 | 0.013 | 0.017 | 0.015 | 0.007 | 0.016 | 0.058 |
| Ssu | 0.006 | 0.135 | 0.046 | 0.014 | 0 | 0.029 | 0.012 | 0.016 | 0.014 | 0.004 | 0 | 0.017 |
| Gap | Explains % | Gap | Explains % | Non-Gap | Explains % | Non-Gap | Explains % | |
|---|---|---|---|---|---|---|---|---|
| ST10 | 16.2 | SOC | 8.6 | ST10 | 33.6 | CWC | 6.5 | |
| ST5 | 15.6 | SWMC | 7.9 | AT | 32.6 | MMC | 6.4 | |
| AP | 15.4 | MWC | 7.9 | ST5 | 27.8 | pH | 5.6 | |
| LI | 13.0 | AK | 7.4 | ST0 | 26.8 | TN | 5.2 | |
| AT | 12.4 | SBD | 7.3 | AP | 21.1 | CP | 5.1 | |
| HN | 12.3 | TK | 6.7 | LI | 20.6 | TP | 4.5 | |
| ST0 | 12.1 | MMC | 6.3 | C/N | 15.8 | SWMC | 4.3 | |
| CP | 11.2 | N/CP | 5.7 | SOC | 15.6 | MWC | 4.3 | |
| CWC | 10.2 | NCP | 5.6 | AH | 14.9 | N/CP | 4.3 | |
| STP | 10.2 | pH | 5.6 | SOM | 14.8 | NCP | 3.9 | |
| SAP | 9.9 | C/N | 5.4 | AK | 10.9 | STP | 3.8 | |
| SVMC | 8.8 | SWC | 4.9 | SBD | 8.1 | SWC | 3.7 | |
| AH | 8.8 | SOM | 4.7 | TK | 7.4 | SVMC | 3.5 | |
| SOC | 8.6 | TN | 4.0 | HN | 7.2 | SAP | 2.8 |
| Gap | Explains % | Non-Gap | Explains % | |
|---|---|---|---|---|
| ST10 | 16.2 | ST10 | 33.7 | |
| LI | 12.7 | AT | 10.8 | |
| ST5 | 12.5 | TN | 7.7 | |
| HN | 12.2 | HN | 7.5 | |
| SAP | 12.6 | AK | 6.9 | |
| C/N | 9.1 | AP | 10.4 | |
| TP | 9.8 | ST5 | 6.9 | |
| AH | 5.3 | MWC | 4.7 | |
| NCP | 4.6 | C/N | 5.9 | |
| MWC | 4.1 | SOC | 2.9 |
| Gap | Non-Gap | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Axis 1 | Axis 2 | Axis 3 | Axis 4 | Axis 1 | Axis 2 | Axis 3 | Axis 4 | ||
| Eigenvalues | 0.1413 | 0.0592 | 0.0482 | 0.0139 | 0.1314 | 0.0495 | 0.0299 | 0.0119 | |
| Cumulative percentage variance of species data | 40.16 | 56.98 | 70.68 | 74.64 | 44.07 | 60.67 | 70.71 | 74.7 | |
| Species-environment correlations | 0.9987 | 0.9985 | 0 | 0 | 0.9996 | 0.9984 | 0 | 0 | |
| Cumulative percentage variance (%) | 40.39 | 57.31 | 45.32 | 62.4 | |||||
| LI | TP | MWC | HN | C/N | ST5 | ST10 | NCP | AH | SAP | |
|---|---|---|---|---|---|---|---|---|---|---|
| Aja | −8.52▼ | −2.44 | −3.19 | −2.01 | −1.53 | −3.88 | −3.11 | 3.46 | 6.38● | 6.12● |
| Apu | 3.28 | 1.3 | 2.52 | 3.16 | 3.15 | 7.54● | 8.03● | −0.3 | −3.64▼ | −5.82▼ |
| Cfa | −8.48▼ | −2.43 | −3.19 | −2.01 | −1.54 | −3.82 | −3.11 | 3.49 | 6.39● | 6.12● |
| Cka | −6.67▼ | −1.55 | −0.67 | 1.84 | 2.53 | 5.71● | 7.36● | 4.26 | 3.2 | −0.17 |
| Coc | −8.96▼ | −2.52 | 3.14 | 1.65 | 1.11 | 2.93 | 2.05 | 3.89 | 6.54● | 5.82● |
| Cth | −6.17▼ | −1.79 | −3.04 | −2.77 | −2.53 | −6.17▼ | −6.09▼ | 1.95 | 5.31● | 6.38● |
| Ddu | 6.42● | 1.49 | 0.56 | −1.93 | −2.6 | −5.85▼ | −7.5▼ | −4.2 | −3.15 | 0.33 |
| Fol | 5.91● | 1.3 | 0.06 | −2.1 | −2.75 | −6.26▼ | −7.88▼ | 0.6 | −2.79 | −5.24▼ |
| Ich | 6.42● | 1.98 | 3.08 | 2.73 | 2.43 | 5.95● | 5.79● | −2.52 | −5.09▼ | −6.39▼ |
| Lsu | 0.17 | −0.05 | −1.4 | −3.06 | −3.39 | −7.95▼ | −9.09▼ | −1.88 | 0.86 | 4.07 |
| Mgr | 4.13 | 1.42 | 2.7 | 3.09 | 3 | 7.22● | 7.59● | −0.77 | −4.2▼ | −6.05▼ |
| Phi | 2.65 | 1.06 | 2.38 | 3.18 | 3.22 | 7.69● | 8.28● | 0 | −3.3 | −5.7▼ |
| Rco | 6.55● | 2.02 | 3.09 | 2.68 | 2.4 | 5.9● | 5.73● | −2.52 | −5.5▼ | −6.4▼ |
| Sbu | 0.75 | −0.14 | −1.45 | −3.08 | −3.4 | −7.96▼ | −9.09▼ | −1.76 | 1.03 | 4.2 |
| Sgr | 2 | 0.91 | 2.26 | 3.2 | 3.28 | 7.8● | 8.5● | 0.3 | −2.94 | −5.46▼ |
| Sla | 7.41● | 1.79 | 1.04 | −1.55 | −2.23 | −4.98▼ | −6.66▼ | −4.51 | −3.97 | −0.56 |
| Ssu | −2.484 | −0.3169 | 0.9355 | 2.9001 | 3.3168 | 7.727● | 9.0415● | 2.5236 | 0.11518 | −3.296 |
| HN | MWC | C/N | AP | ST5 | AT | ST10 | SOC | TN | AK | |
|---|---|---|---|---|---|---|---|---|---|---|
| Sgl | −2.466 | −0.711 | 3.7083 | 5.8113● | 7.443● | 8.4002● | 9.3972● | 6.0411● | 1.470687 | −1.527 |
| Sbu | 0.524 | −0.439 | −6.204▼ | −7.503▼ | −9.11▼ | −9.791▼ | −10.02▼ | −5.989▼ | 0.672668 | 4.0985 |
| Mja | 1.1852 | −0.04 | −5.595▼ | −7.164▼ | −8.821▼ | −9.62▼ | −10.09▼ | −6.166▼ | 2.63 × 10−16 | 3.3418 |
| Cka | 1.4354 | 1.3842 | 7.0677● | 7.0614● | 8.0851● | 8.1246● | 7.3867● | 3.9018 | −3.04056 | −5.81▼ |
| Ddu | −1.113 | 0.0803 | 5.6737● | 7.1886● | 8.8436● | 9.6359● | 10.097● | 6.1601● | 2.63 × 10−16 | −3.426 |
| Sla | 2.15 | 0.4782 | −4.383 | −6.307▼ | −7.967▼ | −9.209▼ | −9.709▼ | −6.147▼ | −1.04026 | 2.1624 |
| Lsu | −4.144 | −1.951 | −1.002 | 1.2709 | 2.3035 | 3.3518 | 4.8966 | 3.7743 | 3.685819 | 2.3997 |
| Mgr | 0.4868 | 0.9355 | 6.8854● | 7.7● | 9.165● | 9.6359● | 9.5207● | 5.4995● | −1.54098 | −4.891 |
| Ich | 1.6455 | 1.4784 | 7.1989● | 7.4017● | 8.5889● | 8.7704● | 8.2226● | 4.4599 | −2.61767 | −5.627▼ |
| Coc | 3.1448 | 0.9538 | −2.986 | −5.102▼ | −6.728▼ | −7.723▼ | −8.834▼ | −5.807▼ | −1.91865 | 0.8721 |
| Cth | −1.919 | −1.723 | −7.187▼ | −7.189▼ | −8.269▼ | −8.356▼ | −7.68▼ | −4.068 | 2.932593 | 5.7601● |
| Ssu | −0.931 | 0.3796 | 6.2037● | 7.3231● | 8.9819● | 9.7162● | 10.094● | 6.1058● | −0.29995 | −3.673 |
| Cfa | 4.0407 | 1.8369 | 0.4395 | −1.863 | −2.995 | −4.064 | −5.575▼ | −4.149 | −3.5007 | −1.969 |
| Aja | −3.913 | −2.278 | −5.266▼ | −3.85 | −3.815 | −3.191 | −1.754 | −0.216 | 4.252768 | 5.2801● |
| Nca | −4.144 | −2.298 | −4.433 | −2.759 | −2.536 | −1.786 | −0.264 | 0.7019 | 4.299836 | 4.8033 |
| Sgr | −1.919 | −0.38 | 4.6281 | 6.4578● | 8.1605● | 9.054● | 9.7783● | 6.1764● | 0.820479 | −2.447 |
| Apu | −0.188 | 0.5369 | 6.4713● | 7.6154● | 9.1828● | 9.794● | 9.9144● | 5.8622● | −1.00382 | −4.385 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Wang, L.; Jiang, L.; Wang, Z.; Liu, J.; Xu, D.; Hong, W. Effect of Microenvironment on Species Distribution Patterns in the Regeneration Layer of Forest Gaps and Non-Gaps in a Subtropical Natural Forest, China. Forests 2019, 10, 90. https://doi.org/10.3390/f10020090
He Z, Wang L, Jiang L, Wang Z, Liu J, Xu D, Hong W. Effect of Microenvironment on Species Distribution Patterns in the Regeneration Layer of Forest Gaps and Non-Gaps in a Subtropical Natural Forest, China. Forests. 2019; 10(2):90. https://doi.org/10.3390/f10020090
Chicago/Turabian StyleHe, Zhongsheng, Lijing Wang, Lan Jiang, Zhe Wang, Jinfu Liu, Daowei Xu, and Wei Hong. 2019. "Effect of Microenvironment on Species Distribution Patterns in the Regeneration Layer of Forest Gaps and Non-Gaps in a Subtropical Natural Forest, China" Forests 10, no. 2: 90. https://doi.org/10.3390/f10020090
APA StyleHe, Z., Wang, L., Jiang, L., Wang, Z., Liu, J., Xu, D., & Hong, W. (2019). Effect of Microenvironment on Species Distribution Patterns in the Regeneration Layer of Forest Gaps and Non-Gaps in a Subtropical Natural Forest, China. Forests, 10(2), 90. https://doi.org/10.3390/f10020090





