Where Did You Come From? Where Did You Go? Investigating the Origin of Invasive Leptocybe Species Using Distribution Modelling
Abstract
1. Introduction
“Know from whence you came. If you know whence you came, there are absolutely no limitations to where you can go.”—James Baldwin
2. Materials and Methods
2.1. MaxEnt Modelling
2.2. Niche Modelling
2.3. Testing for Niche Shifts
2.4. Effects of Environmental Factors
2.5. CLIMEX Modelling
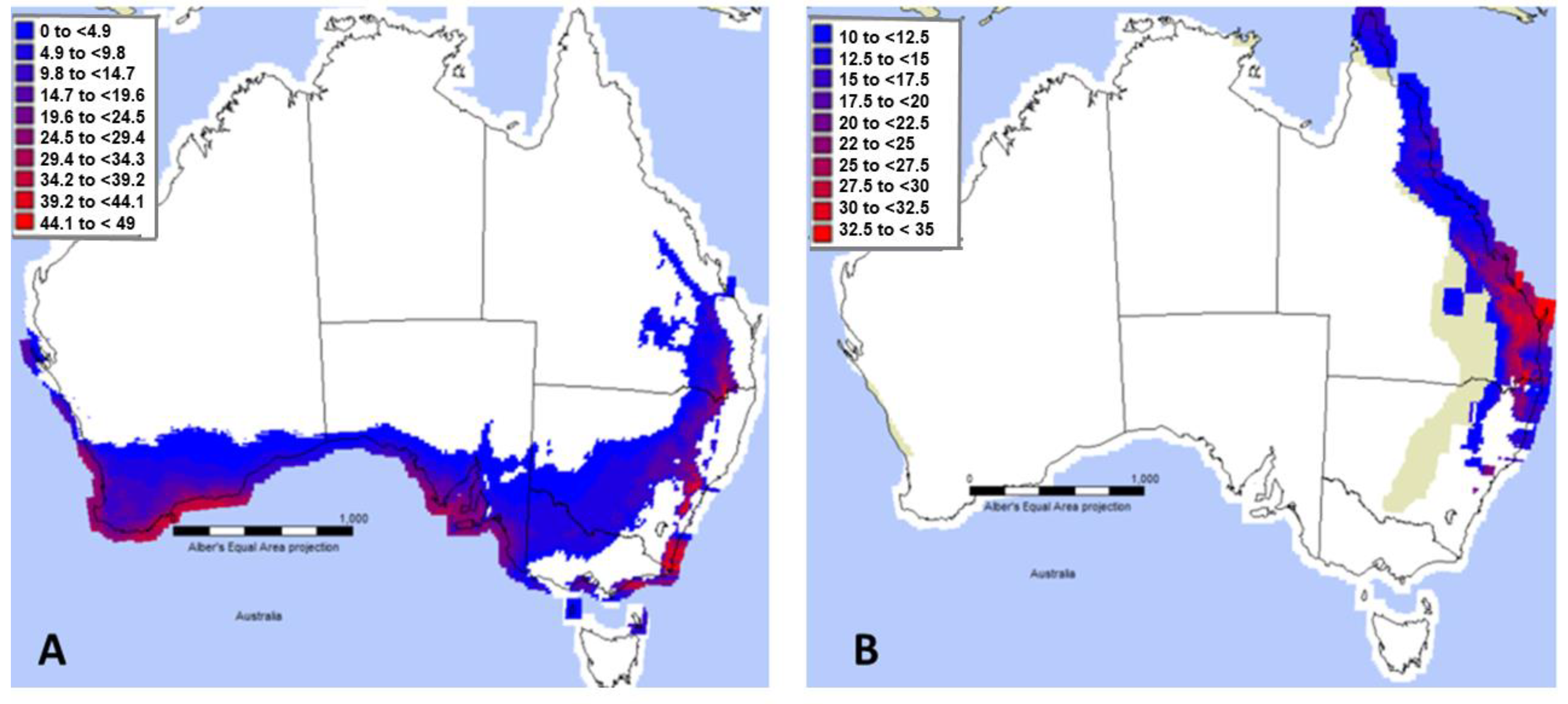
3. Results
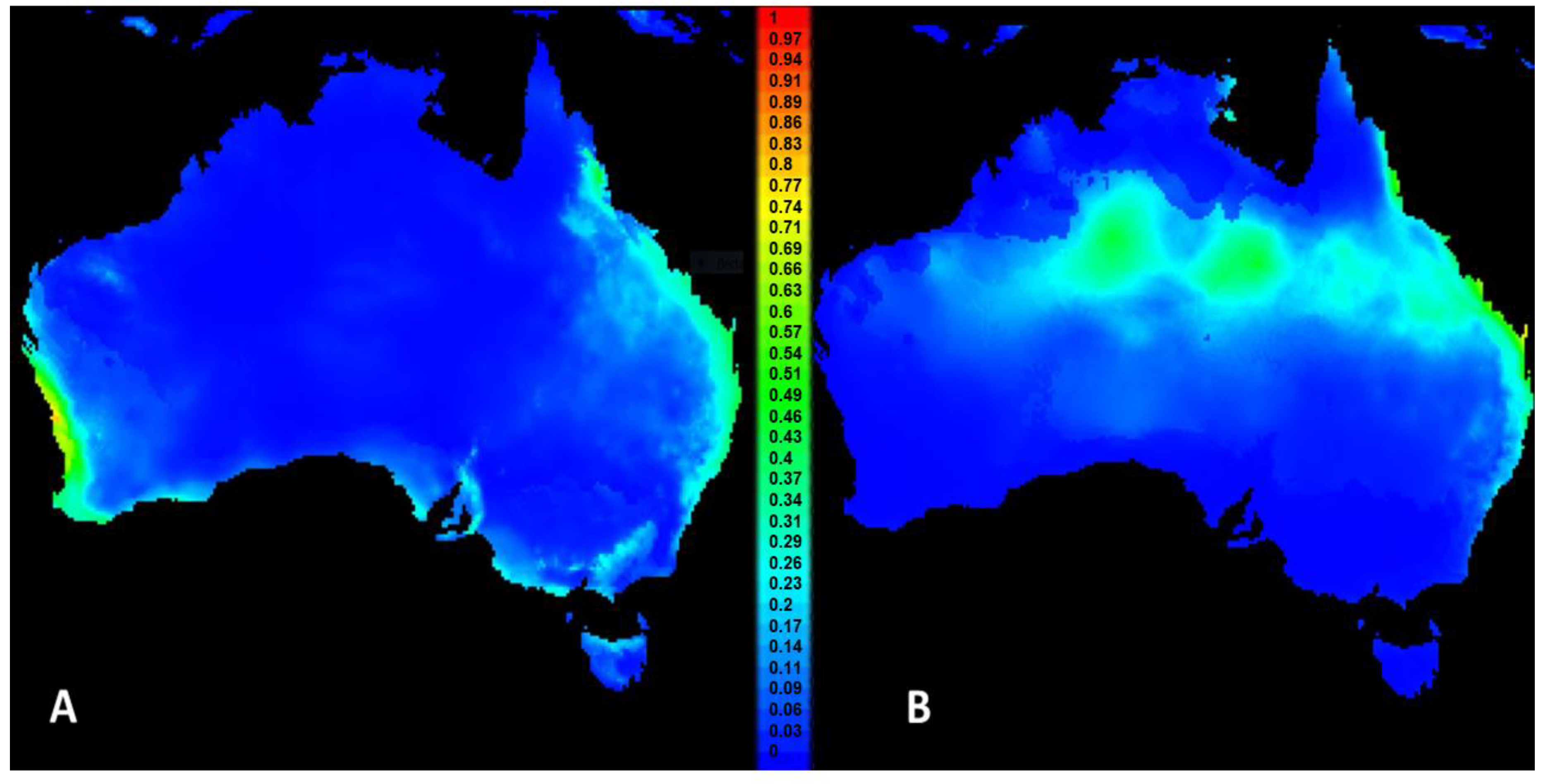
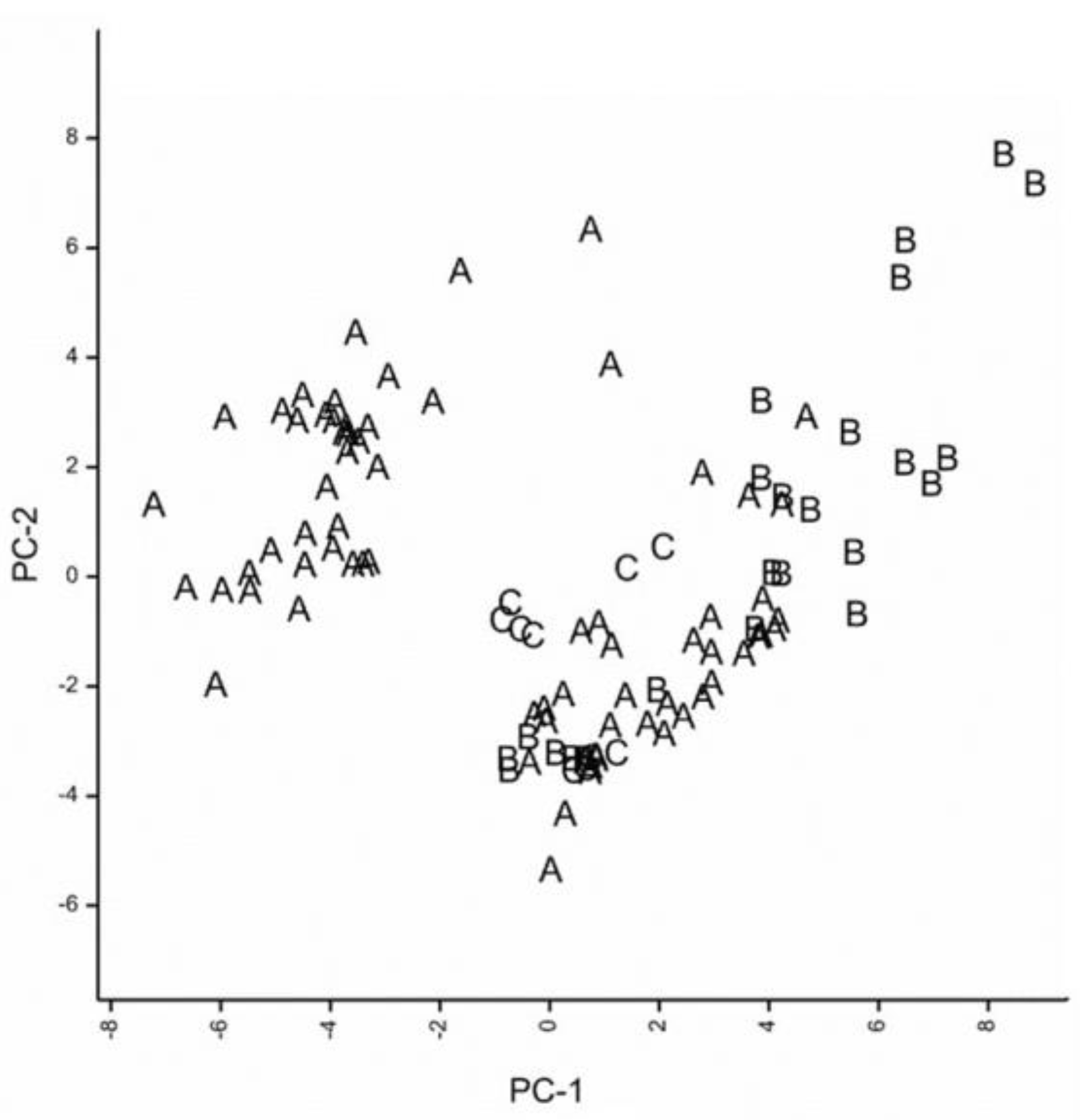
3.1. Niche Modelling and Projections in MaxEnt
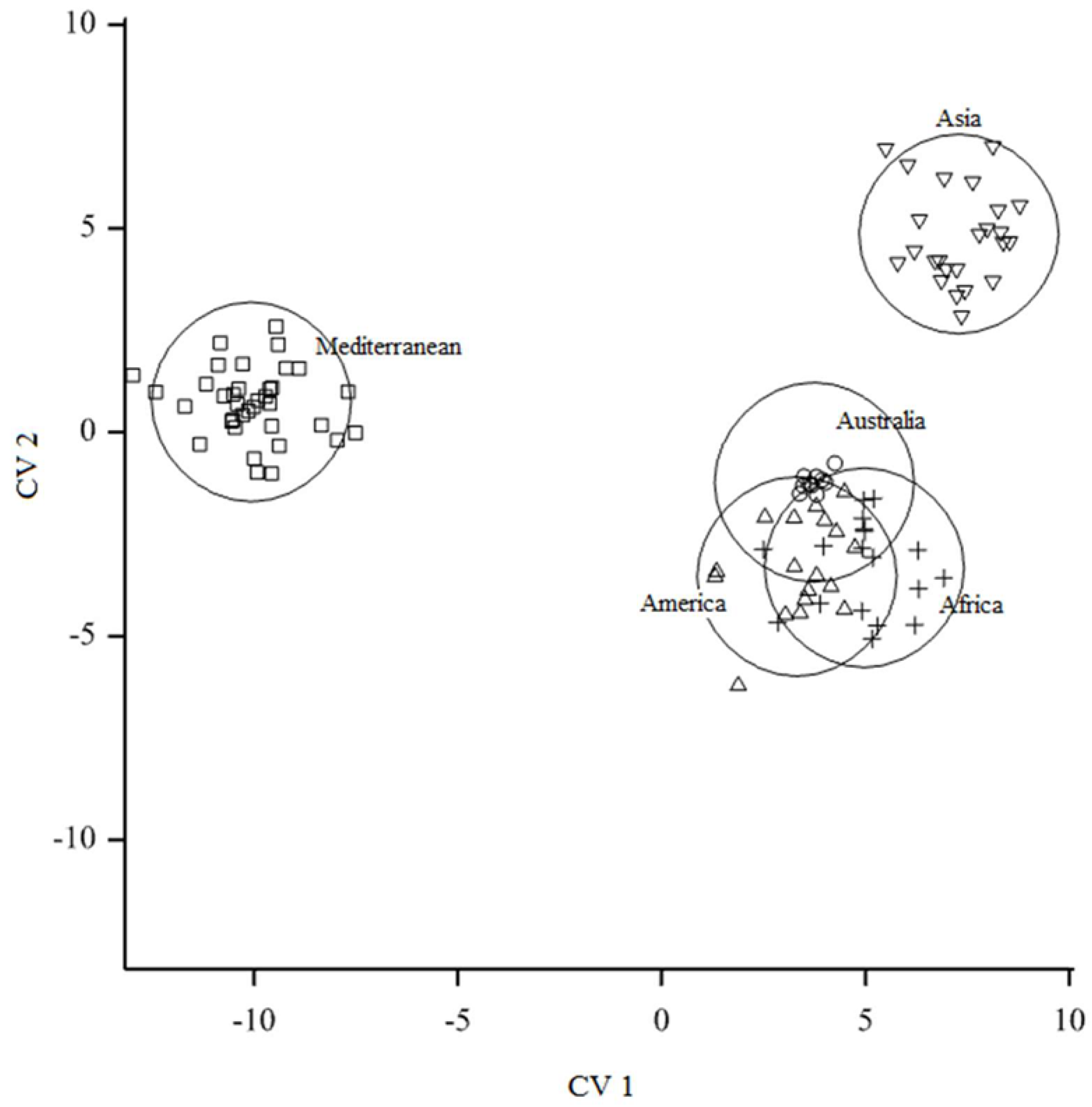
3.2. Testing for Niche Overlap
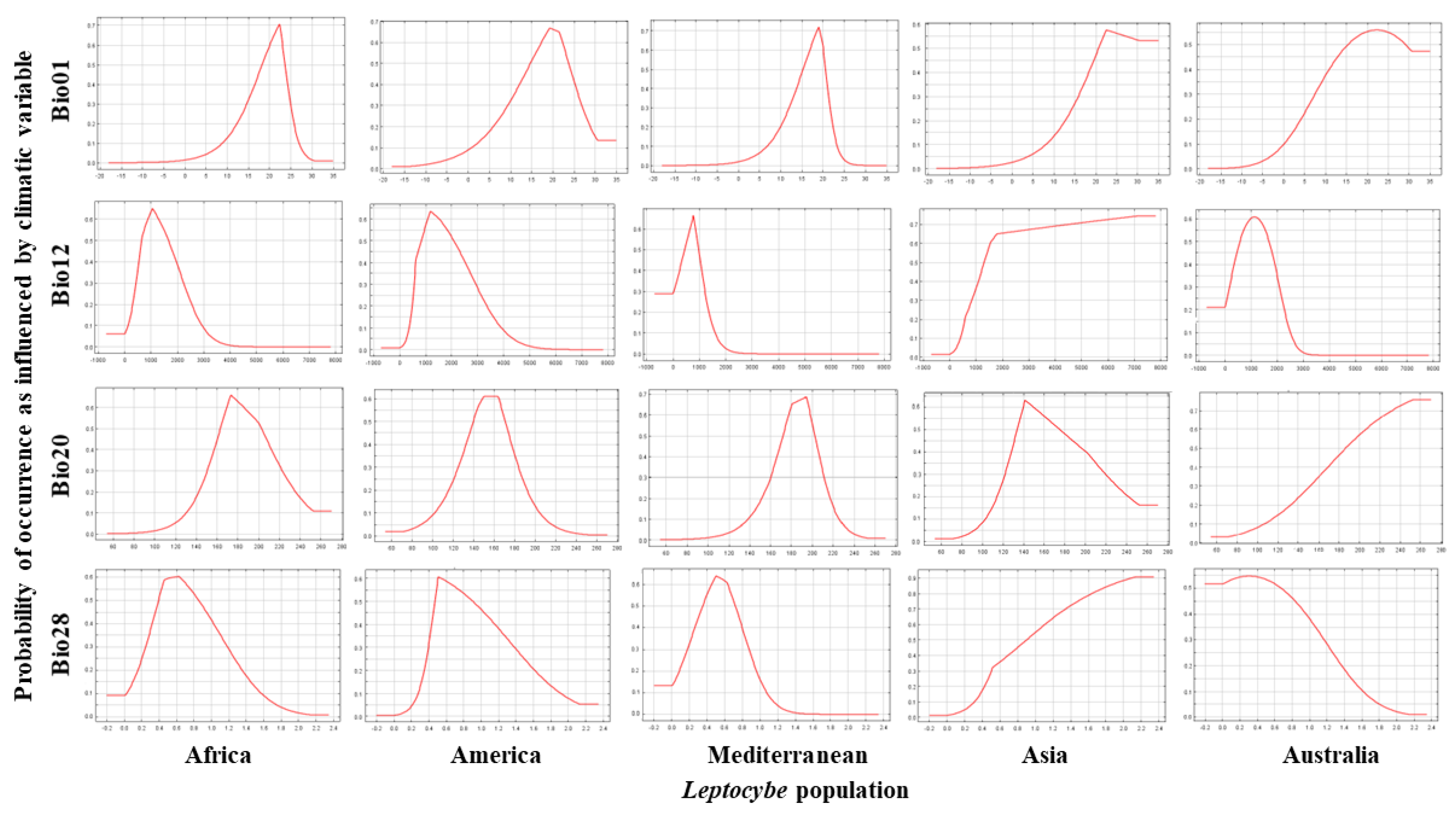
3.3. Contribution of Environmental Variables to the Probability of Occurrence of L. invasa
3.4. CLIMEX Modelling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fox, L.R.; Morrow, P.A. Estimates of damage by herbivorous insects on Eucalyptus trees. Aust. J. Ecol. 1983, 8, 139–147. [Google Scholar] [CrossRef]
- Ohmart, C.P.; Edwards, P.B. Insect herbivory on Eucalyptus. Annu. Rev. Entomol. 1991, 36, 637–657. [Google Scholar] [CrossRef]
- Heatwole, H.; Lowman, M.D.; Abbott, K.L. Grazing on Australian eucalypt leaves by insects. Selbyana 1999, 20, 299–323. [Google Scholar]
- Semple, W.S.; Koen, T.B. Observations of insect damage to leaves of woodland eucalypts on the central western slopes of New South Wales: 1990 to 2004. Proc. Linn. Soc. N. S. W. 2007, 128, 99–110. [Google Scholar]
- Nahrung, H.F.; Loch, A.D.; Matsuki, M. Invasive insects in Mediterranean forest systems: Australia. In Insects and Diseases of Mediterranean Forest Systems; Paine, T.D., Lieutier, F., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 475–498. ISBN 978-3-319-24744-1. [Google Scholar]
- Steinbauer, M.J.; Nahrung, H.F. Native defoliators of Australian Mediterranean forest trees. In Insects and Diseases of Mediterranean Forest Systems; Paine, T.D., Lieutier, F., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 431–454. ISBN 978-3-319-24744-1. [Google Scholar]
- Steinbauer, M.J.; Salminen, J.-P.; Watson, S.J. Yellow, red, dead: the nutritional consequences for Cardiaspina densitexta (Hemiptera: Aphalaridae) nymphs of inducing senescence in old Eucalyptus fasciculosa leaves. Aust. Entomol. 2018, 57, 265–278. [Google Scholar] [CrossRef]
- Wingfield, M.J.; Slippers, B.; Hurley, B.P.; Coutinho, T.A.; Wingfield, B.D.; Roux, J. Eucalypt pests and diseases: Growing threats to plantation productivity. South. For. 2008, 70, 139–144. [Google Scholar] [CrossRef]
- Paine, T.D.; Steinbauer, M.J.; Lawson, S.A. Native and exotic pests of eucalyptus: A worldwide perspective. Annu. Rev. Entomol. 2011, 56, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Nahrung, H.F.; Swain, A.J. Strangers in a strange land: Do life history traits differ for alien and native colonisers of novel environments? Biol. Invasions 2015, 17, 699–709. [Google Scholar] [CrossRef]
- Bush, S.J.; Slippers, B.; Neser, S.; Harney, M.; Dittrich-Schröder, G.; Hurley, B.P. Six recently recorded Australian insects associated with Eucalyptus in South Africa. Afr. Entomol. 2016, 24, 539–544. [Google Scholar] [CrossRef]
- Hurley, B.P.; Garnas, J.; Wingfield, M.J.; Branco, M.; Richardson, D.M.; Slippers, B. Increasing numbers and intercontinental spread of invasive insects on eucalypts. Biol. Invasions 2016, 18, 921–933. [Google Scholar] [CrossRef]
- Mendel, Z.; Protasov, A.; Fisher, N.; La Salle, J. Taxonomy and biology of Leptocybe invasa gen. & sp. n. (Hymenoptera: Eulophidae), an invasive gall inducer on Eucalyptus. Aust. J. Entomol. 2004, 43, 101–113. [Google Scholar] [CrossRef]
- Nugnes, F.; Gebiola, M.; Monti, M.M.; Gualtieri, L.; Giorgini, M.; Wang, J.G.; Bernardo, U. Genetic diversity of the invasive gall wasp Leptocybe invasa (Hymenoptera: Eulophidae) and of its Rickettsia endosymbiont, and associated sex-ratio differences. PLoS ONE 2015, 10, e0124660. [Google Scholar] [CrossRef] [PubMed]
- Le, N.H.; Nahrung, H.F.; Griffiths, M.; Lawson, S.A. Invasive Leptocybe spp. and their natural enemies: Global movement of an insect fauna on eucalypts. Biol. Control 2018, 125, 7–14. [Google Scholar] [CrossRef]
- Austin, A.D.; Yeates, D.K.; Cassis, G.; Fletcher, M.J.; La Salle, J.; Lawrence, J.F.; McQuillan, P.B.; Mound, L.A.; Bickel, D.J.; Gullan, P.J.; et al. Insects ‘Down Under’–diversity, endemism and evolution of the Australian insect fauna: Examples from select orders. Aust. Entomol. 2004, 43, 216–234. [Google Scholar] [CrossRef]
- Cranston, P.S. Biodiversity of Australasian insects. In Insect Biodiversity: Science and Society; Footitt, R.G., Adler, P.H., Eds.; Wiley-Blackwell: Chichester, UK, 2009; pp. 83–105. ISBN 9781405151429. [Google Scholar]
- Dittrich-Schröder, G.; Hoareau, T.B.; Hurley, B.P.; Wingfield, M.J.; Lawson, S.; Nahrung, H.F.; Slippers, B. Population genetic analyses of complex global insect invasions in managed landscapes: a Leptocybe invasa (Hymenoptera) case study. Biol. Invasions 2018, 20, 2395–2420. [Google Scholar] [CrossRef]
- Kim, I.-K.; Mendel, Z.; Protasov, A.; Blumberg, D.; La Salle, J. Taxonomy, biology, and efficacy of two Australian parasitoids of the eucalyptus gall wasp, Leptocybe invasa Fisher & La Salle (Hymenoptera: Eulophidae: Tetrastichinae). Zootaxa 2008, 1910, 1–20. [Google Scholar]
- Kelly, J.; La Salle, J.; Harney, M.; Dittrich-Schröder, G.; Hurley, B. Selitrichodes neseri n. sp., a new parasitoid of the eucalyptus gall wasp Leptocybe invasa Fisher & La Salle (Hymenoptera: Eulophidae: Tetrastichinae). Zootaxa 2012, 3333, 50–57. [Google Scholar]
- Kim, I.-K. Evolution of gall inducing Eulophidae (Hymenoptera: Chalcidoidea) on Myrtaceae in Australia. Ph.D. Thesis, The Australian National University, Canberra ACT, Australia, 2008. [Google Scholar]
- Zheng, X.L.; Li, J.; Yang, Z.D.; Xian, Z.H.; Wei, J.G.; Fei, C.L.; Wang, X.P.; Lu, W. A review of invasive biology, prevalence and management of Leptocybe invasa Fisher & La Salle (Hymenoptera: Eulophidae: Tetrastichinae). Afr. Entomol. 2014, 22, 68–79. [Google Scholar] [CrossRef]
- Nyeko, P.; Mutitu, K.E.; Otieno, B.O.; Ngae, G.N.; Day, R.K. Variations in Leptocybe invasa (Hymenoptera: Eulophidae) population intensity and infestation on eucalyptus germplasms in Uganda and Kenya. Int. J. Pest Manag. 2010, 56, 137–144. [Google Scholar] [CrossRef]
- Petro, R.; Madoffe, S.S.; Iddi, S. Infestation density of eucalyptus gall wasp, Leptocybe invasa Fisher and La Salle (Hymenoptera: Eulophidae) on five commercially grown Eucalyptus species in Tanzania. J. Sustain. For. 2014, 33, 276–297. [Google Scholar] [CrossRef]
- Aytar, F. Natural history, distribution and hosts of Eucalyptus gall wasps in Turkey. Presented at the VIIIth European Congress of Entomology, Izmir, Turkey, September 2006. [Google Scholar]
- Steinbauer, M.J.; Kriticos, D.J.; Lukacs, Z.; Clarke, A.R. Modelling a forest lepidopteran: Phenological plasticity determines voltinism which influences population dynamics. For. Ecol. Manag. 2004, 198, 117–1142. [Google Scholar] [CrossRef]
- Jaworski, T.; Hilszczański, J. The effect of temperature and humidity changes on insects development their impact on forest ecosystems in the expected climate change. Leśne Prace Badawcze (For. Res. Pap.) 2013, 74, 345–355. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Shirriffs, J.; Scott, M. Relative importance of plastic vs genetic factors in adaptive differentiation: geographical variation for stress resistance in Drosophila melanogaster from eastern Australia. Funct. Ecol. 2005, 19, 222–227. [Google Scholar] [CrossRef]
- Hill, M.P.; Chown, S.L.; Hoffmann, A.A. A predicted niche shift corresponds with increased thermal resistance in an invasive mite, Halotydeus destructor. Glob. Ecol. Biogeogr. 2013, 22, 942–951. [Google Scholar] [CrossRef]
- PengFei, H.; JianHui, W.; BaoLi, Q.; ShunXiang, R. Extreme low temperature tolerance of Leptocybe invasa. Chin. J. Appl. Entomol. 2012, 49, 1553–1558. [Google Scholar]
- Lu, T.C.; Zhang, Y.Z.; Huang, Z.H.; Huang, J. The effect on the longevity of Leptocybe invasa Fisher & LaSalle (Hymenoptera: Eulophidae) studied by microcalorimetry and traditional methods. J. Therm. Anal. Calorim. 2014, 116, 461–467. [Google Scholar] [CrossRef]
- Yang, Z.D.; Zhao, Y.Y.; Yu, S.Z.; Zheng, X.L.; Xu, L. Rapid cold-hardening response of adult Leptocybe invasa Fisher & La Salle (Hymenoptera: Eulophidae: Tetrastichinae). Afr. Entomol. 2015, 23, 139–146. [Google Scholar] [CrossRef]
- Zhu, F.L.; Ren, S.X.; Qiu, B.L.; Wu, J.H. Effect of temperature on life table parameters of Leptocybe invasa (Hymenoptera: Eulophidae). Aust. Entomol. 2015, 54, 71–78. [Google Scholar] [CrossRef]
- Broennimann, O.; Guisan, A. Predicting current and future biological invasions: Both native and invaded ranges matter. Biol. Lett. 2008, 4, 585–589. [Google Scholar] [CrossRef]
- Byeon, D.-H.; Jung, S.; Lee, W.-H. Review of CLIMEX and MaxEnt for studying species distribution in South Korea. J. Asia-Pac. Biodivers. 2018, 11, 325–333. [Google Scholar] [CrossRef]
- Kriticos, D.J.; Webber, B.L.; Leriche, A.; Ota, N.; Macadam, I.; Bathols, J.; Scott, J.K. CliMond: Global high-resolution historical and future scenario climate surfaces for bioclimatic modelling. Method. Ecol. Evol. 2012, 3, 53–64. [Google Scholar] [CrossRef]
- Soberon, J.; Peterson, A.T. Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodivers. Inform. 2005, 2, 1–10. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudik, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Pearson, R.G.; Raxworthy, C.J.; Nakamura, M.; Peterson, A.T. Predicting species distributions from small numbers of occurrence records: A test case using cryptic geckos in Madagascar. J. Biogeogr. 2007, 34, 102–117. [Google Scholar] [CrossRef]
- Wisz, M.S.; Hijmans, R.J.; Li, J.; Peterson, A.T.; Graham, C.H.; Guisan, A.; NCEAS Predicting Species Distributions Working Group. Effects of sample size on the performance of species distribution models. Divers. Distrib. 2008, 14, 763–773. [Google Scholar] [CrossRef]
- Hunt, R.; Zobel, B. Frost-hardy Eucalyptus grow well in the southeast. South. J. Appl. For. 1978, 2, 6–10. [Google Scholar] [CrossRef]
- Kellison, R.; Lea, R.; Marsh, P. Introduction of Eucalyptus spp. into the United States with special emphasis on the southern United States. Int. J. For. Res. 2013, 189393. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Elith, J.; Graham, C.H.; Lehmann, A.; Leathwick, J.; Ferrier, S. Sample selection bias and presence-only distribution models: Implications for background and pseudo-absence data. Ecol. Appl. 2009, 19, 181–197. [Google Scholar] [CrossRef]
- VanDerWal, J.; Shoo, L.P.; Graham, C.; William, S.E. Selecting pseudo-absence data for presence-only distribution modeling: how far should you stray from what you know? Ecol. Model. 2009, 220, 589–594. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A. A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- Robertson, M.P.; Caithness, N.; Villet, M.H. A PCA-based modelling technique for predicting environmental suitability for organisms from presence records. Divers. Distrib. 2001, 7, 15–27. [Google Scholar] [CrossRef]
- Janžekovič, F.; Novak, T. PCA—A powerful method for analyze ecological niches. In Principal Component Analysis-Multidisciplinary Applications; Open access peer-reviewed Edited Volume; Sanguansat, P., Ed.; IntechOpen: Rijeka, Croatia, 2012; pp. 127–142. ISBN 978-953-51-0129-1. [Google Scholar]
- Anderson, M.J.; Willis, T.J. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Kriticos, D.J.; Maywald, G.F.; Yonow, T.; Zurcher, E.J.; Herrmann, N.I.; Sutherst, R.W. CLIMEX. Version 4: Exploring the Effects of Climate on Plants, Animals and Diseases. CSIRO: Canberra, Australia, 2015; 184p. [Google Scholar]
- Hill, K.D.; Johnson, L.A.S. Systematic studies in the eucalypts. 10. New tropical and subtropical eucalypts from Australia and New Guinea (Eucalyptus, Myrtaceae). Telopea 2000, 8, 503–539. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.549.127&rep=rep1&type=pdf (accessed on 22 January 2019). [CrossRef]
- West-Eberhard, M.J. Phenotypic plasticity and the origins of diversity. Annu. Rev. Ecol. Syst. 1989, 20, 249–278. [Google Scholar] [CrossRef]
- Agrawal, A.F. Spatial heterogeneity and the evolution of sex in diploids. Am. Nat. 2009, 174, S54–S70. [Google Scholar] [CrossRef] [PubMed]
- Luijckx, P.; Ho, E.K.H.; Gasim, M.; Chen, S.; Stanic, A.; Yanchus, C.; Kim, Y.S.; Agrawal, A.F. Higher rates of sex evolve during adaptation to more complex environments. Proc. Nat. Acad. Sci. USA 2017, 114, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Lavanchy, G.; Strehler, M.; Roman, M.N.L.; Lessard-Therrien, M.; Humbert, J.Y.; Dumas, Z.; Jalvingh, K.; Ghali, K.; Garćia-Cuenca, A.F.; Zijlstra, B.; et al. Habitat heterogeneity favors asexual reproduction in natural populations of grassthrips. Evolution 2016, 70, 1780–1790. [Google Scholar] [CrossRef] [PubMed]
- Damos, P.; Savopoulou-Soultani, M. Temperature-driven models for insect development and vital thermal requirements. Psyche: J. Entomol. 2011, 2012, 123405. [Google Scholar] [CrossRef]
- Padmavathi, C.; Katti, G.; Sailaja, V.; Padmakumari, A.P.; Jhansilakshmi, V.; Prabhakar, M.; Prasad, Y.G. Temperature thresholds and thermal requirements for the development of the rice leaf folder, Cnaphalocrocis medinalis. J. Insect Sci. 2013, 13, 1–14. [Google Scholar] [CrossRef]
- Mapondera, T.S.; Burgess, T.; Matsuki, M.; Oberprieler, R.G. Identification and molecular phylogenetics of the cryptic species of the Gonipterus scutellatus complex (Coleoptera: Curculionidae: Gonipterini). Aust. J. Entomol. 2012, 51, 175–188. [Google Scholar] [CrossRef]
- Protasov, A.; Doğanlar, M.; La Salle, J.; Mendel, Z. Occurrence of two local Megastigmus species parasitic on the Eucalyptus gall wasp Leptocybe invasa in Israel and Turkey. Phytoparasitica 2008, 36, 449–459. [Google Scholar] [CrossRef]
- Kulkarni, H.; Kumari, N.K.; Vastrad, A.; Basavanagoud, K. Release and recovery of parasitoids in eucalyptus against gall wasp, Leptocybe invasa (Hymenoptera: Eulophidae) under green house. Karnataka J. Agric. Sci. 2010, 23, 91–92. [Google Scholar]
- Sangtongpraow, B.; Charernsom, K. Evaluation of parasitism capacity of Megastigmus thitipornae Dogănlar & Hassan (Hymenoptera: Torymidae), the local parasitoid of Eucalyptus gall wasp, Leptocybe invasa Fisher & La Salle (Hymenoptera: Eulophidae). Kasetsart J. (Nat. Sci.) 2013, 47, 191–204. [Google Scholar]
- Otieno, B.A. Genetic and environmental influences on the utilization of Eucalyptus camaldulensis (Dehnh.) by Leptocybe invasa Fisher & LaSalle (Hymenoptera: Eulophidae). Ph.D. Thesis, La Trobe University, Melbourne, Austria, May 2017. Available online: http://hdl.handle.net/1959.9/563309 (accessed on 22 January 2019).
| Parameters | Code | Lineage A (Mediterranean Default) [50] | Lineage B Modified Model [33] |
|---|---|---|---|
| Temperature (°C) | |||
| Lower temperature threshold | DV0 | 10 | 19.1 |
| Lower optimum temperature | DV1 | 16 | 26 |
| Upper optimum temperature | DV2 | 24 | 29 |
| Upper temperature threshold | DV3 | 28 | 32 |
| Soil moisture (SM) | |||
| Lower soil moisture threshold | SM0 | 0.1 | 0.1 |
| Lower optimum soil moisture | SM1 | 0.4 | 0.4 |
| Upper optimum soil moisture | SM2 | 0.7 | 0.7 |
| Upper soil moisture threshold | SM3 | 1.5 | 1.5 |
| Cold stress (CS) | |||
| CS temperature threshold | TTCS | 0 | −1 |
| CS temperature rate | THCS | −0.005 | −0.005 |
| CS degree-day threshold | DTCS | 15 | 0 |
| CS degree-day rate | DHCS | −0.001 | 0 |
| Heat stress (HS) | |||
| HS temperature threshold (°C) | TTHS | 30 | 32 |
| HS temperature rate | THHS | 0.002 | 0.002 |
| HS degree-day threshold | DTHS | 0 | 0 |
| HS degree-day rate | DHHS | 0 | 0 |
| Dry stress (DS) | |||
| DS threshold | SMDS | 0.02 | 0.02 |
| DS rate | HDS | −0.05 | −0.05 |
| Wet stress (WS) | |||
| WS threshold | SMWS | 1.6 | 1.6 |
| WS rate | HWS | 0.0015 | 0.0015 |
| Cold-dry stress | not used | not used | |
| Cold-wet stress | not used | not used | |
| Hot-dry stress | not used | not used | |
| Hot-wet stress (HW) | |||
| Hot-wet stress threshold | TTHW | 23 | 32 |
| Hot-wet stress moisture threshold | MTHW | 0.5 | 1.5 |
| Hot-wet stress rate | PHW | 0.075 | 0.075 |
| Light index | not used | not used | |
| Diapause index | not used | not used | |
| Radiation index | not used | not used | |
| Physical substrate index | not used | not used | |
| Biotic substrate index | not used | not used | |
| Degree-day accumulation | PDD | 600 | 563 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otieno, B.A.; Nahrung, H.F.; Steinbauer, M.J. Where Did You Come From? Where Did You Go? Investigating the Origin of Invasive Leptocybe Species Using Distribution Modelling. Forests 2019, 10, 115. https://doi.org/10.3390/f10020115
Otieno BA, Nahrung HF, Steinbauer MJ. Where Did You Come From? Where Did You Go? Investigating the Origin of Invasive Leptocybe Species Using Distribution Modelling. Forests. 2019; 10(2):115. https://doi.org/10.3390/f10020115
Chicago/Turabian StyleOtieno, Beryn A., Helen F. Nahrung, and Martin J. Steinbauer. 2019. "Where Did You Come From? Where Did You Go? Investigating the Origin of Invasive Leptocybe Species Using Distribution Modelling" Forests 10, no. 2: 115. https://doi.org/10.3390/f10020115
APA StyleOtieno, B. A., Nahrung, H. F., & Steinbauer, M. J. (2019). Where Did You Come From? Where Did You Go? Investigating the Origin of Invasive Leptocybe Species Using Distribution Modelling. Forests, 10(2), 115. https://doi.org/10.3390/f10020115





