Temperature and Rainfall Are Separate Agents of Selection Shaping Population Differentiation in a Forest Tree
Abstract
1. Introduction
2. Materials and Methods
2.1. Common-Garden Experiments and Measurements
2.2. Data Analysis
2.2.1. (Co)Variance Components and Genetic Parameters
2.2.2. Estimation of Marginal Means
2.2.3. Climate-Trait Associations
2.2.4. Selection Analysis
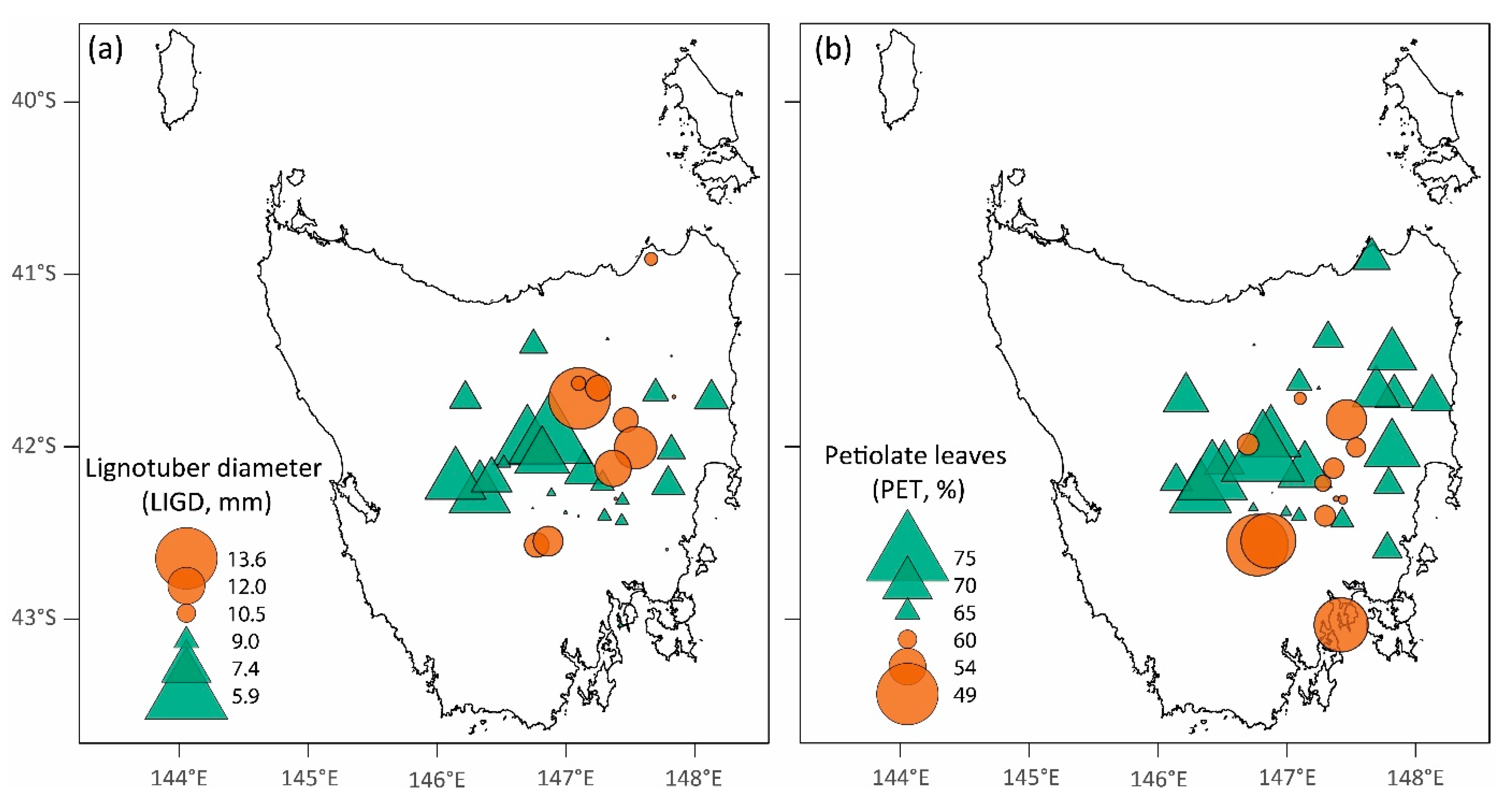
3. Results
3.1. (Co)Variance Components and Genetic Parameters
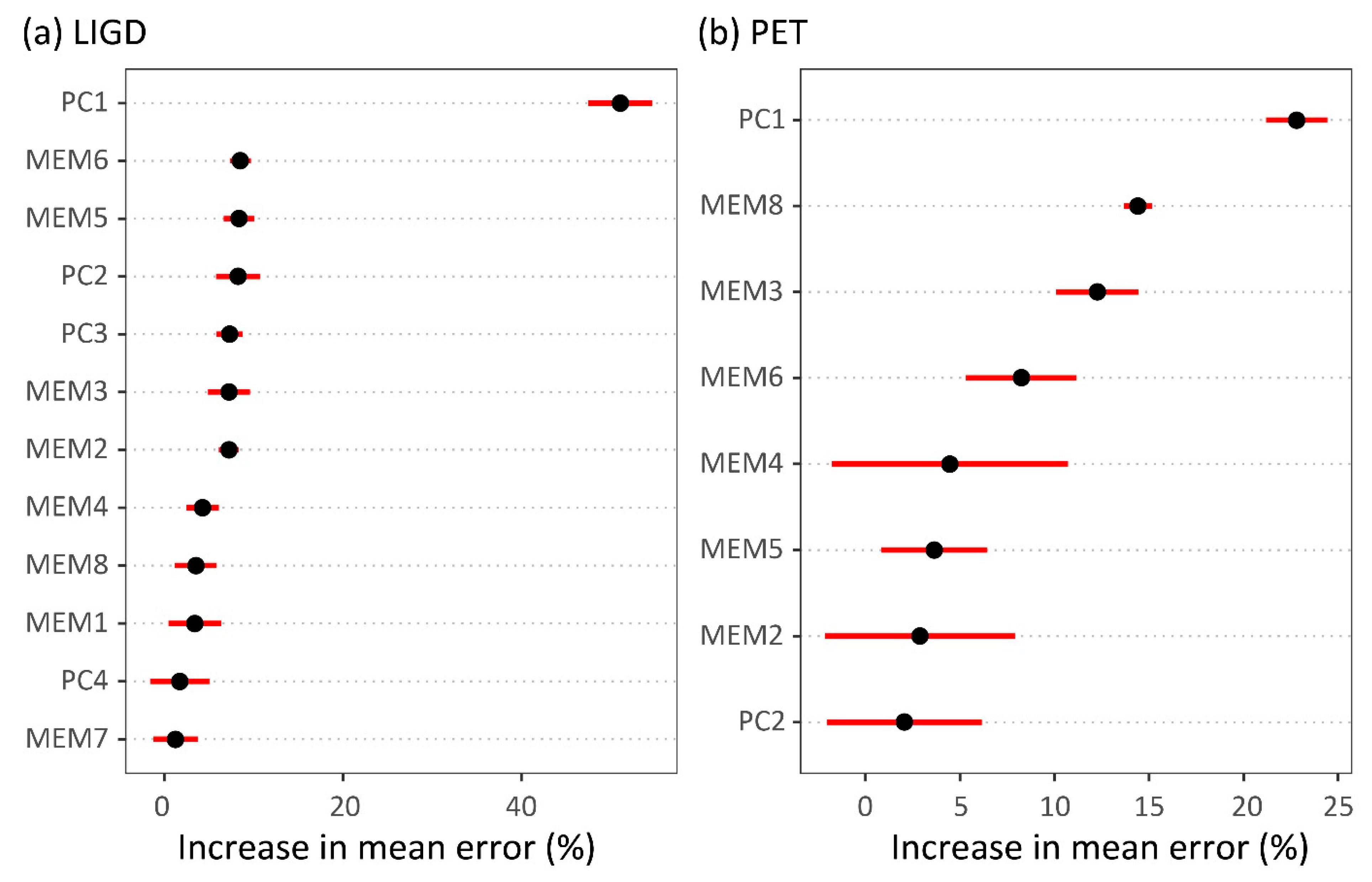
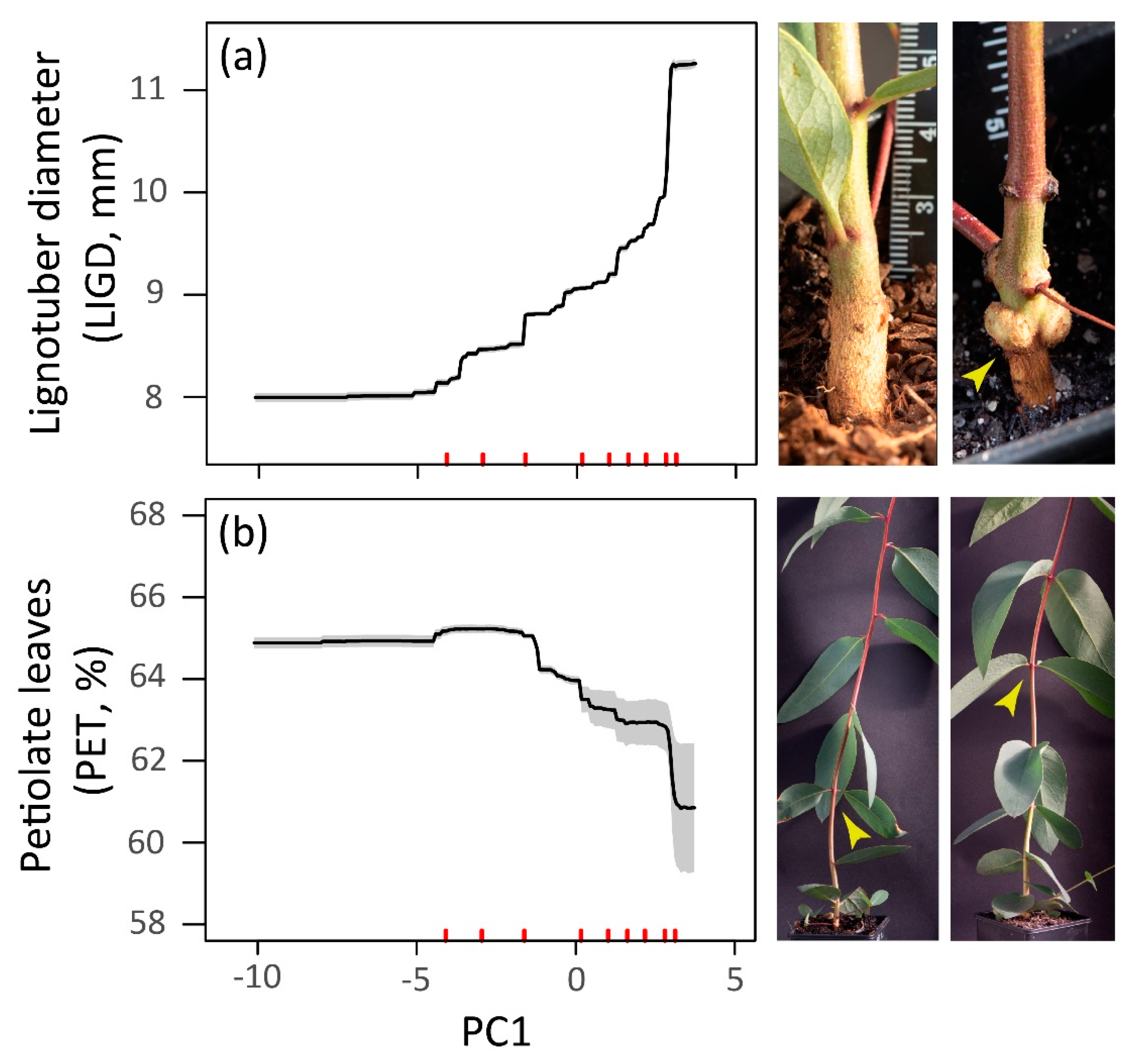
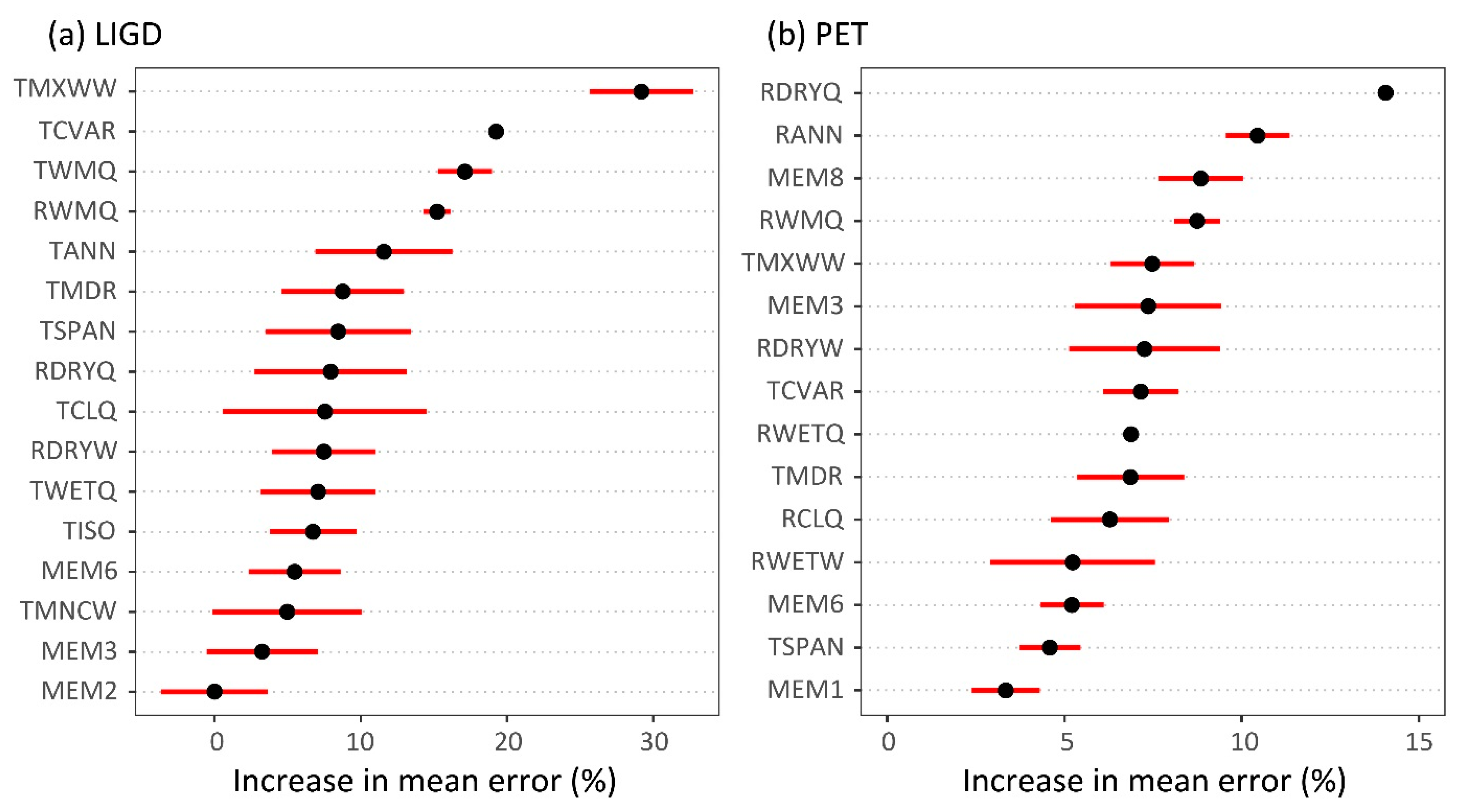
3.2. Climate-Trait Associations
3.3. Selection Differentials and Gradients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murren, C.J. Phenotypic integration in plants. Plant Species Biol. 2002, 17, 89–99. [Google Scholar] [CrossRef]
- Armbruster, W.S.; Pélabon, C.; Bolstad Geir, H.; Hansen Thomas, F. Integrated phenotypes: Understanding trait covariation in plants and animals. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130245. [Google Scholar] [CrossRef]
- Conner, J.K.; Cooper, I.A.; La Rosa, R.J.; Pérez, S.G.; Royer, A.M. Patterns of phenotypic correlations among morphological traits across plants and animals. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130246. [Google Scholar] [CrossRef]
- Klingenberg, C.P. Studying morphological integration and modularity at multiple levels: Concepts and analysis. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130249. [Google Scholar] [CrossRef]
- Raffard, A.; Lecerf, A.; Cote, J.; Buoro, M.; Lassus, R.; Cucherousset, J. The functional syndrome: Linking individual trait variability to ecosystem functioning. Proc. R. Soc. B Biol. Sci. 2017, 284, 20171893. [Google Scholar] [CrossRef]
- Endler, J.A. Multiple-trait coevolution and environmental gradients in guppies. Trends Ecol. Evol. 1995, 10, 22–29. [Google Scholar] [CrossRef]
- Peiman, K.S.; Robinson, B.W. Comparative analyses of phenotypic trait covariation within and among populations. Am. Nat. 2017, 190, 451–468. [Google Scholar] [CrossRef]
- Wright, J.W. Introduction to Forest Genetics; Academic Press: New York, NY, USA, 1976. [Google Scholar]
- Morgenstern, E.K. Geographic Variation in Forest Trees: Genetic Basis and Application of Knowledge in Silviculture; BC Press: Vancouver, QC, Canada, 1996. [Google Scholar]
- Alberto, F.J.; Aitken, S.N.; Alía, R.; González-Martínez, S.C.; Hänninen, H.; Kremer, A.; Lefèvre, F.; Lenormand, T.; Yeaman, S.; Whetten, R.; et al. Potential for evolutionary responses to climate change—Evidence from tree populations. Glob. Chang. Biol. 2013, 19, 1645–1661. [Google Scholar] [CrossRef]
- Eldridge, K.; Davidson, J.; Harwood, C.; van Wyk, G. Eucalypt Domestication and Breeding; Clarendon Press: Oxford, UK, 1993. [Google Scholar]
- Kremer, A.; Potts, B.M.; Delzon, S. Genetic divergence in forest trees: Understanding the consequences of climate change. Funct. Ecol. 2014, 28, 22–36. [Google Scholar] [CrossRef]
- Armbruster, W.S.; Schwaegerle, K.E. Causes of covariation of phenotypic traits among populations. J. Evol. Biol. 1996, 9, 261–276. [Google Scholar] [CrossRef]
- Armbruster, W.S.; Pélabon, C.; Hansen, T.; Mulder, C. Floral integration, modularity, and accuracy: Distinguishing complex adaptations from genetic constraints. In Phenotypic Integration: Studying the Ecology and Evolution of Complex Phenotypes; Pigliucci, M., Preston, K., Eds.; Oxford University Press: Oxford, UK, 2004; pp. 23–49. [Google Scholar]
- Chenoweth, S.F.; Rundle, H.D.; Blows, M.W. The contribution of selection and genetic constraints to phenotypic divergence. Am. Nat. 2010, 175, 186–196. [Google Scholar] [CrossRef]
- Colautti, R.I.; Barrett, S.C.H. Population divergence along lines of genetic variance and covariance in the invasive plant Lythrum salicaria in eastern North America. Evolution 2011, 65, 2514–2529. [Google Scholar] [CrossRef]
- Bolstad, G.H.; Hansen, T.F.; Pélabon, C.; Falahati-Anbaran, M.; Pérez-Barrales, R.; Armbruster, W.S. Genetic constraints predict evolutionary divergence in Dalechampia blossoms. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130255. [Google Scholar] [CrossRef]
- Leinonen, T.; McCairns, R.J.S.; O’Hara, R.B.; Merilä, J. QST–FST comparisons: Evolutionary and ecological insights from genomic heterogeneity. Nat. Rev. Genet. 2013, 14, 179. [Google Scholar] [CrossRef]
- Thomassen, H.A.; Buermann, W.; Milá, B.; Graham, C.H.; Cameron, S.E.; Schneider, C.J.; Pollinger, J.P.; Saatchi, S.; Wayne, R.K.; Smith, T.B. Modeling environmentally associated morphological and genetic variation in a rainforest bird, and its application to conservation prioritization. Evol. Appl. 2010, 3, 1–16. [Google Scholar] [CrossRef]
- Fitzpatrick, M.C.; Keller, S.R. Ecological genomics meets community-level modelling of biodiversity: Mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 2015, 18, 1–16. [Google Scholar] [CrossRef]
- Lande, R.; Arnold, S.J. The measurement of selection on correlated characters. Evolution 1983, 37, 1210–1226. [Google Scholar] [CrossRef]
- Phillips, P.C.; Arnold, S.J. Visualizing multivariate selection. Evolution 1989, 43, 1209–1222. [Google Scholar] [CrossRef]
- Conner, J.K.; Hartl, D.L. A Primer of Ecological Genetics; Sinauer Associates: Sunderland, MA, USA, 2004; p. 304. [Google Scholar]
- Kingsolver, J.G.; Diamond, S.E.; Siepielski, A.M.; Carlson, S.M. Synthetic analyses of phenotypic selection in natural populations: Lessons, limitations and future directions. Evol. Ecol. 2012, 26, 1101–1118. [Google Scholar] [CrossRef]
- Costa e Silva, J.; Harrison, P.A.; Wiltshire, R.; Potts, B.M. Evidence that divergent selection shapes a developmental cline in a forest tree species complex. Ann. Bot. 2018, 122, 181–194. [Google Scholar] [CrossRef]
- Gauli, A.; Vaillancourt, R.E.; Bailey, T.G.; Steane, D.A.; Potts, B.M. Evidence for local climate adaptation in early-life traits of Tasmanian populations of Eucalyptus pauciflora. Tree Genet. Genomes 2015, 11, 104. [Google Scholar] [CrossRef]
- Prober, S.M.; Potts, B.M.; Bailey, T.; Byrne, M.; Dillon, S.; Harrison, P.A.; Hoffmann, A.A.; Jordan, R.; Mclean, E.H.; Steane, D.A.; et al. Climate adaptation and ecological restoration in eucalypts. Proc. R. Soc. Vic. 2016, 128, 40–53. [Google Scholar] [CrossRef]
- Prober, S.M.; Byrne, M.; McLean, E.H.; Steane, D.A.; Potts, B.M.; Vaillancourt, R.E.; Stock, W.D. Climate-adjusted provenancing: A strategy for climate-resilient ecological restoration. Front. Ecol. Evol. 2015, 3, 65. [Google Scholar] [CrossRef]
- Harrison, P.A.; Vaillancourt, R.E.; Harris, R.M.B.; Potts, B.M. Integrating climate change and habitat fragmentation to identify candidate seed sources for ecological restoration. Restor. Ecol. 2017, 25, 524–531. [Google Scholar] [CrossRef]
- Gauli, A.; Steane, D.A.; Vaillancourt, R.E.; Potts, B.M. Molecular genetic diversity and population structure in Eucalyptus pauciflora subsp pauciflora (Myrtaceae) on the island of Tasmania. Aust. J. Bot. 2014, 62, 175–188. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Davidson, A. Adaptive phenotypic plasticity and plant water use. Funct. Plant Biol. 2010, 37, 117–127. [Google Scholar] [CrossRef]
- Ramírez-Valiente, J.A.; Valladares, F.; Delgado, A.; Nicotra, A.B.; Aranda, I. Understanding the importance of intrapopulation functional variability and phenotypic plasticity in Quercus suber. Tree Genet. Genomes 2015, 11, 35. [Google Scholar] [CrossRef]
- Borralho, N.M.G.; Kanowski, P.J.; Cotterill, P.P. Genetic control of growth of Eucalyptus globulus in Portugal I. Genetic and phenotypic parameters. Silvae Genet. 1992, 41, 39–45. [Google Scholar]
- Mora, F.; Gleadow, R.; Perret, S.; Scapim, C.A. Genetic variation for early flowering, survival and growth in sugar gum (Eucalyptus cladocalyx F. Muell) in southern Atacama Desert. Euphytica 2009, 169, 335–344. [Google Scholar] [CrossRef]
- Stackpole, D.J.; Vaillancourt, R.E.; Aguigar, M.; Potts, B.M. Age trends in genetic parameters for growth and wood density in Eucalyptus globulus. Tree Genet. Genomes 2010, 6, 179–193. [Google Scholar] [CrossRef]
- Nickolas, H.; Harrison, P.A.; Tilyard, P.; Vaillancourt, R.E.; Potts, B.M. Inbreeding depression and differential maladaptation shape the fitness trajectory of two co-occurring Eucalyptus species. Ann. For. Sci. 2019, 76, 10. [Google Scholar] [CrossRef]
- Peet, R.K.; Christensen, N.L. Competition and tree death. BioScience 1987, 37, 586–595. [Google Scholar] [CrossRef]
- Gilmour, A.R.; Gogel, B.J.; Cullis, B.R.; Welham, S.J.; Thompson, R. ASReml User Guide Release 4.1; VSN International Ltd.: Hemel Hempstead, UK, 2015; p. 372. [Google Scholar]
- SAS. SAS/STAT® 14.1. User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2015. [Google Scholar]
- R-Core-Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Gilmour, A.R.; Thompson, R.; Cullis, B.R. Average information REML: An efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 1995, 51, 1440–1450. [Google Scholar] [CrossRef]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates Inc.: Sunderland, MA, USA, 1998; p. 971. [Google Scholar]
- Stram, D.O.; Lee, J.W. Variance components testing in the longitudinal mixed effects model. Biometrics 1994, 50, 1171–1177. [Google Scholar] [CrossRef]
- Kenward, M.G.; Roger, J.H. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 1997, 53, 983–997. [Google Scholar] [CrossRef]
- Xu, T.; Hutchinson, M.F. New developments and applications in the ANUCLIM spatial climatic and bioclimatic modelling package. Environ. Model. Softw. 2013, 40, 267–279. [Google Scholar] [CrossRef]
- Jones, D.A.; Wang, W.; Fawcett, R. High-quality spatial climate data-sets for Australia. Aust. Meteorol. Oceanogr. J. 2009, 58, 233–248. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Strobl, C.; Boulesteix, A.-L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef]
- Ellis, N.; Smith, S.J.; Pitcher, C.R. Gradient forests: Calculating importance gradients on physical predictors. Ecology 2012, 93, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Dray, S.; Legendre, P.; Peres-Neto, P.R. Spatial modelling: A comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol. Model. 2006, 196, 483–493. [Google Scholar] [CrossRef]
- Dray, S.; Bauman, D.; Blanchet, G.; Borcard, D.; Clappe, S.; Guenard, G.; Jombart, T.; Larocque, G.; Legendre, P.; Madi, N.; et al. adespatial: Multivariate Multiscale Spatial Analysis. R Package Version 0.3-4. 2019. Available online: https://cran.r-project.org/package=adespatial (accessed on 12 December 2019).
- Rausher, M.D. The measurement of selection on quantitative traits—Biases due to environmental covariances between traits and fitness. Evolution 1992, 46, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Stinchcombe, J.R.; Rutter, M.T.; Burdick, D.S.; Tiffin, P.; Rausher, M.D.; Mauricio, R. Testing for environmentally induced bias in phenotypic estimates of natural selection: Theory and practice. Am. Nat. 2002, 160, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Stinchcombe, J.R.; Agrawal, A.F.; Hohenlohe, P.A.; Arnold, S.J.; Blows, M.W. Estimating nonlinear selection gradients using quadratic regression coefficients: Double or nothing? Evolution 2008, 62, 2435–2440. [Google Scholar] [CrossRef] [PubMed]
- Endler, J.A. Natural Selection in the Wild; Princeton University Press: Princeton, NJ, USA, 1986; p. 336. [Google Scholar]
- Mitchell-Olds, T.; Shaw, R.G. Regression analysis of natural selection: Statistical inference and biological interpretation. Evolution 1987, 41, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Steppan, S.J.; Phillips, P.C.; Houle, D. Comparative quantitative genetics: Evolution of the G matrix. Trends Ecol. Evol. 2002, 17, 320–327. [Google Scholar] [CrossRef]
- Arnold, S.J.; Bürger, R.; Hohenlohe, P.A.; Ajie, B.C.; Jones, A.G. Understanding the evolution and stability of the G-matrix. Evolution 2008, 62, 2451–2461. [Google Scholar] [CrossRef]
- Karschon, R. Lignotuber occurrence in Eucalyptus camaldulensis Dehn. and its phylogenetic significance. Flora 1971, 160, 495–510. [Google Scholar] [CrossRef]
- Ammitzboll, H.; Vaillancourt, R.E.; Potts, B.M.; Harrison, P.A.; Brodribb, T.; Sussmilch, F.C.; Freeman, J.S. Independent genetic control of drought resistance, recovery, and growth of Eucalyptus globulus seedlings. Plant Cell Environ. 2019, in press. [Google Scholar] [CrossRef]
- Hudson, C.J.; Freeman, J.S.; Jones, R.C.; Potts, B.M.; Wong, M.M.L.; Weller, J.L.; Hecht, V.F.G.; Poethig, R.S.; Vaillancourt, R.E. Genetic control of heterochrony in Eucalyptus globulus. G3 Genes Genomes Genet. 2014, 4, 1235–1245. [Google Scholar] [CrossRef][Green Version]
- Ladiges, P.Y. Differentiation in some populations of Eucalyptus viminalis Labill. in relation to factors affecting seedling establishment. Aust. J. Bot. 1974, 22, 471–487. [Google Scholar] [CrossRef]
- Ladiges, P.Y.; Ashton, D.H. Variation in some central Victorian populations of Eucalyptus viminalis Labill. Aust. J. Bot. 1974, 22, 81–102. [Google Scholar] [CrossRef]
- Walters, J.R.; Bell, T.L.; Read, S. Intra-specific variation in carbohydrate reserves and sprouting ability in Eucalyptus obliqua seedlings. Aust. J. Bot. 2005, 53, 195–203. [Google Scholar] [CrossRef]
- Coyne, J.A.; Beecham, E. Heritability of two morphological characters within and among natural populations of Drosophila melanogaster. Genetics 1987, 117, 727–737. [Google Scholar]
- Clucas, R.; Ladiges, P. Variations in populations of Eucalyptus ovata Labill., and the effects of waterlogging on seedling growth. Aust. J. Bot. 1979, 27, 301–315. [Google Scholar] [CrossRef]
- Jahnke, R.; Carr, D.; Carr, S. Lignotuber development and growth parameters in Eucalyptus camaldulensis (Dehnh.): Effects of phosphorus and nitrogen levels. Aust. J. Bot. 1983, 31, 283–292. [Google Scholar] [CrossRef]
- Williams, D.R.; Potts, B.M.; Smethurst, P.J. Phosphorus fertiliser can induce earlier vegetative phase change in Eucalyptus nitens. Aust. J. Bot. 2004, 52, 281–284. [Google Scholar] [CrossRef]
- Jaya, E.; Kubien, D.S.; Jameson, P.E.; Clemens, J. Vegetative phase change and photosynthesis in Eucalyptus occidentalis: Architectural simplification prolongs juvenile traits. Tree Physiol. 2010, 30, 393–403. [Google Scholar] [CrossRef]
- Potts, B.M. Variation in the Eucalyptus gunnii-archeri complex. III. Reciprocal transplant trials. Aust. J. Bot. 1985, 33, 687–704. [Google Scholar] [CrossRef]
- Wiltshire, R.J.E.; Potts, B.M.; Reid, J.B. The genetic control of reproductive and vegetative phase change in the Eucalyptus risdonii/E. tenuiramis complex. Aust. J. Bot. 1998, 46, 45–63. [Google Scholar] [CrossRef]
- Hamilton, M.G.; Tilyard, P.A.; Williams, D.R.; Vaillancourt, R.E.; Wardlaw, T.J.; Potts, B.M. The genetic variation in the timing of heteroblastic transition in Eucalyptus globulus is stable across environments. Aust. J. Bot. 2011, 59, 170–175. [Google Scholar] [CrossRef]
- James, S.A.; Bell, D.T. Influence of light availability on leaf structure and growth of two Eucalyptus globulus ssp globulus provenances. Tree Physiol. 2000, 20, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Pasanen, A.E.J. Genetic Variation in the Phenotypic Plasticity of Eucalyptus pauciflora subsp. pauciflora Sieb. ex. Spreng. Ph.D. Thesis, University of Tasmania, Hobart, Tasmania, 2014. [Google Scholar]
- Falconer, D.S.; MacKay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longman Scientific and Technical: New York, NY, USA, 1996; p. 438. [Google Scholar]
- Teplitsky, C.; Tarka, M.; Møller, A.P.; Nakagawa, S.; Balbontín, J.; Burke, T.A.; Doutrelant, C.; Gregoire, A.; Hansson, B.; Hasselquist, D.; et al. Assessing multivariate constraints to evolution across ten long-term avian studies. PLoS ONE 2014, 9, e90444. [Google Scholar] [CrossRef] [PubMed]
- Etterson, J.R. Evolutionary potential of Chamaecrista fasciculata in relation to climate change. I. Clinal patterns of selection along an environmental gradient in the Great Plains. Evolution 2004, 58, 1446–1458. [Google Scholar] [CrossRef]
- Dudley, S.A. Differing selection on plant physiological traits in response to environmental water availability: A test of adaptive hypotheses. Evolution 1996, 50, 92–102. [Google Scholar] [CrossRef]
- Simonsen, A.K.; Stinchcombe, J.R. Quantifying evolutionary genetic constraints in the ivyleaf morning glory, Ipomoea hederacea. Int. J. Plant Sci. 2010, 171, 972–986. [Google Scholar] [CrossRef][Green Version]
- Körner, C. The use of altitude in ecological research. Trends Ecol. Evol. 2007, 22, 569–574. [Google Scholar] [CrossRef]
- De Frenne, P.; Graae, B.J.; Rodríguez-Sánchez, F.; Kolb, A.; Chabrerie, O.; Decocq, G.; De Kort, H.; De Schrijver, A.; Diekmann, M.; Eriksson, O.; et al. Latitudinal gradients as natural laboratories to infer species’ responses to temperature. J. Ecol. 2013, 101, 784–795. [Google Scholar] [CrossRef]
- Montesinos-Navarro, A.; Wig, J.; Xavier Pico, F.; Tonsor, S.J. Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol. 2011, 189, 282–294. [Google Scholar] [CrossRef]
- Kooyers, N.J.; Greenlee, A.B.; Colicchio, J.M.; Oh, M.; Blackman, B.K. Replicate altitudinal clines reveal that evolutionary flexibility underlies adaptation to drought stress in annual Mimulus guttatus. New Phytol. 2015, 206, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Pratt, J.D.; Mooney, K.A. Clinal adaptation and adaptive plasticity in Artemisia californica: Implications for the response of a foundation species to predicted climate change. Glob. Chang. Biol. 2013, 19, 2454–2466. [Google Scholar] [CrossRef] [PubMed]
- Steane, D.A.; Mclean, E.H.; Potts, B.M.; Prober, S.M.; Stock, W.D.; Stylianou, V.M.; Vaillancourt, R.E.; Byrne, M. Evidence for adaptation and acclimation in a widespread eucalypt of semi-arid Australia. Biol. J. Linn. Soc. 2017, 121, 484–500. [Google Scholar] [CrossRef]
- Prunier, J.; Laroche, J.; Beaulieu, J.; Bousquet, J. Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Mol. Ecol. 2011, 20, 1702–1716. [Google Scholar] [CrossRef]
- Jordan, R.; Hoffmann, A.A.; Dillon, S.K.; Prober, S.M. Evidence of genomic adaptation to climate in Eucalyptus microcarpa: Implications for adaptive potential to projected climate change. Mol. Ecol. 2017, 26, 6002–6020. [Google Scholar] [CrossRef]
- Frachon, L.; Bartoli, C.; Carrère, S.; Bouchez, O.; Chaubet, A.; Gautier, M.; Roby, D.; Roux, F. A genomic map of climate adaptation in Arabidopsis thaliana at a micro-geographic scale. Front. Plant Sci 2018, 9, 967. [Google Scholar] [CrossRef]
- Nicolle, D. A classification and census of regenerative strategies in the eucalypts (Angophora, Corymbia and Eucalyptus-Myrtaceae), with special reference to the obligate seeders. Aust. J. Bot. 2006, 54, 391–407. [Google Scholar] [CrossRef]
- Gosper, C.R.; Hopley, T.; Byrne, M.; Hopper, S.D.; Prober, S.M.; Yates, C.J. Phylogenomics shows lignotuber state is taxonomically informative in closely related eucalypts. Mol. Phylogenet. Evol. 2019, 135, 236–248. [Google Scholar] [CrossRef]
- Graham, A.W.; Wallwork, M.A.; Sedgley, M. Lignotuber bud development in Eucalyptus cinerea (F. Muell. ex benth). Int. J. Plant Sci. 1998, 159, 979–988. [Google Scholar] [CrossRef]
- Karschon, R. Ecotypic variation in Eucalyptus camaldulensis Dehn. In Contributions on Eucalypts in Israel, III; National and University Institute of Agriculture, Ilanot, and Land Development Authority: Qiryat Hayim, Israel, 1967; Volume III, pp. 35–53. [Google Scholar]
- Ashton, D.H. Ecology of eucalypt regeneration. In Diseases and Pathogens of Eucalypts; Keane, P.J., Kile, G.A., Podger, F.D., Brown, B.N., Eds.; CSIRO Publishing: Collingwood, ON, USA, 2000; pp. 47–60. [Google Scholar]
- Smith, M.G.; Arndt, S.K.; Miller, R.E.; Kasel, S.; Bennett, L.T. Trees use more non-structural carbohydrate reserves during epicormic than basal resprouting. Tree Physiol. 2018, 38, 1779–1791. [Google Scholar] [CrossRef]
- Myers, B. The Influence of the lignotuber on hydraulic conductance and leaf conductance in Eucalyptus behriana seedlings. Funct. Plant Biol. 1995, 22, 857–863. [Google Scholar] [CrossRef]
- Richardson, A.D.; Carbone, M.S.; Keenan, T.F.; Czimczik, C.I.; Hollinger, D.Y.; Murakami, P.; Schaberg, P.G.; Xu, X. Seasonal dynamics and age of stemwood nonstructural carbohydrates in temperate forest trees. New Phytol. 2013, 197, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Eyles, A.; Pinkard, E.A.; Mohammed, C. Shifts in biomass and resource allocation patterns following defoliation in Eucalyptus globulus growing with varying water and nutrient supplies. Tree Physiol. 2009, 29, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Quentin, A.G.; Beadle, C.L.; O’Grady, A.P.; Pinkard, E.A. Effects of partial defoliation on closed canopy Eucalyptus globulus Labilladiere: Growth, biomass allocation and carbohydrates. For. Ecol. Manag. 2011, 261, 695–702. [Google Scholar] [CrossRef]
- Atkinson, R.R.L.; Burrell, M.M.; Rose, K.E.; Osborne, C.P.; Rees, M. The dynamics of recovery and growth: How defoliation affects stored resources. Proc. R. Soc. B Biol. Sci. 2014, 281, 20133355. [Google Scholar] [CrossRef]
- Mitchell, P.J.; O’Grady, A.P.; Tissue, D.T.; White, D.A.; Ottenschlaeger, M.L.; Pinkard, E.A. Drought response strategies define the relative contributions of hydraulic dysfunction and carbohydrate depletion during tree mortality. New Phytol. 2013, 197, 862–872. [Google Scholar] [CrossRef]
- Ruehr, N.K.; Gast, A.; Weber, C.; Daub, B.; Arneth, A. Water availability as dominant control of heat stress responses in two contrasting tree species. Tree Physiol. 2015, 36, 164–178. [Google Scholar] [CrossRef]
- Williams, K.J.; Potts, B.M. The natural distribution of Eucalyptus species in Tasmania. Tasforests 1996, 8, 39–165. [Google Scholar]
- Nürnberger, B.; Barton, N.; MacCallum, C.; Gilchrist, J.; Appleby, M. Natural selection on quantitative traits in the Bombina hybrid zone. Evolution 1995, 49, 1224–1238. [Google Scholar] [CrossRef]
- Hochberg, U.; Albuquerque, C.; Rachmilevitch, S.; Cochard, H.; David-Schwartz, R.; Brodersen, C.R.; McElrone, A.; Windt, C.W. Grapevine petioles are more sensitive to drought induced embolism than stems: Evidence from in vivo MRI and microcomputed tomography observations of hydraulic vulnerability segmentation. Plant Cell Environ. 2016, 39, 1886–1894. [Google Scholar] [CrossRef]
- Wason, J.W.; Anstreicher, K.S.; Stephansky, N.; Huggett, B.A.; Brodersen, C.R. Hydraulic safety margins and air-seeding thresholds in roots, trunks, branches and petioles of four northern hardwood trees. New Phytol. 2018, 219, 77–88. [Google Scholar] [CrossRef] [PubMed]
- James, S.A.; Bell, D.T. Leaf morphological and anatomical characteristics of heteroblastic Eucalyptus globulus ssp globulus (Myrtaceae). Aust. J. Bot. 2001, 49, 259–269. [Google Scholar] [CrossRef]
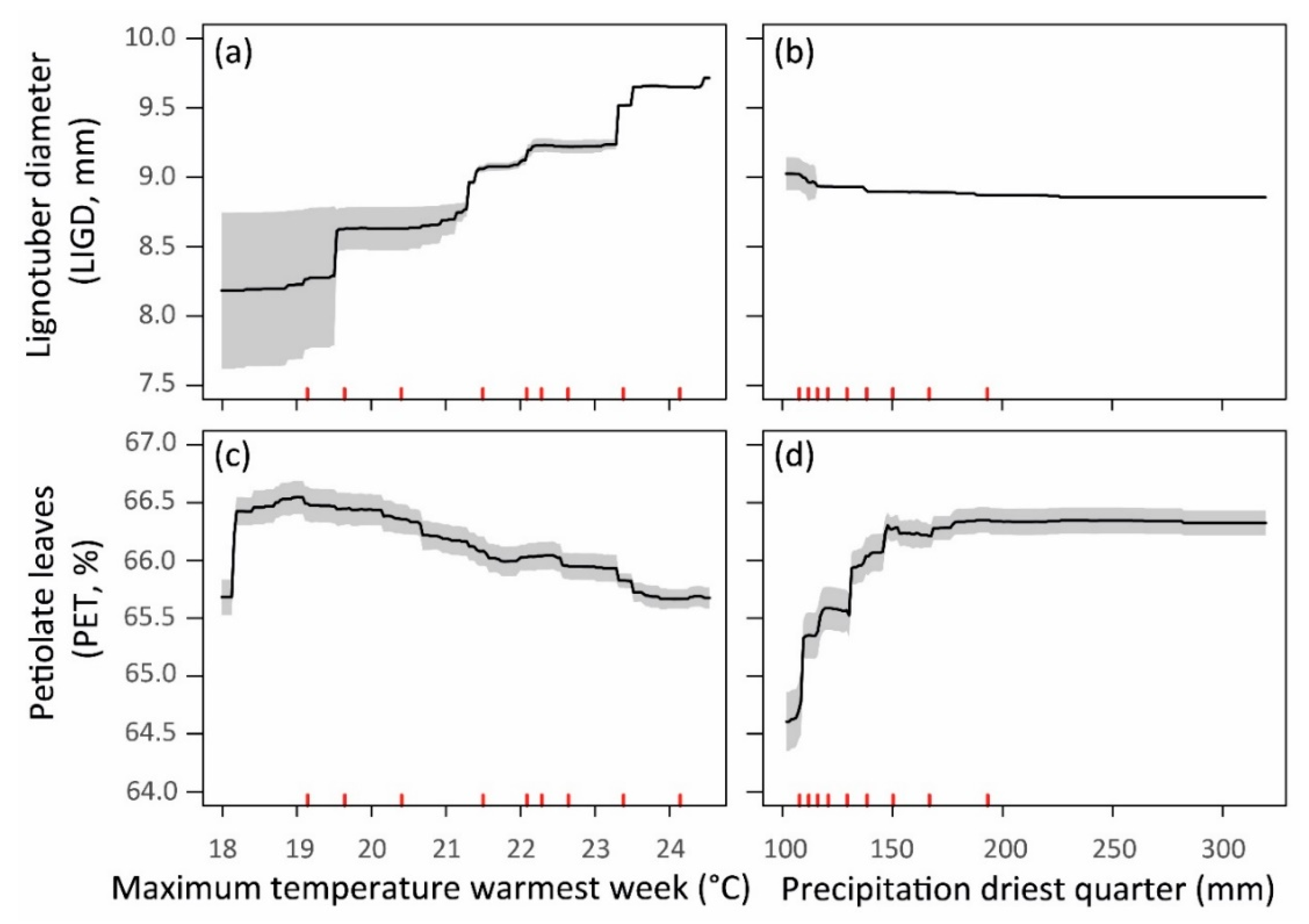
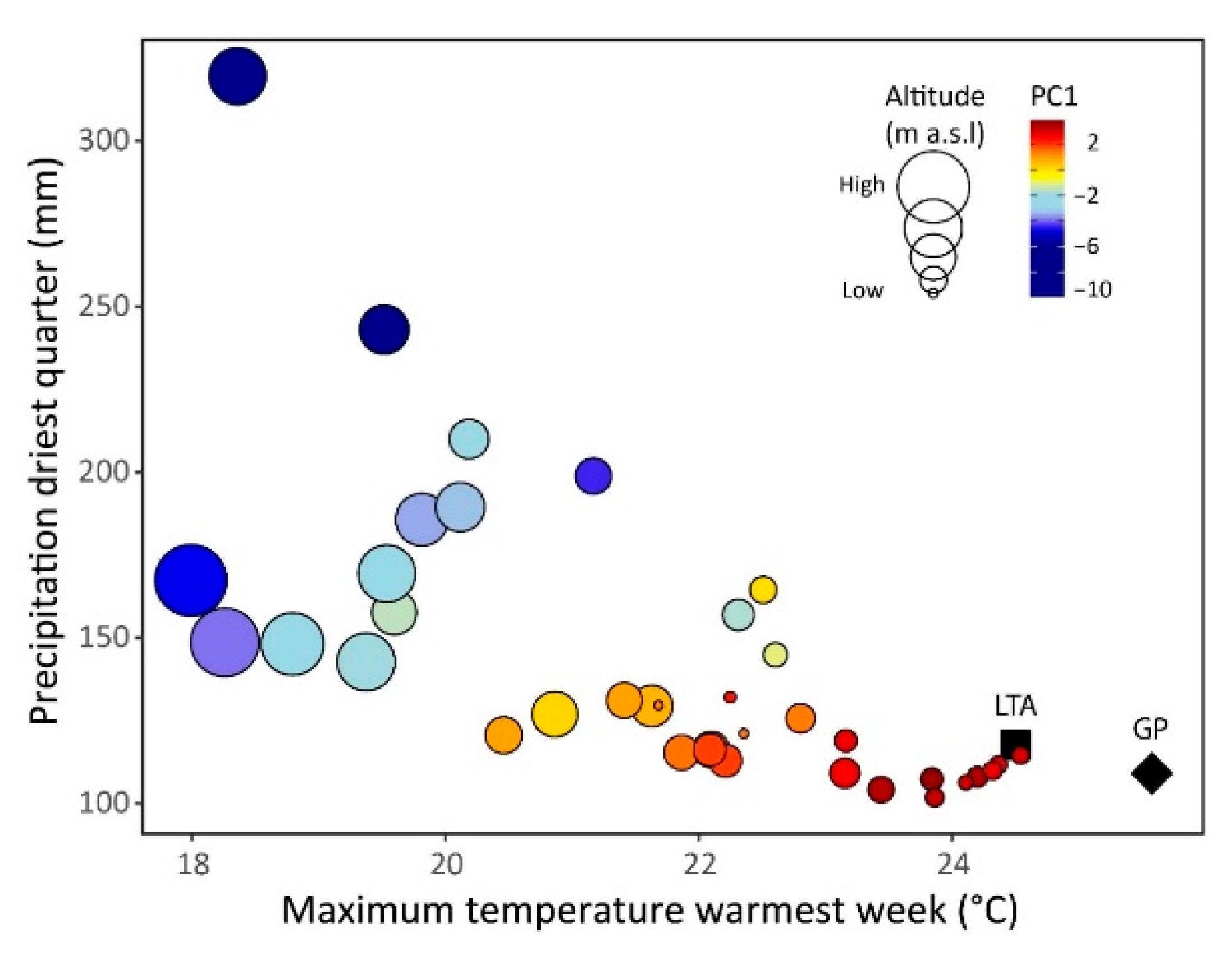
| LIGD | 1.31 ± 0.54 | 2.56 ± 0.66 | 0.203 ± 0.085 |
| (p = 0.003) | (p < 0.001) | ||
| PET | 91.54 ± 27.65 | 26.87 ± 9.46 | 0.425 ± 0.127 |
| (p < 0.001) | (p < 0.001) | ||
| HT | 0.59 ± 0.12 | 0.33 ± 0.09 | 0.399 ± 0.078 |
| (p < 0.001) | (p < 0.001) |
| Linear Selection Differentials | Linear and Non-Linear Selection Gradients | |||
|---|---|---|---|---|
| LIGD | PET | |||
| LIGD | 0.141 ± 0.013 (p < 0.001) | 0.135 ± 0.014 (p < 0.001) | −0.012 ± 0.020 (p > 0.05) | |
| PET | −0.062 ± 0.015 (p < 0.001) | −0.020 ± 0.014 (p > 0.05) | −0.013 ± 0.015 (p > 0.05) | −0.029 ± 0.020 (p > 0.05) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa e Silva, J.; Potts, B.; Harrison, P.A.; Bailey, T. Temperature and Rainfall Are Separate Agents of Selection Shaping Population Differentiation in a Forest Tree. Forests 2019, 10, 1145. https://doi.org/10.3390/f10121145
Costa e Silva J, Potts B, Harrison PA, Bailey T. Temperature and Rainfall Are Separate Agents of Selection Shaping Population Differentiation in a Forest Tree. Forests. 2019; 10(12):1145. https://doi.org/10.3390/f10121145
Chicago/Turabian StyleCosta e Silva, João, Brad Potts, Peter A. Harrison, and Tanya Bailey. 2019. "Temperature and Rainfall Are Separate Agents of Selection Shaping Population Differentiation in a Forest Tree" Forests 10, no. 12: 1145. https://doi.org/10.3390/f10121145
APA StyleCosta e Silva, J., Potts, B., Harrison, P. A., & Bailey, T. (2019). Temperature and Rainfall Are Separate Agents of Selection Shaping Population Differentiation in a Forest Tree. Forests, 10(12), 1145. https://doi.org/10.3390/f10121145






