Comprehensive Transcriptome Analysis Revealed the Effects of the Light Quality, Light Intensity, and Photoperiod on Phlorizin Accumulation in Lithocarpus polystachyus Rehd.
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Material
2.2. RNA Extraction and Illumina Sequencing
2.3. Transcript Splicing, Assembly, and Functional Annotation
2.4. Screening of Differentially Expressed Genes
2.5. Quantitative Real-Time PCR
2.6. Determination of the Phlorizin Content and Phenylalanine Ammonia Lyase Activity
3. Results
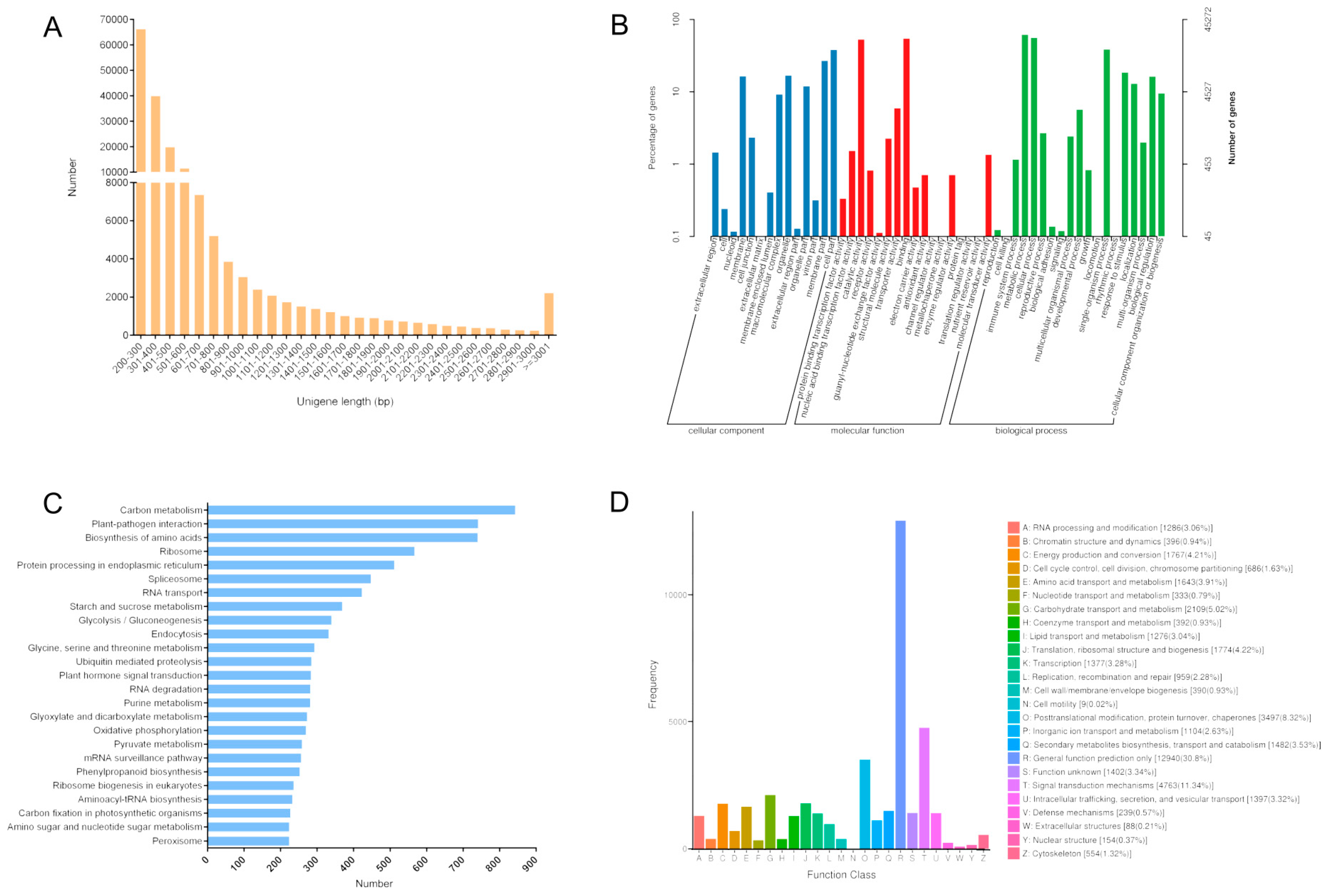
3.1. Transcriptome Sequencing and Sequence Assembly
3.2. Annotation and Classification of Transcripts
3.3. Analysis of Transcript Abundance in L. polystachyus Plants Exposed to Various Light Conditions
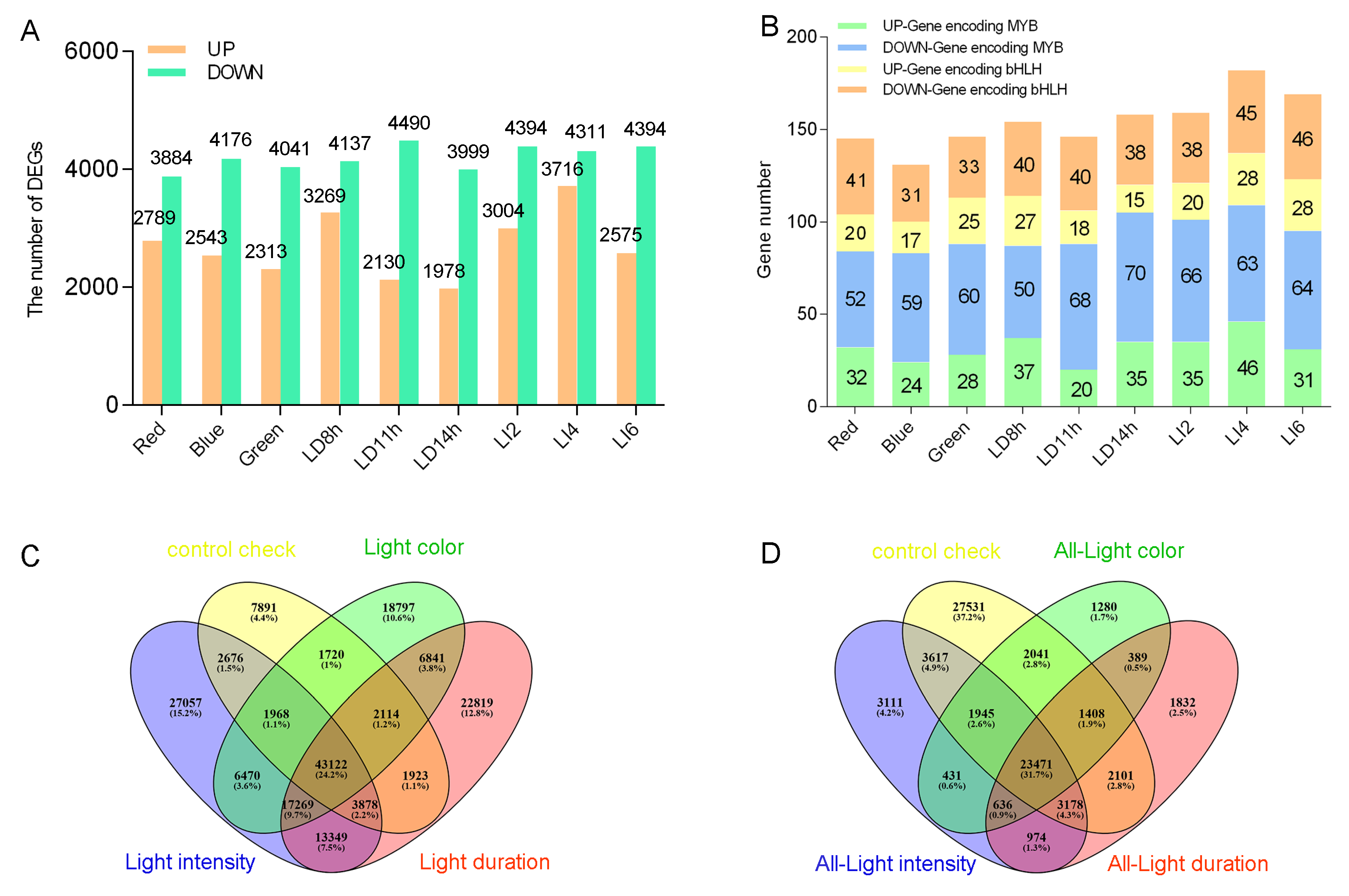
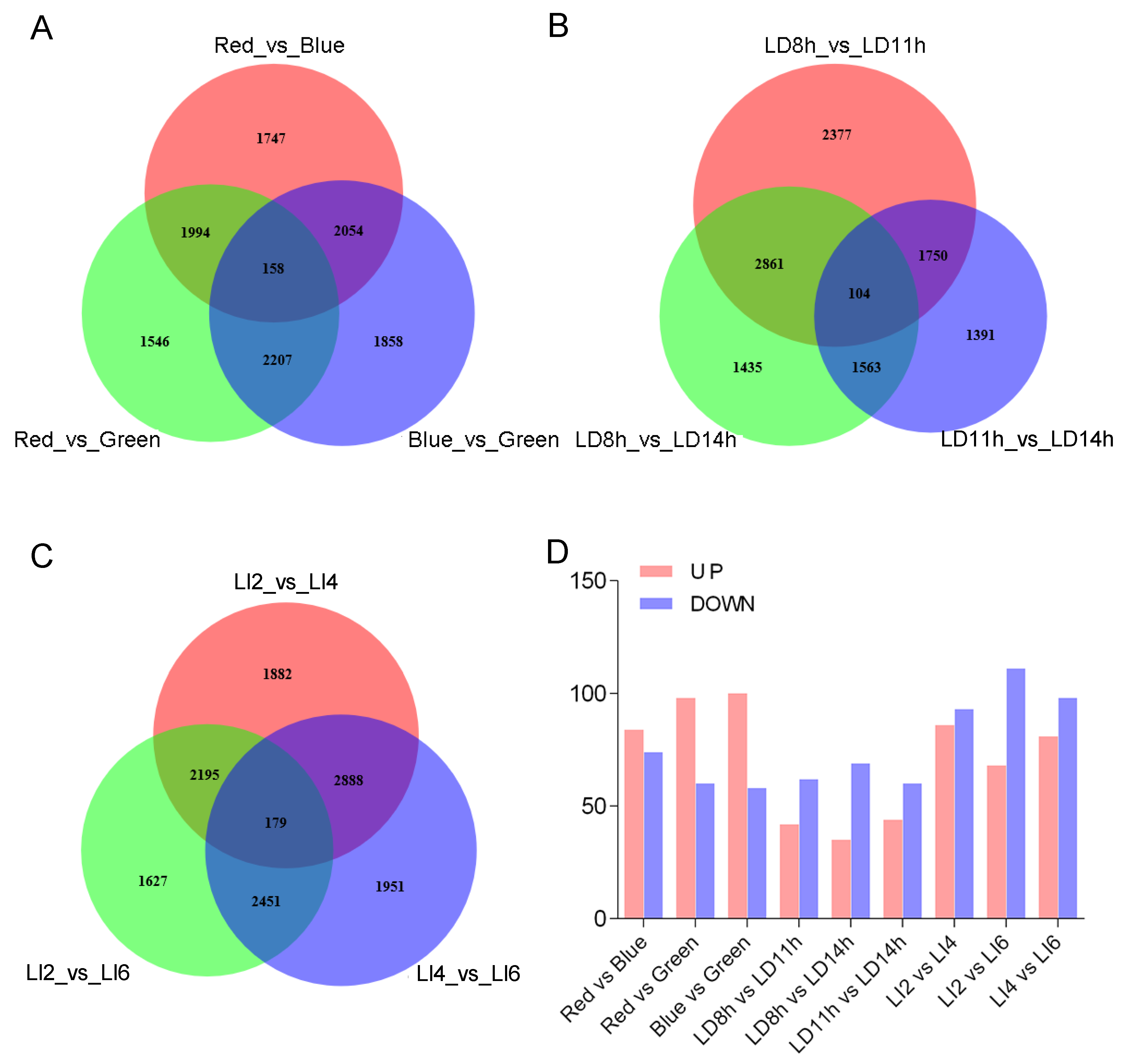
3.4. Differences in the Transcript Abundance of L. polystachyus Plants Exposed to Various Light Conditions
3.5. Identification of Differentially Expressed Genes Associated with Light-Stress-Responsive Transcription Factors
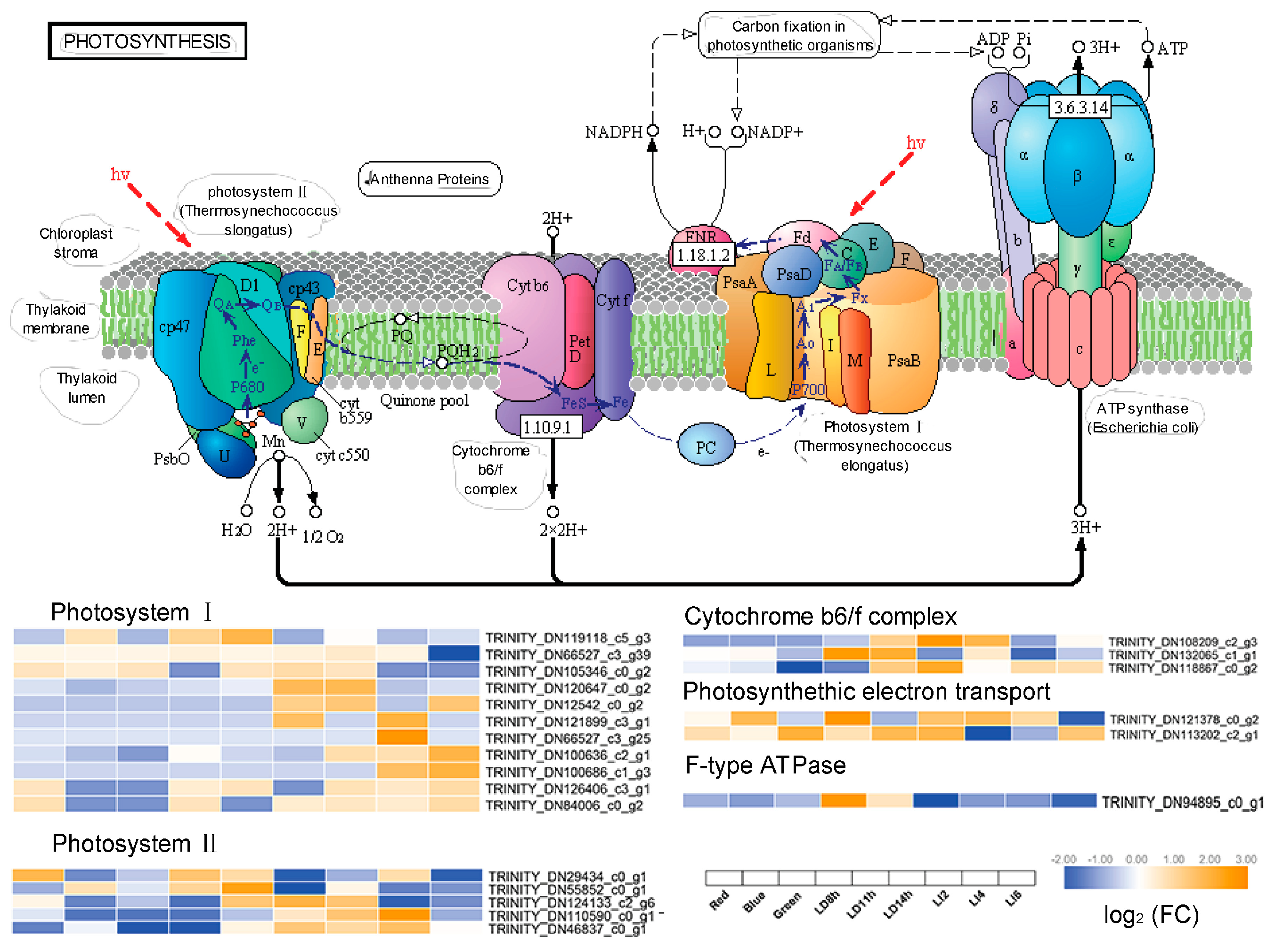
3.6. Expression of Genes Involved in Secondary Metabolite Synthesis and Photosynthesis
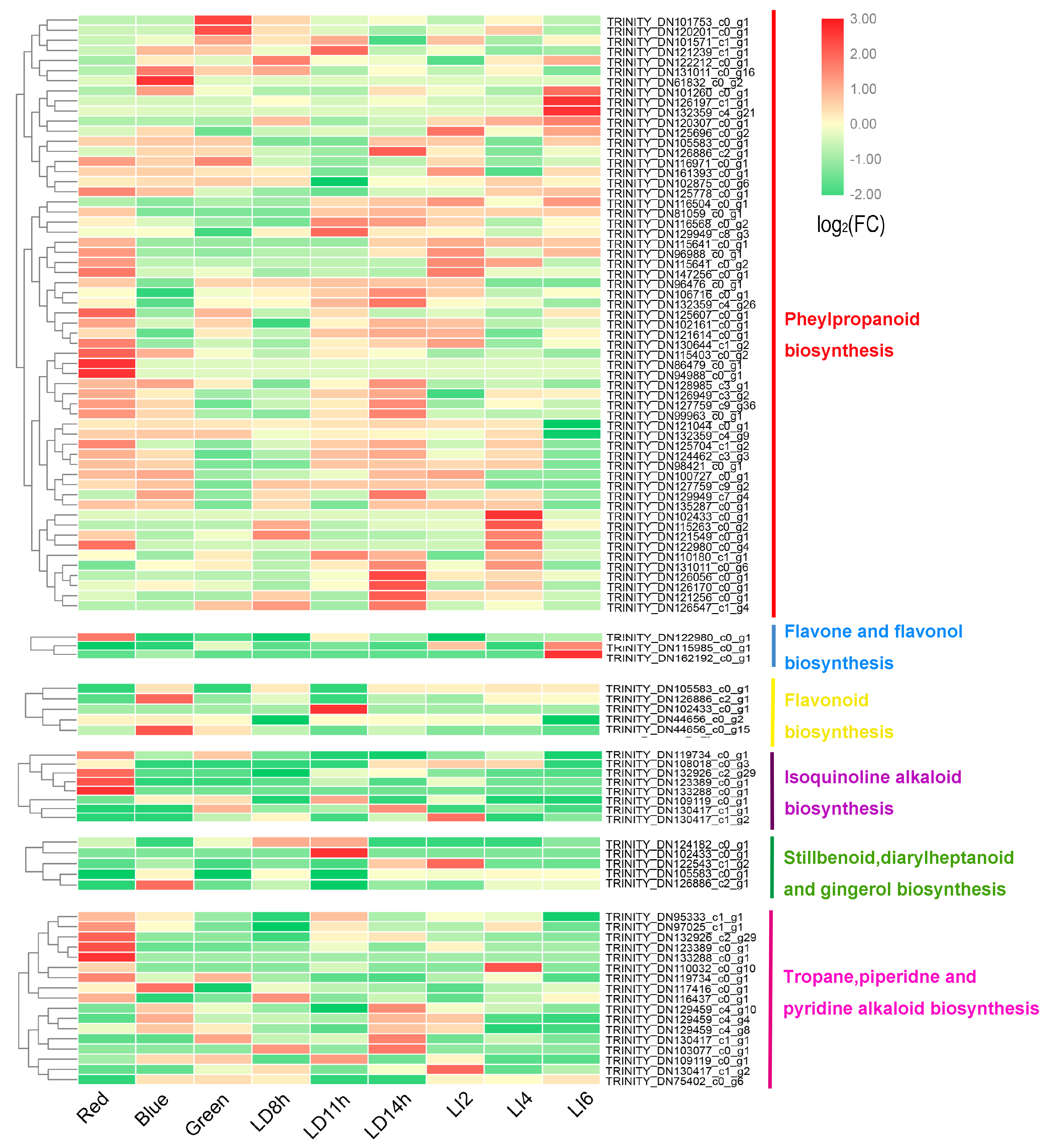
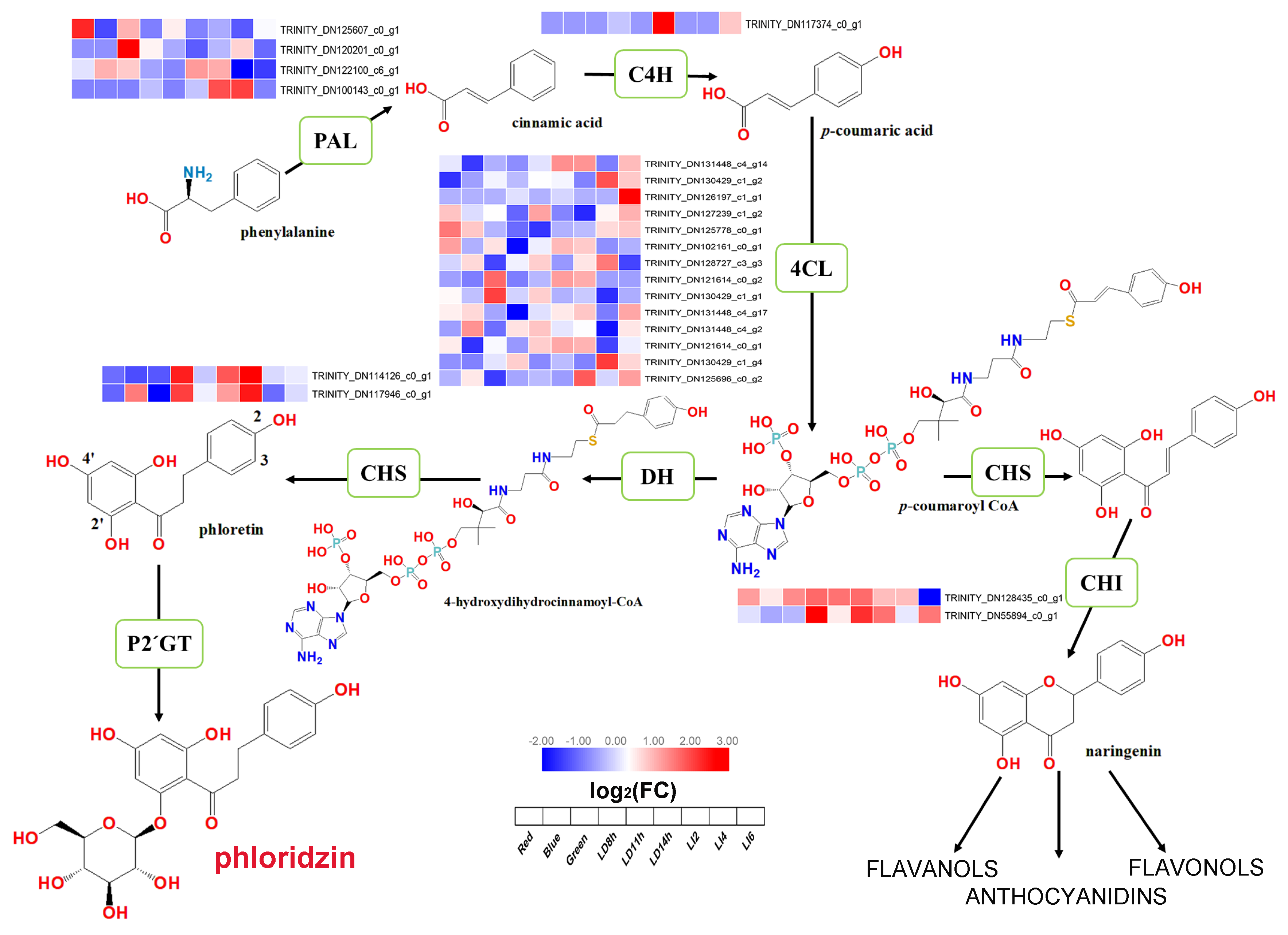
3.7. Changes in the Expression of Phlorizin Biosynthesis-Related Genes in L. polystachyus under Various Light Conditions
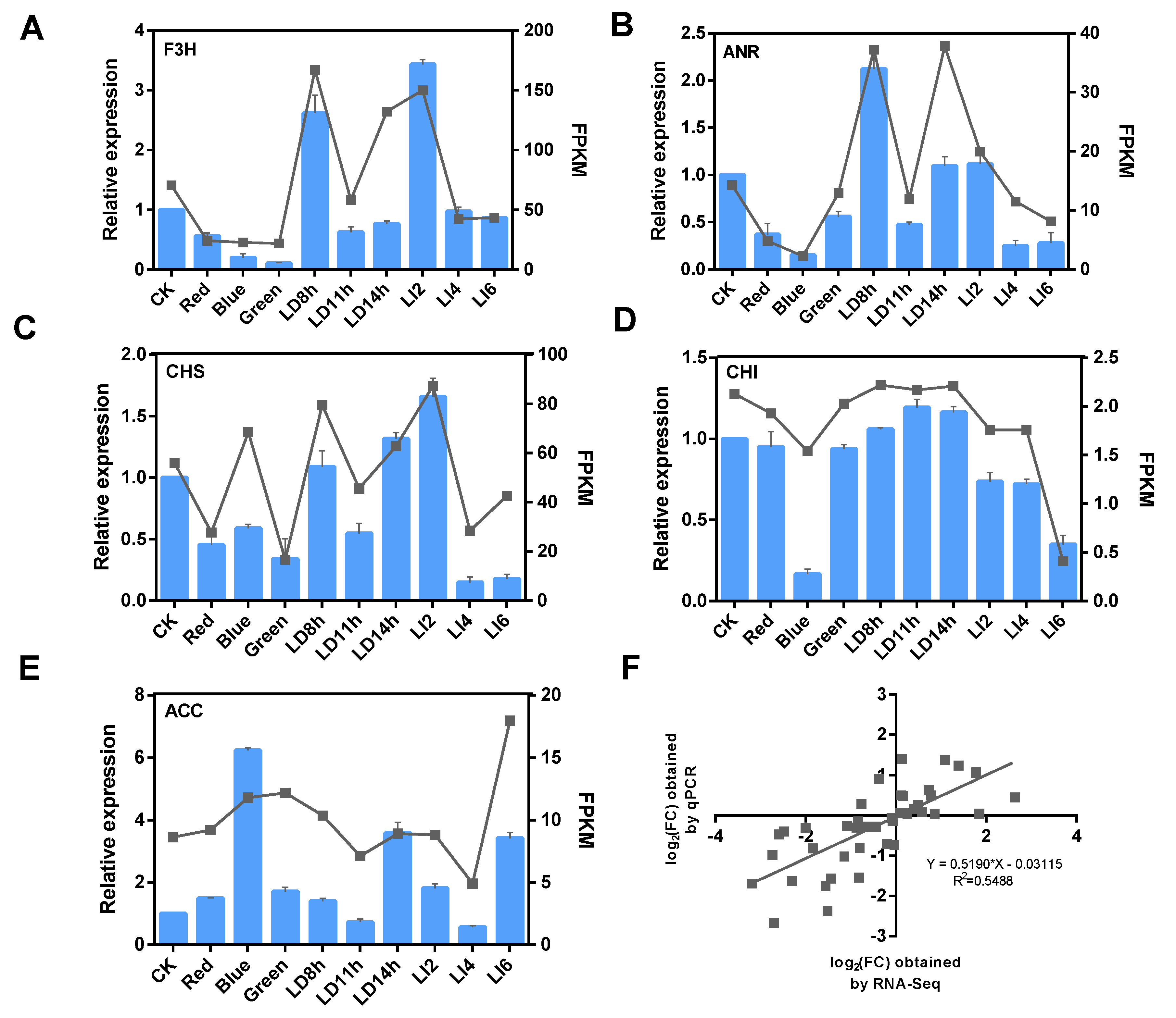
3.8. Verification of Gene Expression Levels in a qRT-PCR Assay
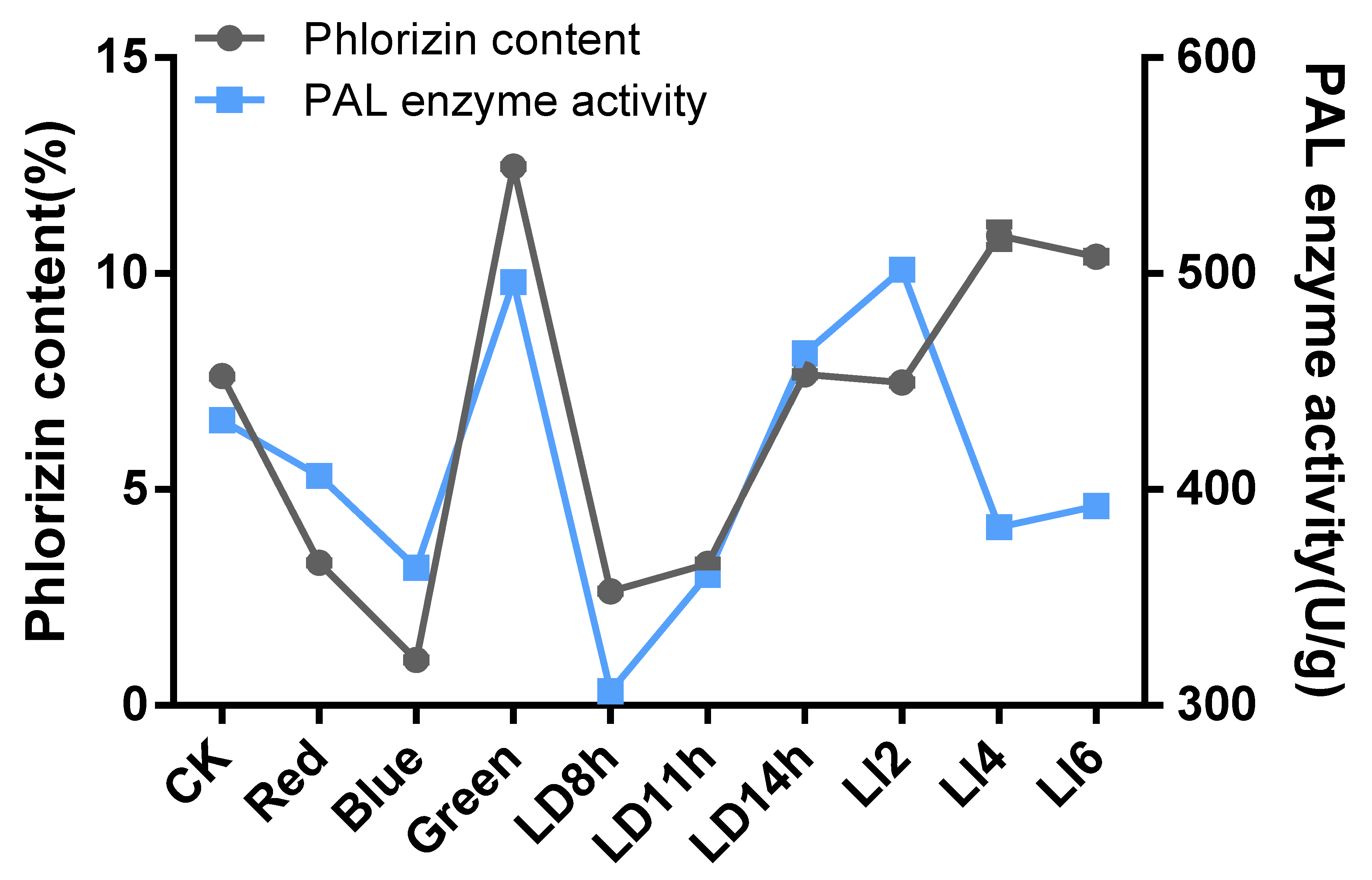
3.9. Changes to the Phlorizin Content and Phenylalanine Ammonia Lyase Activity
4. Discussion
4.1. Transcription Factors as Potential Regulators of Flavonoid Accumulation
4.2. Effects of Different Light Treatments on the Enzymes of the Secondary Metabolism Pathways and Photosystems of L. polystachyus
4.3. Molecular Characteristics of the Enrichment of the Phlorizin Biosynthesis Pathway in L. polystachyus Leaves under Different Light Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, J.F.; Huang, Y.M.; Li, K.X.; Chen, Y.Y.; Vanegas, D.; McLamore, E.S.; Shen, Y.B. Leaf Extract from Lithocarpus polystachyus Rehd. Promote Glycogen Synthesis in T2DM Mice. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Mariadoss, A.V.A.; Vinyagam, R.; Rajamanickam, V.; Senthilkumar, V.; Venkatesan, S.; David, E. Pharmacological aspects and potential use of phloretin: A systemic review. Mini-Rev. Med. Chem. 2019, 19, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Gosch, C.; Halbwirth, H.; Kuhn, J.; Miosic, S.; Stich, K. Biosynthesis of phloridzin in apple (Malus domestica Borkh.). Plant Sci. 2009, 176, 223–231. [Google Scholar] [CrossRef]
- Batista, D.S.; Felipe, S.H.S.T.; Silva, D.; Castro, K.M.; Mamedes-Rodrigues, T.C.; Miranda, N.A.; Ríos-Ríos, A.M.; Fortini, E.A.; Chagas, K.; Torres-Silva, G.; et al. Light quality in plant tissue culture: Does it matter? Vitr. Cell. Dev. Biol. 2018, 54, 195–215. [Google Scholar] [CrossRef]
- Plantenga, F.D.; Bergonzi, S.; Bachem, C.W.; Visser, R.G.; Heuvelink, E.; Marcelis, L.F. High light accelerates potato flowering independently of the FT-like flowering signal StSP3D. Environ. Exp. Bot. 2019, 160, 35–44. [Google Scholar] [CrossRef]
- Xu, J.M.; Yan, Z.M.; Xu, Z.G.; Wang, Y.H.; Xie, Z.Q. Transcriptome analysis and physiological responses of the potato plantlets in vitro under red, blue, and white light conditions. 3 Biotechnol. 2018, 8, 394. [Google Scholar] [CrossRef]
- Li, A.M.; Li, S.H.; Wu, X.J.; Zhang, J.; He, A.N.; Zhao, G.; Yang, X. Effect of Light Intensity on Leaf Photosynthetic Characteristics and Accumulation of Flavonoids in Lithocarpus litseifolius (Hance) Chun. (Fagaceae). Open J. For. 2016, 6, 445–459. [Google Scholar] [CrossRef]
- Ni, J.; Dong, L.X.; Jiang, Z.F.; Yang, X.L.; Chen, Z.Y.; Wu, Y.H.; Xu, M.J. Comprehensive transcriptome analysis and flavonoid profiling of Ginkgo leaves reveals flavonoid content alterations in day-night cycles. PLoS ONE 2018, 13, 0193897. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.N.; Liu, J.H. Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf) in response to cold stress. BMC Genom. 2015, 16, 555. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- Chen, Y.; Yin, L.Z.; Zhao, L.; Shu, G.; Yuan, Z.X.; Fu, H.L.; Lv, C.; Lin, J.C. Optimization of the ultrasound-assisted extraction of antioxidant phloridzin from Lithocarpus polystachyus Rehd. using response surface methodology. J. Sep. Sci. 2017, 40, 4329–4337. [Google Scholar] [CrossRef]
- Chen, S.Y.; Zhang, X.Q.; Ma, X.; Huang, L.K. Assessment of genetic diversity and differentiation of Elymus nutans indigenous to Qinghai-Tibet plateau using simple sequence repeats markers. Can. J. Plant Sci. 2013, 93, 1089–1096. [Google Scholar] [CrossRef]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef]
- Neugart, S.; Krumbein, A.; Zrenner, R. Influence of light and temperature on gene expression leading to accumulation of specific flavonol glycosides and hydroxycinnamic acid derivatives in Kale (Brassica oleracea var. sabellica). Front. Plant Sci. 2016, 7, 326. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, S.Z.; Yang, W.X.; Shang, X.L.; Fu, X.X. Light quality affects flavonoid production and related gene expression in Cyclocarya paliurus. J. Photochem. Photobiol. B 2018, 179, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.J.; Gao, S.; Yang, Y.J.; Huang, M.Y.; Cheng, L.N.; Wei, Q.; Fei, Z.J.; Gao, J.P.; Hong, B. Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genom. 2013, 14, 662. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wei, W.; Pang, X.; Wang, X.; Zhang, H.; Dong, B.; Xing, Y.; Li, X.; Wang, M. Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BMC Genom. 2014, 15, 671. [Google Scholar] [CrossRef] [PubMed]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. BBA-Gene Regul. Mech. 2012, 1819, 137–148. [Google Scholar] [CrossRef]
- Boyko, A.; Kovalchuk, I. Epigenetic control of plant stress response. Environ. Mol. Mutagen 2018, 49, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Sazegari, S.; Niazi, A. Isolation and molecular characterization of wheat (Triticum aestivum) dehydration responsive element binding factor (DREB) isoforms. Aust. J. Crop Sci. 2012, 6, 1037–1044. [Google Scholar]
- Lata, C.; Yadav, A.; Prasad, M. Role of plant transcription factors in abiotic stress tolerance. In Abiotic Stress Response in Plants—Physiological, Biochemical and Genetic Perspectives; Shanker, A., Venkateshwarulu, B., Eds.; InTechOpen: London, UK, 2011. [Google Scholar]
- Liu, J.H.; Peng, T.; Dai, W.S. Critical cis-acting elements and interacting transcription factors: Key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 2014, 32, 303–317. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Misra, P.; Trivedi, P.K. Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 2015, 34, 1515–1528. [Google Scholar] [CrossRef]
- Huang, W.; Khaidun, A.B.M.; Chen, J.; Zhang, C.; Lv, H.; Yuan, L.; Wang, Y. A R2R3-MYB Transcription Factor Regulates the Flavonol Biosynthetic Pathway in a Traditional Chinese Medicinal Plant, Epimedium sagittatum. Front Plant Sci. 2016, 7, 1089. [Google Scholar] [CrossRef]
- Chen, Z.X.; Liu, G.H.; Tang, N.; Li, Z.G. Transcriptome Analysis Reveals Molecular Signatures of Luteoloside Accumulation in Senescing Leaves of Lonicera macranthoides. Int. J. Mol. Sci. 2018, 19, 1012. [Google Scholar] [CrossRef] [PubMed]
- Adato, A.; Mandel, T.; Mintz-Oron, S.; Venger, I.; Levy, D.; Yativ, M.; Dominguez, E.; Wang, Z.; De Vos, R.C.H.; Jetter, R.; et al. Fruit-Surface Flavonoid Accumulation in Tomato is Controlled by a SIMYB12-Regulated Transcriptional Network. PLoS Genet. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.J.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thevenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Chiu, L.W.; Zhou, X.J.; Burke, S.; Wu, X.L.; Prior, R.L.; Li, L. The Purple Cauliflower Arises from Activation of a MYB Transcription Factor. Plant Physiol. 2010, 154, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.C.; Chen, C.H.; Chen, S.H.; Lu, I.H.; Chu, M.J.; Huang, L.C.; Lin, C.Y.; Chen, C.Y.; Lo, H.F.; Jeng, S.T.; et al. The effect of red light and far-red light conditions on secondary metabolism in agarwood. BMC Plant Biol. 2015, 15, 139. [Google Scholar] [CrossRef]
- Fu, B.; Ji, X.M.; Zhao, M.Q.; He, F.; Wang, X.L.; Wang, Y.D.; Liu, P.F.; Niu, L. The influence of light quality on the accumulation of flavonoids in tobacco (Nicotiana tabacum L.) leaves. J. Photochem. Photobiol. B 2016, 162, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Ma, J.; Zhang, L.; Chen, F.; Sang, Z.; Jia, Z.; Ma, L. De novo transcriptome sequencing and gene expression profiling of Magnolia wufengensis in response to cold stress. BMC Plant Biol. 2019, 19, 321. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Wang, X.W.; Ban, Q.Y.; Zhu, X.X.; Jiang, C.J.; Wei, C.L.; Bennetzen, J.L. Comparative transcriptomic analysis reveals gene expression associated with cold adaptation in the tea plant Camellia sinensis. BMC Genom. 2019, 20, 624. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Leaves in the dark see the light. Science 1999, 284, 599–601. [Google Scholar] [CrossRef]
- Powlem, S.B. Photoinhibition of photosyntheain induced by viaible light. Annu. Rev. Plant Physiol. 1984, 35, 15–50. [Google Scholar] [CrossRef]
- Ouzounis, T.; Razi Parjikolaei, B.; Fretté, X.; Rosenqvist, E.; Ottosen, C.O. Predawn and high intensity application of supplemental blue light decreases the quantum yield of PSII and enhances the amount of phenolic acids, flavonoids, and pigments in Lactuca sativa. Front. Plant Sci. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Arcas, M.C.; Botia, J.M.; Ortuño, A.M.; Del Río, J.A. UV irradiation alters the levels of flavonoids involved in the defence mechanism of Citrus aurantium fruits against Peniillium digitatum. Eur. J. Plant Pathol. 2000, 106, 617. [Google Scholar] [CrossRef]
- Oyaert, E.; Volckert, E.; Debergh, P.C. Growth of Chrysanthemum under coloured plastic films with different light qualities and quantities. Sci. Hortic. 1999, 79, 195–205. [Google Scholar] [CrossRef]
- Zoratti, L.; Karppinen, K.; Luengo Escobar, A.; Häggman, H.; Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 2014, 5, 534. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.X.; Li, Y.Q.; Bai, M.; He, H.J.; Liang, G.X.; Wu, H. Correlation between the dynamic accumulation of the main effective components and their associated regulatory enzyme activities at different growth stages in Lonicera japonica Thunb. Ind. Crops Prod. 2017, 96, 16–22. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, Z.Y.; Jiang, C.; Wang, X.M.; Huang, L.Q. Exploiting genes and functional diversity of chlorogenic acid and luteolin biosyntheses in Lonicera japonica and their substitutes. Gene 2014, 534, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.Z.; Shen, G.A.; Wu, W.S.; Liu, X.F.; Lin, J.; Tan, F.; Snu, X.F.; Tang, K.X. Characterization and expression of chalcone synthase gene from Ginkgo biloba. Plant Sci. 2005, 168, 1525–1531. [Google Scholar] [CrossRef]

| Sample Number | Light Quality | Light Control Duration (Light/Dark) | Light Intensity (μmol·m−2·s−1/lx) | Temperature (℃) | Soil Moisture (%) | Number of Plants Treated | Processing Type |
|---|---|---|---|---|---|---|---|
| CK | Natural light | 12/12 h | 18–72/1000–4000 | 25 ± 2 | 75 | 10 | Control group |
| Red | Red | 12/12 h | 37.5/3000 | 25 ± 2 | 75 | 10 | Light quality |
| Blue | Blue | ||||||
| Green | Green | ||||||
| LD8h | White | 8/16 h | 37.5/3000 | 25 ± 2 | 75 | 10 | photoperiod |
| LD11h | 11/13 h | ||||||
| LD14h | 14/10 h | ||||||
| LI2 | White | 12/12 h | 12.5/1000 | 25 ± 2 | 75 | 10 | Light intensity |
| LI4 | 25/2000 | ||||||
| LI6 | 37.5/3000 |
| Genes | Primer Name | Primer Sequence (5’–3’) | Fragment Size/bp |
|---|---|---|---|
| acetyl coenzyme A carboxylase (ACC) | DACCrt1S2 | AAGGCAAGCACTTCATCTCATCC | 200 |
| DACCrt1X2 | ATAAGAATCTCGGCTATTGTTCC | ||
| chalcone synthase (CHS) | DCHSrt2S1 | GCAGCAACAAAGGCAATCAAA | 186 |
| DCHSrt2X1 | CACAGTCCCACCAGCAAAGCA | ||
| chalcone isomerase (CHI) | DCHIrt2S1 | TTTATCAGGATCTTCTCAGGGAG | 144 |
| DCHIrt2X1 | AATCAGTGTCTGGGTTTGTCTTC | ||
| anthocyanidin reductase (ANR) | DANRrt1S2 | CACTGGGTTCGTCGCTTCTTT | 276 |
| DANRrt1X2 | CCTTGGATCGCTGGCTTTATC | ||
| flavanone 3-hydroxylase (F3H) | DF3Hrt1S1 | TATTTCGCATCATATTGCCACTC | 229 |
| DF3Hrt1X1 | CAACTTTAGGACGCTCATCTTCG | ||
| glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | DGrtS1 | CCTTCCGTGTTCCTACCCCCAA | 133 |
| DGrtX1 | TAGCCCAAGATGCCCTTCAGT |
| Sample | Total Claen Reads | Total Claen Bases (bp) | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| CK | 20,840,776 | 6,252,232,800 | 45.16% | 98.04% | 94.73% |
| Red | 20,510,984 | 6,153,295,200 | 45.01% | 97.96% | 94.56% |
| Blue | 20,951,216 | 6,285,364,800 | 45.26% | 98.03% | 94.74% |
| Green | 20,922,975 | 6,276,892,500 | 45.34% | 97.97% | 94.59% |
| LD8h | 20,725,909 | 6,217,772,700 | 45.40% | 98.11% | 94.90% |
| LD11h | 20,966,197 | 6,289,859,100 | 45.26% | 97.90% | 94.39% |
| LD14h | 20,793,406 | 6,238,021,800 | 45.29% | 97.98% | 94.60% |
| LI2 | 20,815,319 | 6,244,595,700 | 45.37% | 97.95% | 94.53% |
| LI4 | 20,753,400 | 6,226,020,000 | 46.00% | 97.96% | 94.54% |
| LI6 | 24,266,308 | 7,279,892,400 | 44.34% | 97.67% | 93.47% |
| Type | Unigenes | Transcripts |
|---|---|---|
| Total_sequence number | 177,894 | 206,515 |
| Total_sequence length | 99,790,703 | 124,260,610 |
| Maximum length (bp) | 15,707 | 15,707 |
| Minimum length (bp) | 201 | 201 |
| Average length (bp) | 560 | 601 |
| GC (%) | 39.19% | 39.33% |
| N40 (bp) | 981 | 1111 |
| N50 (bp) | 689 | 786 |
| N60 (bp) | 509 | 572 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Lin, L.; Long, Y.; Guo, H.; Wang, Z.; Cui, M.; Huang, J.; Xing, Z. Comprehensive Transcriptome Analysis Revealed the Effects of the Light Quality, Light Intensity, and Photoperiod on Phlorizin Accumulation in Lithocarpus polystachyus Rehd. Forests 2019, 10, 995. https://doi.org/10.3390/f10110995
Zhang Y, Lin L, Long Y, Guo H, Wang Z, Cui M, Huang J, Xing Z. Comprehensive Transcriptome Analysis Revealed the Effects of the Light Quality, Light Intensity, and Photoperiod on Phlorizin Accumulation in Lithocarpus polystachyus Rehd. Forests. 2019; 10(11):995. https://doi.org/10.3390/f10110995
Chicago/Turabian StyleZhang, Yantong, Limei Lin, Yuehong Long, Hongyu Guo, Zhuo Wang, Minghui Cui, Jian Huang, and Zhaobin Xing. 2019. "Comprehensive Transcriptome Analysis Revealed the Effects of the Light Quality, Light Intensity, and Photoperiod on Phlorizin Accumulation in Lithocarpus polystachyus Rehd." Forests 10, no. 11: 995. https://doi.org/10.3390/f10110995
APA StyleZhang, Y., Lin, L., Long, Y., Guo, H., Wang, Z., Cui, M., Huang, J., & Xing, Z. (2019). Comprehensive Transcriptome Analysis Revealed the Effects of the Light Quality, Light Intensity, and Photoperiod on Phlorizin Accumulation in Lithocarpus polystachyus Rehd. Forests, 10(11), 995. https://doi.org/10.3390/f10110995




