Phylogeographical Structure of Liquidambar formosana Hance Revealed by Chloroplast Phylogeography and Species Distribution Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. Chloroplast DNA Amplification, Sequencing, and Sequence Alignment
2.3. Genetic Diversity, Haplotype Network Construction, and Haplotype Distribution
2.4. Phylogenetic Analyses and Divergence Time Estimation
2.5. Population Genetic Structure
2.6. Tests of Expansion
2.7. Ecological Niche Modeling
3. Results
3.1. Chloroplast Haplotype Variation, Haplotype Network, and Distribution
3.2. Genetic Diversity and Genetic Structure
3.3. Population History Dynamic Analysis and Divergence Time Estimation
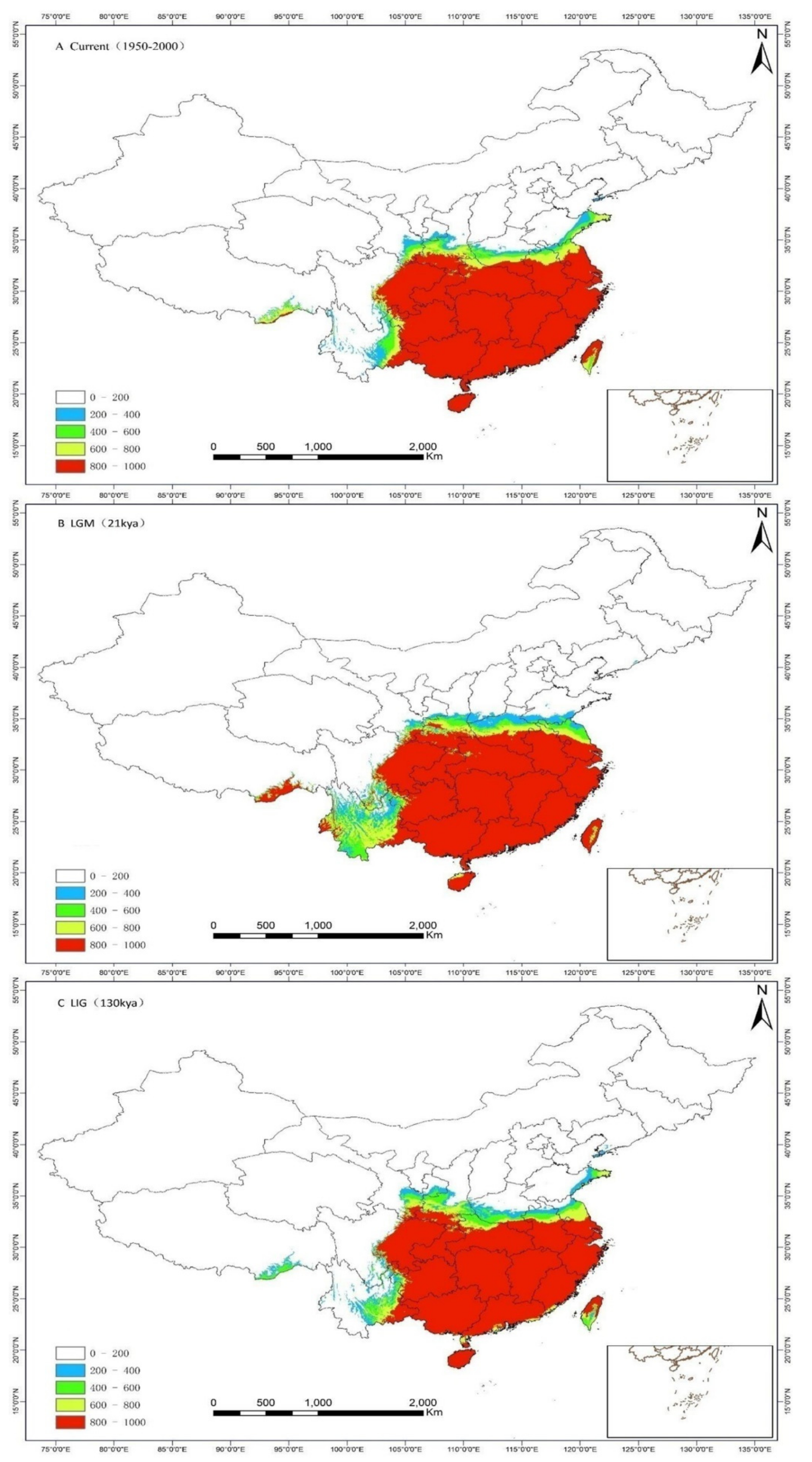
3.4. Ecological Niche Model
4. Discussion
4.1. Genetic Diversity and Genetic Differentiation of cpDNA Haplotype in L. Formosana
4.2. Phylogeographic Structure and Inference of Demographic History
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.T.; Sun, B.; Zhang, Z.S.; Peng, S.Z.; Xiao, G.Q.; Ge, J.Y.; Hao, Q.Z.; Qiao, Y.S.; Liang, M.Y.; Liu, J.F.; et al. A major reorganization of Asian climate by the early Miocene. Clim. Past 2008, 4, 153–174. [Google Scholar] [CrossRef]
- Clift, P.; Lee, J.I.; Clark, M.K.; Blusztajn, J. Erosional response of South China to arc rifting and monsoonal strengthening; a record from the South China Sea. Mar. Geol. 2002, 184, 207–226. [Google Scholar] [CrossRef]
- Guo, Z.T.; Ruddiman, W.F.; Hao, Q.Z.; Wu, H.B.; Qiao, Y.S.; Zhu, R.X.; Peng, S.Z.; Wei, J.J.; Yuan, B.Y.; Liu, T.S. Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 2002, 416, 159. [Google Scholar] [CrossRef]
- Klootwijk, C.T.; Gee, J.S.; Peirce, J.W.; Smith, G.M.; Mcfadden, P.L. An early India-Asia contact: Paleomagnetic constraints from Ninetyeast Ridge, ODP Leg 121. Geology 1992, 20, 395. [Google Scholar] [CrossRef]
- Wan, S.; Li, A.; Clift, P.D.; Stuut, J.B.W. Development of the East Asian monsoon: Mineralogical and sedimentologic records in the northern South China Sea since 20Ma. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2007, 254, 561–582. [Google Scholar] [CrossRef]
- An, Z.S.; Kutzbach, J.E.; Prell, W.L.; Porter, S.C. Evolution of Asian monsoons and phased uplift of Himalaya-Tibetan plateau since Late Miocene times. Nature 2001, 411, 62–66. [Google Scholar]
- Chou, Y.W.; Wang, C.N. Refugia and phylogeography of Taiwania in East Asia. J. Biogeogr. 2011, 38, 1992–2005. [Google Scholar] [CrossRef]
- Kou, Y.; Cheng, S.; Shuang, T.; Bo, L.; Zhang, Z. The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late Early Miocene. J. Biogeogr. 2016, 43, 351–360. [Google Scholar] [CrossRef]
- Wu, Z.Y. Vegetation of China; Science Press: Beijing, China, 1980. [Google Scholar]
- Tian, S.; Lei, S.Q.; Hu, W.; Deng, L.L.; Li, B.; Meng, Q.L.; Soltis, D.E.; Soltis, P.S.; Fan, D.M.; Zhang, Z.Y. Repeated range expansions and inter-/postglacial recolonization routes of Sargentodoxa cuneata (Oliv.) Rehd. et Wils. (Lardizabalaceae) in subtropical China revealed by chloroplast phylogeography. Mol. Phylogenet. Evol. 2015, 85, 238–246. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Xu, H.; Yang, L.; Liu, G.; Sheng, Y.; Zheng, C.; Zheng, W.; Cheng, T.; Shi, J. The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng. Front. Plant Sci. 2015, 6, 447. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.S.; Chen, C.; Comes, H.P.; Sakaguchi, S.; Liu, Y.H.; Tanaka, N.; Sakio, H.; Qiu, Y.X. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol. 2012, 196, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Kuprianova, L.A. Palynological data contributing to the history of Liquidambar. Pollen Spores 1960, 2, 71–88. [Google Scholar]
- The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants AGP II. Bot. J. Linn. Soc. 2003, 141, 399–436. [Google Scholar] [CrossRef]
- Ozturk, M.; Celik, A.; Guvensen, A.; Hamzaoğlu, E. Ecology of tertiary relict endemic Liquidambar orientalis Mill. forests. For. Ecol. Manag. 2008, 256, 510–518. [Google Scholar] [CrossRef]
- Ickert-Bond, S.M.; Pigg, K.B.; Wen, J. Comparative infructescence morphology in Liquidambar (Altingiaceae) and its evolutionary significance. Am. J. Bot. 2005, 92, 1234–1255. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Lin, F.; Huang, P.; Zheng, Y. Moderate Genetic Diversity and Genetic Differentiation in the Relict Tree Liquidambar formosana Hance Revealed by Genic Simple Sequence Repeat Markers. Front. Plant Sci. 2016, 7, 1411. [Google Scholar] [CrossRef]
- Wu, W. Natural Hybridization, Phylogeography and Speciation Patterns of Altingiaceae. Ph.D. Thesis, Sun Yat-sen University, Guangzhou, China, 2009. [Google Scholar]
- Bi, X.Q.; Jin, X.Z.; Li, H.J. Genetic diversity in the natural populations of Liquidambar formosana revealed by ISSR molecular markers. Bull. Bot. Res. 2010, 30, 120–125. [Google Scholar]
- Liu, L.X.; Li, R.; Worth, J.R.P.; Li, X.; Li, P.; Cameron, K.M.; Fu, C.X. The Complete Chloroplast Genome of Chinese Bayberry (Morella rubra, Myricaceae): Implications for Understanding the Evolution of Fagales. Front. Plant Sci. 2017, 8, 968. [Google Scholar] [CrossRef]
- Wen, Z.; Zhe, X.; Zhang, H.; Ying, F. Chloroplast phylogeography of a desert shrub, Calligonum calliphysa (Calligonum, Polygonaceae), in arid Northwest China. Biochem. Syst. Ecol. 2015, 60, 56–62. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Caicedo, A.L.; Schaal, B.A. Population structure and phylogeography of Solanum pimpinellifolium inferred from a nuclear gene. Mol. Ecol. 2004, 13, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Pons, O.; Petit, R.J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 1996, 144, 1237–1245. [Google Scholar]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0 Beta; Sinauer Associates: Sun-derland, MA, USA, 2002. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1. 4.0. A Graphical Viewer of Phylogenetic Trees; Institute of Evolutionary Biology University of Edinburgh: Edinburgh, UK, 2012. [Google Scholar]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.B.; Ickert-Bond, S.M.; Brunson, D.B.; Soltis, D.E.; Soltis, P.S. Phylogeographical structure and temporal complexity in American sweetgum (Liquidambar styraciflua; Altingiaceae). Mol. Ecol. 2008, 17, 3889–3900. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. 2014. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 31 January 2017).
- Excoffier, L.; Lischer, H. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, M.M.; Wilder, J.A.; Mendez, F.L.; Cox, M.P.; Woerner, A.; Angui, T.; Kingan, S.; Mobasher, Z.; Batini, C.; Destro-Bisol, G.; et al. Contrasting signatures of population growth for mitochondrial DNA and Y chromosomes among human populations in Africa. Mol. Biol. Evol. 2008, 25, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.T.; Nakazawa, Y. Environmental data sets matter in ecological niche modeling: An example with. Glob. Ecol. Biogeogr. 2010, 17, 135–144. [Google Scholar]
- Petit, R.J.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Vendramin, G.G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chiang, Y.C.; Schaal, B.A.; Chou, C.H.; Chiang, T.Y. Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Mol. Ecol. 2001, 10, 2669–2681. [Google Scholar] [CrossRef]
- Su, Y.; Wang, T.B.; Jiang, Y.; Chen, G.; Ouyang, P.; Sun, Y. Genetic differentiation of relictual populations of Alsophila spinulosa in southern China inferred from cpDNA trnL-F noncoding sequences. Mol. Phylogenet. Evol. 2005, 34, 323–333. [Google Scholar] [CrossRef]
- Yuan, Q.J.; Zhang, Z.Y.; Peng, H.; Ge, S. Chloroplast phylogeography of Dipentodon (Dipentodontaceae) in southwest China and northern Vietnam. Mol. Ecol. 2008, 17, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Moore, M.J.; Yue, L.; Feng, T.; Chu, H.; Chen, S.; Ji, Y.; Wang, H.; Li, J. Chloroplast phylogeography of the East Asian Arcto-Tertiary relict Tetracentron sinense (Trochodendraceae). J. Biogeogr. 2014, 41, 1721–1732. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Effects of Life History Traits on Genetic Diversity in Plant Species. Philos. Trans. Biol. Sci. 1996, 351, 1291–1298. [Google Scholar]
- Petit, R.J.; Itziar, A.; Jacques-Louis, D.B.; Christiane, B.; Simon, B.; Rachid, C.; Richard, E.; Silvia, F.; Delphine, G.; Martin, L. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science 2003, 300, 1563–1565. [Google Scholar] [CrossRef] [PubMed]
- Nuttle, T.; Haefner, J.W. Seed dispersal in heterogeneous environments: Bridging the gap between mechanistic dispersal and forest dynamics models. Am. Nat. 2005, 165, 336–349. [Google Scholar] [CrossRef] [PubMed]
- Ozdilek, A.; Cengel, B.; Kandemir, G.; Tayanc, Y.; Velioglu, E.; Kaya, Z. Molecular phylogeny of relict-endemic Liquidambar orientalis Mill based on sequence diversity of the chloroplast-encoded mat K gene. Plant Syst. Evol. 2012, 298, 337–349. [Google Scholar] [CrossRef]
- Koizumi, I.; Yamamoto, S.; Maekawa, K. Decomposed pairwise regression analysis of genetic and geographic distances reveals a metapopulation structure of stream-dwelling Dolly Varden charr. Mol. Ecol. 2006, 15, 3175–3189. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.K. A survey of the Liquidambaroideae (Hamamelidaceae) with a view to elucidating its fossil record. Evol. Syst. Foss. Hist. Hamamelidae 1989, 1, 249–272. [Google Scholar]
- Sun, X. Palynofloristical inveatgation on the late cretaceous and paleocene of China. Acta Phytotaxon. Sin. 1979, 17, 8–23. [Google Scholar]
- Avise, J.C. Phylogeography: Retrospect and prospect. J. Biogeogr. 2010, 36, 3–15. [Google Scholar] [CrossRef]
- Royden, L.H.; Burchfiel, B.C.; Van Der Hilst, R.D. The geological evolution of the Tibetan Plateau. Science 2008, 321, 1054–1058. [Google Scholar] [CrossRef]
- An, Z.; Huang, Y.; Liu, W.; Guo, Z.; Clemens, S.; Li, L.; Prell, W.; Ning, Y.; Cai, Y.; Zhou, W. Multiple expansions of C4 plant biomass in East Asia since 7 Ma coupled with strengthened monsoon circulation. Geology 2005, 33, 705–708. [Google Scholar]
- Liu, L.; Chen, W.; Yan, D.T.; Li, J.; Liu, L.; Wang, Y.L. Molecular phylogeography and paleodistribution modeling of the boreal tree species Ulmus lamellosa (T. Wang et S.L. Chang) (Ulmaceae) in China. Tree Genet. Genomes 2017, 13, 11. [Google Scholar] [CrossRef]
- Xu, J.; Deng, M.; Jiang, X.L.; Westwood, M.; Song, Y.G.; Turkington, R. Phylogeography of Quercus glauca (Fagaceae), a dominant tree of East Asian subtropical evergreen forests, based on three chloroplast DNA interspace sequences. Tree Genet. Genomes 2015, 11, 1–17. [Google Scholar] [CrossRef]
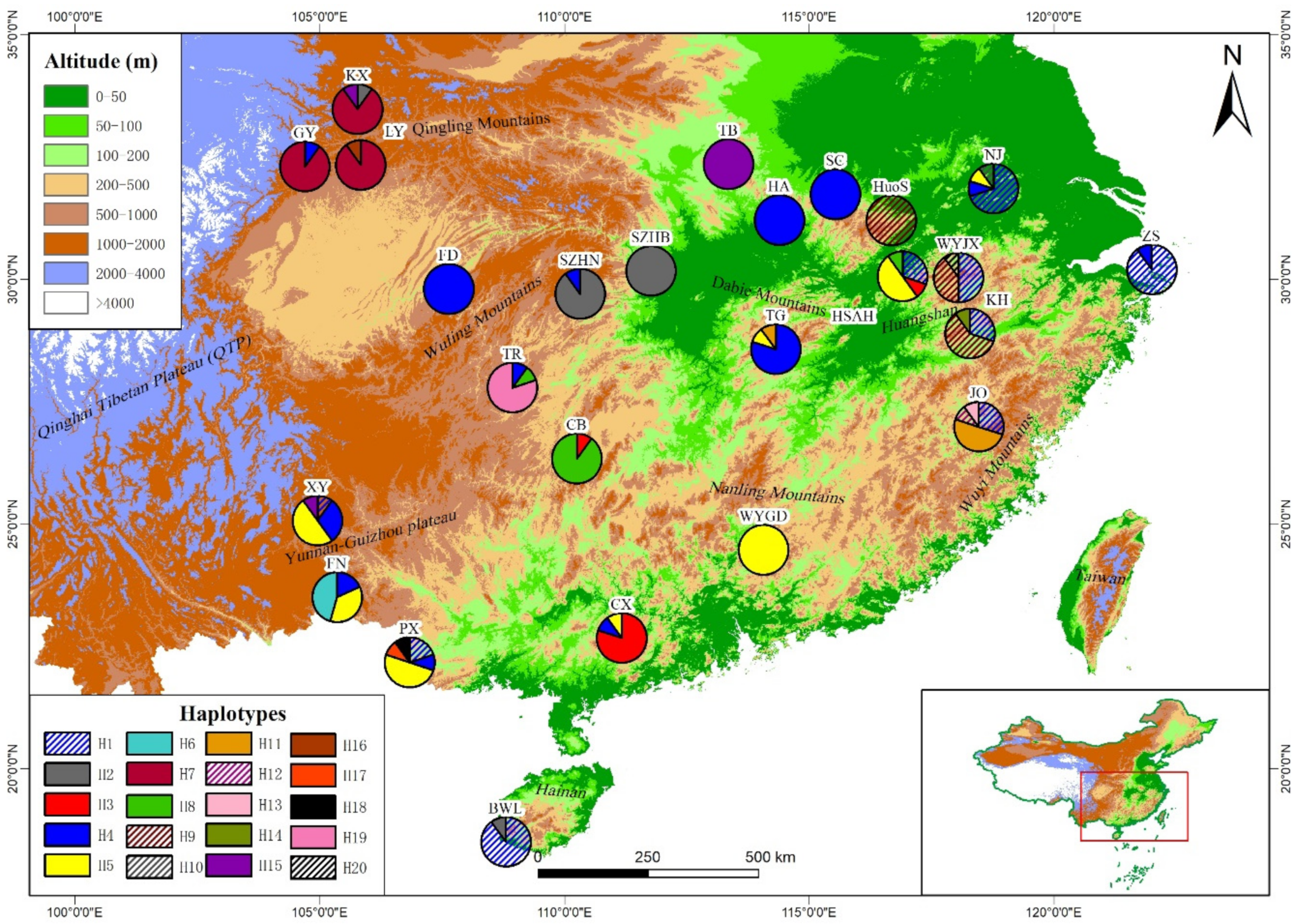
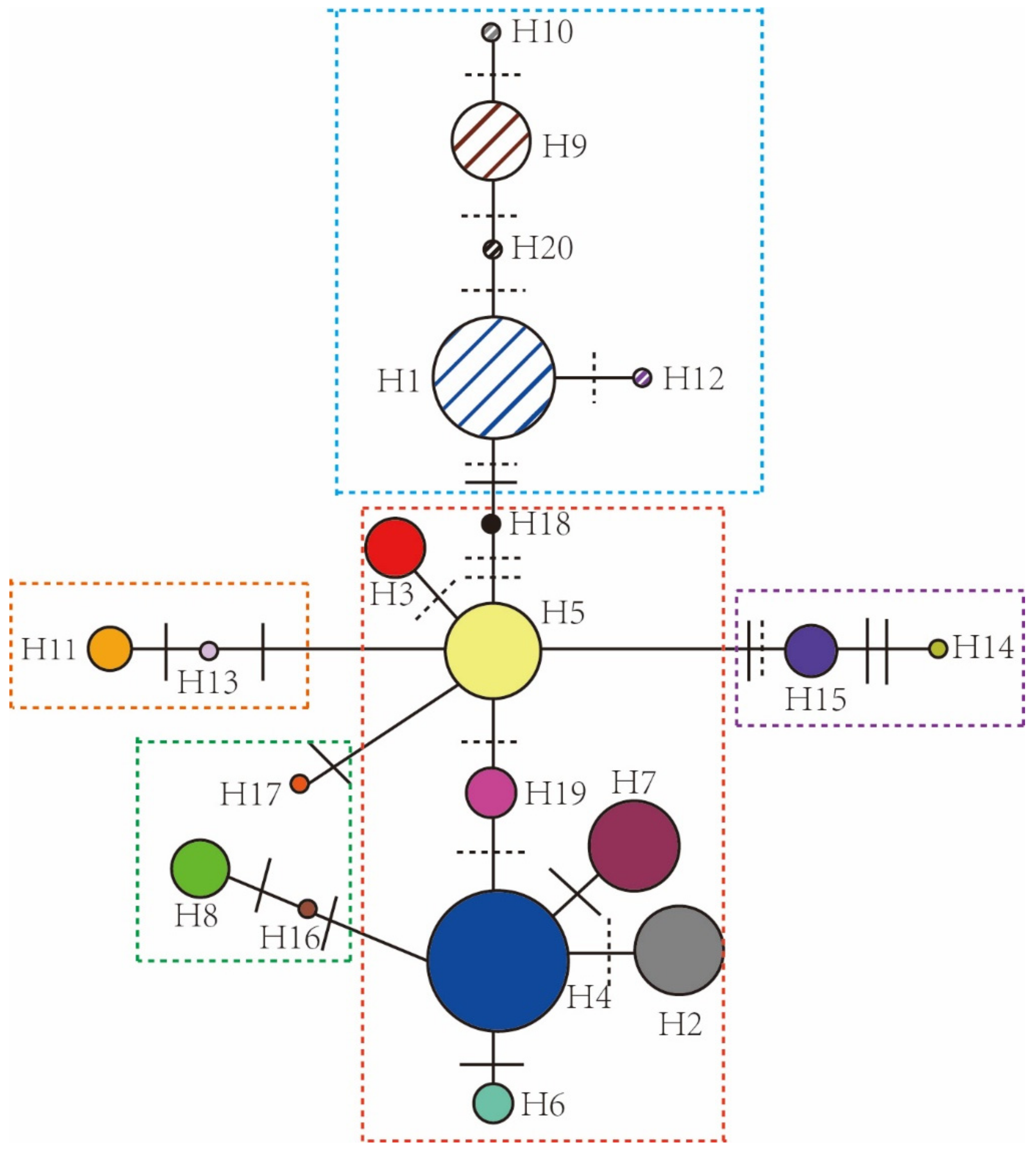
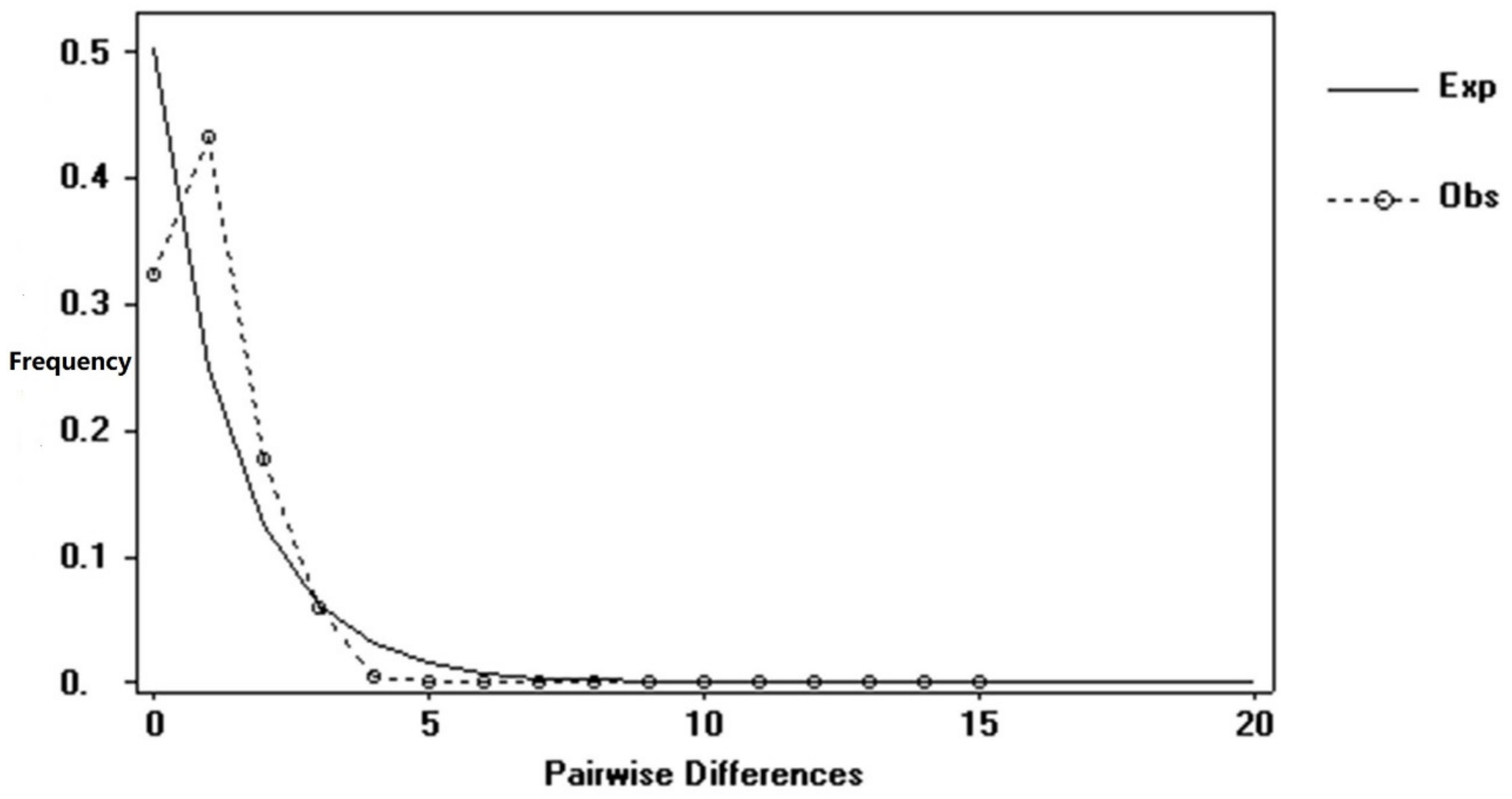
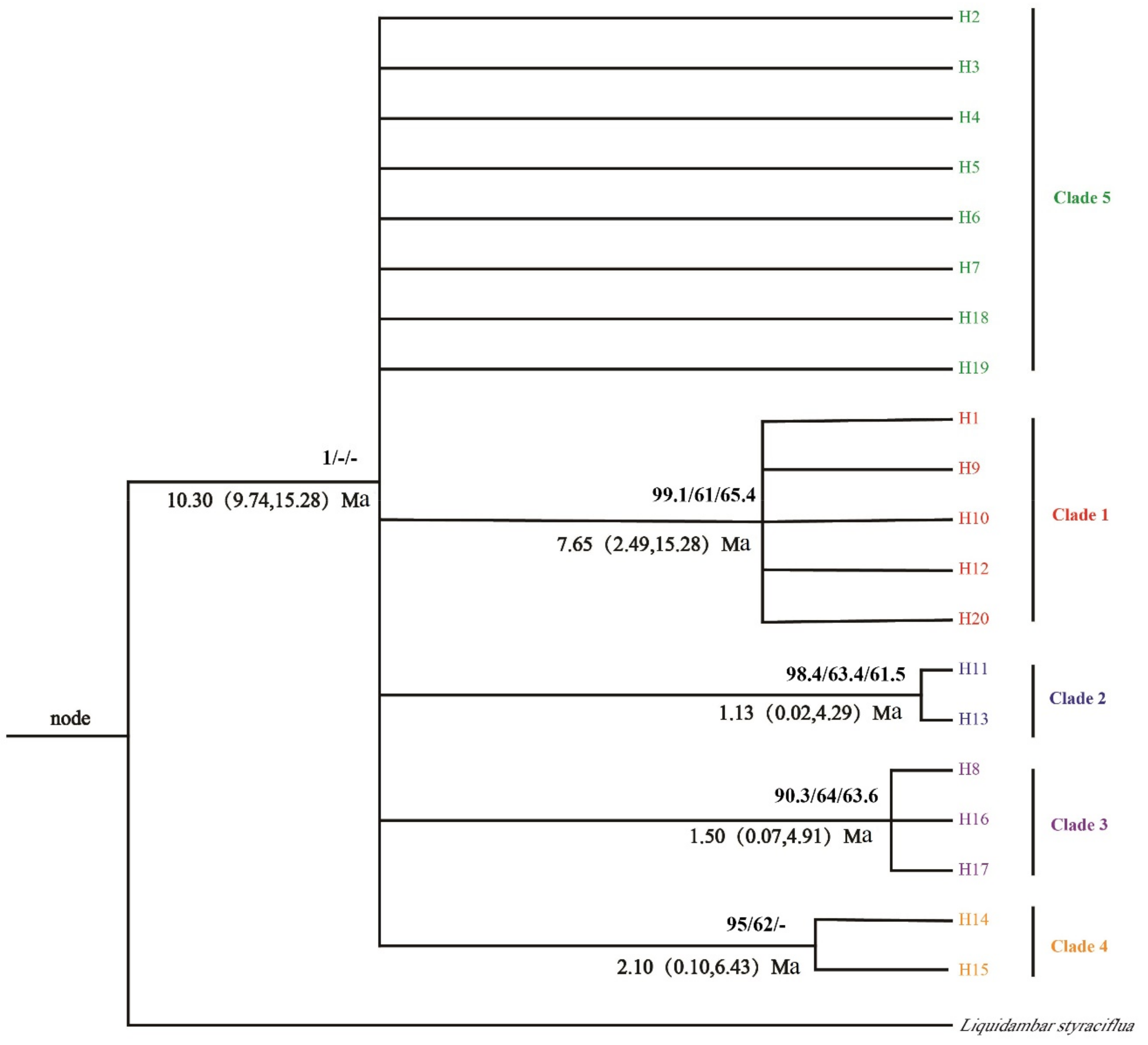
| Population Code | Sampling Location | Number of Sample Trees | Longitude (E) | Latitude (N) | Altitude (m) |
|---|---|---|---|---|---|
| BWL | Bawangling, Hainan | 10 | 108°48′ | 18°30′ | 655 |
| PX | Pingxiang, Guangxi | 10 | 106°50′ | 22°10′ | 420 |
| TR | Tongren, Guizhou | 10 | 108°56′ | 27°47′ | 400 |
| XY | Xingyi, Guizhou | 10 | 104°57′ | 25°04′ | 1100 |
| HSAH | Huangshan, Anhui | 10 | 117°36′ | 30°08′ | 150 |
| KX | Kangxian, Gansu | 10 | 105°46′ | 33°09′ | 1020 |
| LY | Lueyang, Shaanxi | 10 | 105°50′ | 32°20′ | 700 |
| TG | Tonggu, Jiangxi | 10 | 114°19′ | 28°34′ | 550 |
| WYJX | Wuyuan, Jiangxi | 10 | 117°53′ | 30°02′ | 831 |
| CB | Chengbu, Hunan | 10 | 110°15′ | 26°20′ | 610 |
| KH | Kaihua, Zhejiang | 10 | 118°17′ | 29°14′ | 1453 |
| WYGD | Wengyuan, Guangdong | 10 | 114°04′ | 24°28′ | 215 |
| FN | Funing, Yunnan | 11 | 105°21′ | 23°30′ | 1100 |
| FD | Fengdu, Chongqing | 10 | 107°38′ | 29°48′ | 560 |
| JO | Jianou, Fujian | 10 | 118°28′ | 26°59′ | 204 |
| ZS | Zhoushan, Zhejiang | 10 | 121°60′ | 30°12′ | 350 |
| SZHN | Sangzhi, Hunan | 10 | 110°19′ | 29°42′ | 700 |
| SZHB | Songzi, Hubei | 10 | 111°46′ | 30°10′ | 424 |
| NJ | Nanjing, Jiangsu | 10 | 118°46′ | 31°51′ | 28 |
| GY | Guangyuan, Sichuan | 10 | 105°36′ | 32°18′ | 890 |
| CX | Cenxi, Guangxi | 10 | 111°10′ | 22°40′ | 230 |
| HuoS | Huoshan, Anhui | 10 | 116°15′ | 31°12′ | 150 |
| SC | Shangcheng, Henan | 10 | 115°32′ | 31°44′ | 410 |
| TB | Tongbai, Henan | 10 | 113°21′ | 32°21′ | 300 |
| HA | Hongan, Hubei | 10 | 114°38′ | 31°32′ | 300 |
| Population | Haplotype (Number) | Hd | π |
|---|---|---|---|
| BWL | H1(9), H2(1) | 0.20000 | 0.00053 |
| PX | H1(2), H4(1), H5(5), H17(1), H18(1) | 0.75556 | 0.00085 |
| TR | H4(1), H8(1), H19(8) | 0.37778 | 0.00029 |
| XY | H1(1), H4(3), H5(5), H15(1) | 0.71111 | 0.00081 |
| HSAH | H1(3), H3(1), H5(5), H8(1) | 0.71111 | 0.00109 |
| KX | H2(1), H7(8), H15(1) | 0.37778 | 0.00051 |
| LY | H7(9), H16(1) | 0.20000 | 0.00015 |
| TG | H4(8), H5(1), H11(1) | 0.37778 | 0.00042 |
| WYJX | H1(5), H9(4), H20(1) | 0.64444 | 0.00041 |
| CB | H3(1), H8(9) | 0.20000 | 0.00038 |
| KH | H1(3), H9(6), H14(1) | 0.60000 | 0.00101 |
| WYGD | H5(10) | 0.00000 | 0.00000 |
| FN | H4(2), H5(4), H6(5) | 0.69091 | 0.00059 |
| FD | H4(10) | 0.00000 | 0.00000 |
| JO | H1(3), H11(5), H12(1), H13(1) | 0.71111 | 0.00120 |
| ZS | H1(9), H4(1) | 0.20000 | 0.00046 |
| SZHN | H2(9), H4(1) | 0.20000 | 0.00008 |
| SZHB | H2(10) | 0.00000 | 0.00000 |
| NJ | H1(7), H4(1), H5(1), H10(1) | 0.53333 | 0.00088 |
| GY | H4(1), H7(9) | 0.20000 | 0.00008 |
| CX | H3(8), H4(1), H5(1) | 0.37778 | 0.00029 |
| HuoS | H9(10) | 0.00000 | 0.00000 |
| SC | H4(10) | 0.00000 | 0.00000 |
| TB | H15(10) | 0.00000 | 0.00000 |
| HA | H4(10) | 0.00000 | 0.00000 |
| Total | 20(10) | 0.88762 | 0.00144 |
| Source of Variation | Degree of Freedom | Sum of Squares | Variance Components | Percentage of Variation (%) |
|---|---|---|---|---|
| Among populations | 24 | 923.722 | 3.71250 Va | 75.34 |
| Within populations | 226 | 274.700 | 1.21549 Vb | 24.66 |
| Total | 250 | 1198.422 | 4.92799 | Fst = 0.75 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, R.; Lin, F.; Huang, P.; Ye, X.; Lai, J.; Zheng, Y. Phylogeographical Structure of Liquidambar formosana Hance Revealed by Chloroplast Phylogeography and Species Distribution Models. Forests 2019, 10, 858. https://doi.org/10.3390/f10100858
Sun R, Lin F, Huang P, Ye X, Lai J, Zheng Y. Phylogeographical Structure of Liquidambar formosana Hance Revealed by Chloroplast Phylogeography and Species Distribution Models. Forests. 2019; 10(10):858. https://doi.org/10.3390/f10100858
Chicago/Turabian StyleSun, Rongxi, Furong Lin, Ping Huang, Xuemin Ye, Jiuxin Lai, and Yongqi Zheng. 2019. "Phylogeographical Structure of Liquidambar formosana Hance Revealed by Chloroplast Phylogeography and Species Distribution Models" Forests 10, no. 10: 858. https://doi.org/10.3390/f10100858
APA StyleSun, R., Lin, F., Huang, P., Ye, X., Lai, J., & Zheng, Y. (2019). Phylogeographical Structure of Liquidambar formosana Hance Revealed by Chloroplast Phylogeography and Species Distribution Models. Forests, 10(10), 858. https://doi.org/10.3390/f10100858





