The Relic Trochodendron aralioides Siebold & Zucc. (Trochodendraceae) in Taiwan: Ensemble Distribution Modeling and Climate Change Impacts
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. SDM
2.3. Accuracy Evaluation and Ensemble Forecasting
3. Results and Discussion
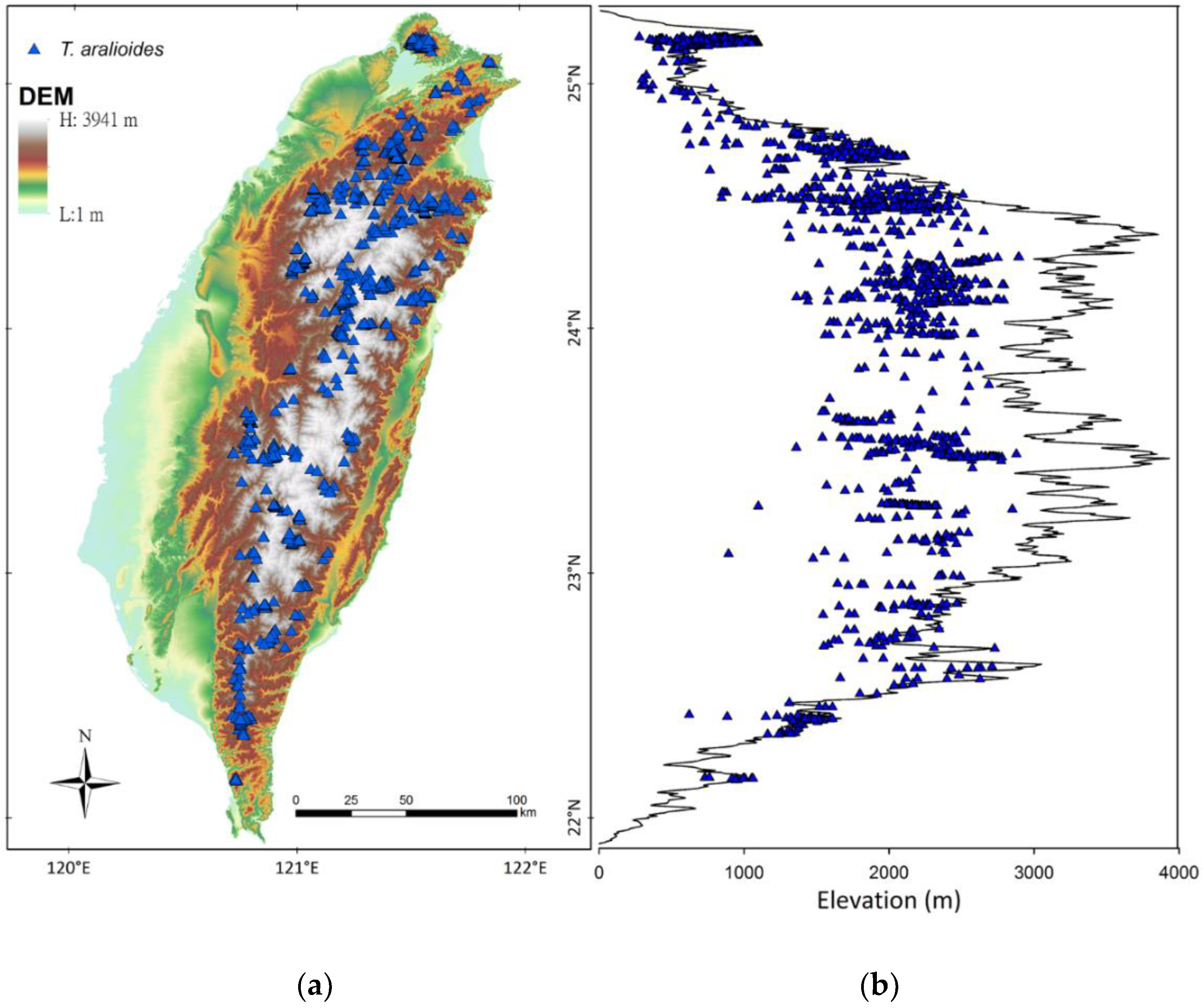
3.1. Occurrence Data of T. aralioides
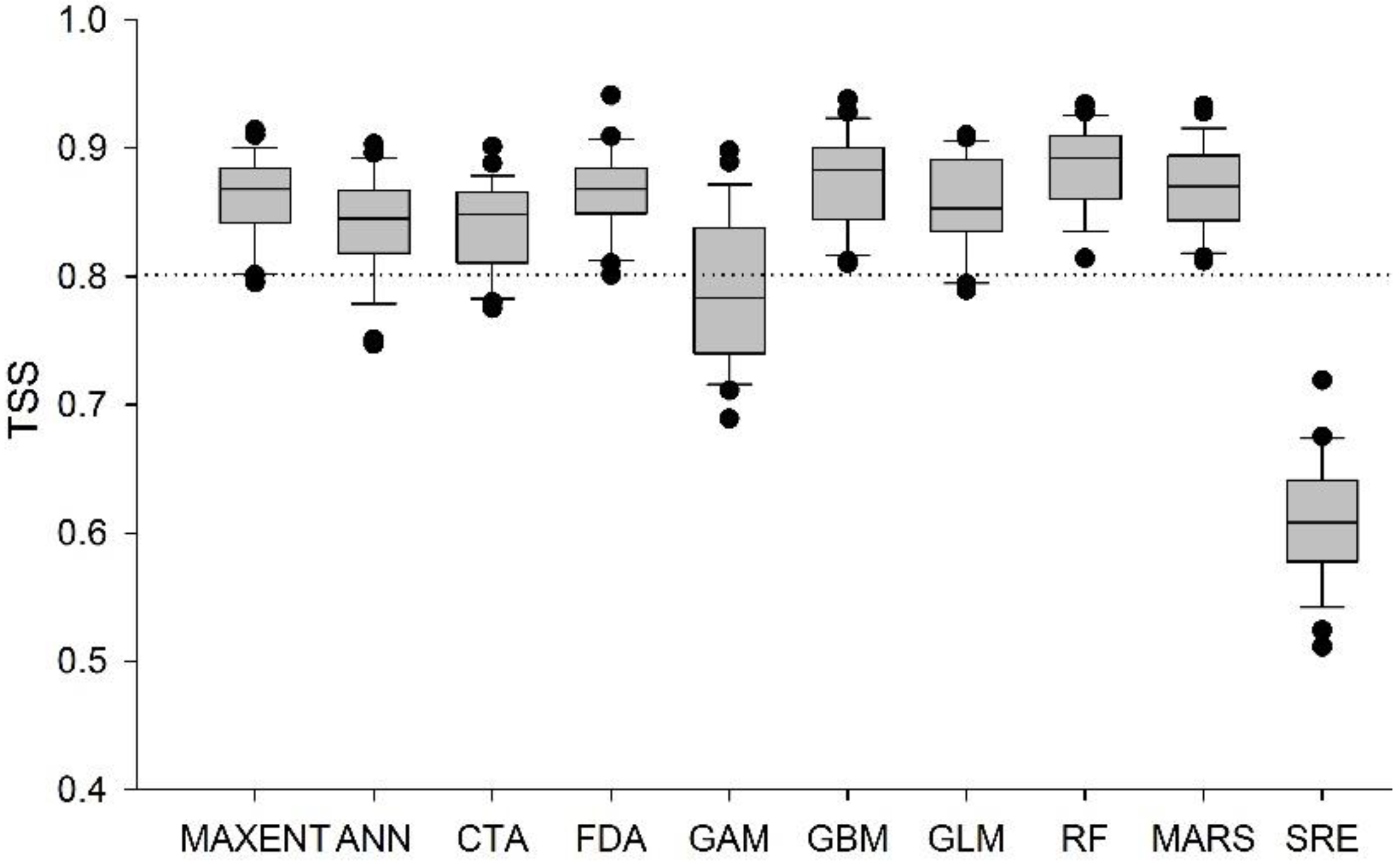
3.2. Ensemble Distribution Modeling
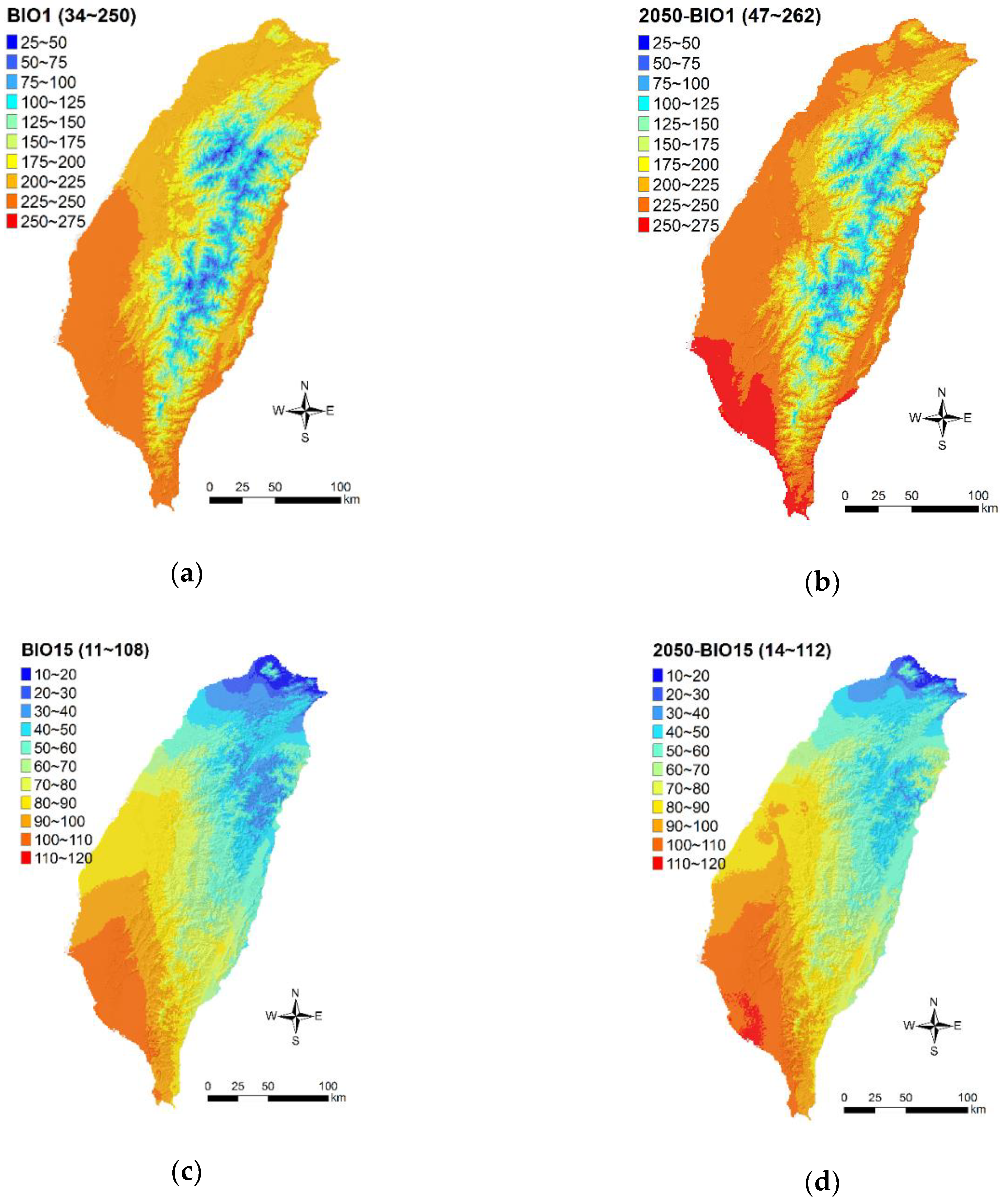
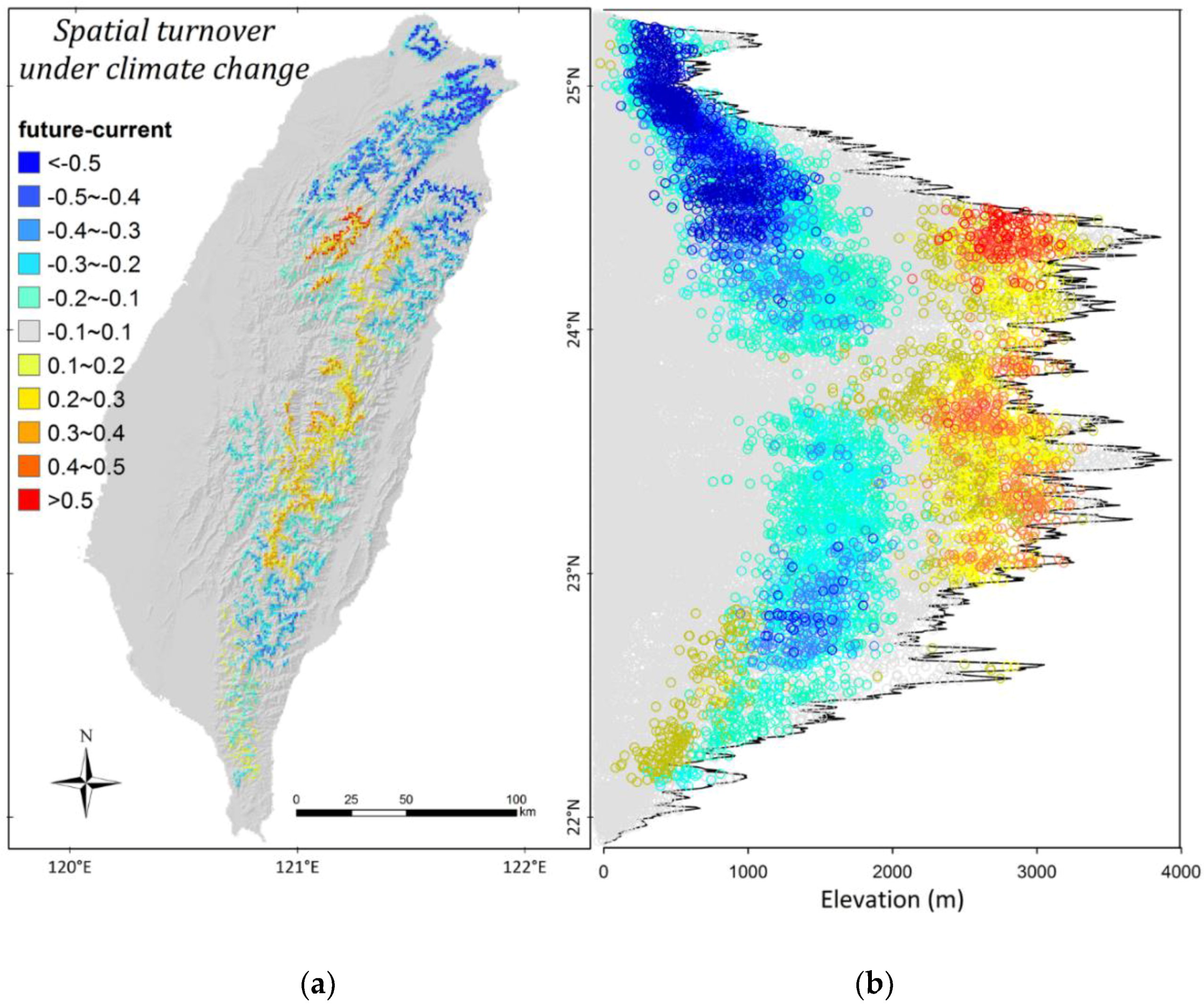
3.3. Climate Change Effects
4. Conclusion
Author Contributions
Funding
Conflicts of Interest
References
- Pigg, K.B.; Wehr, W.C.; Ickert-Bond, S.M. Trochodendron and Nordenskioldia (Trochodendraceae) from the middle eocene of Washington State, U.S.A. Int. J. Plant Sci. 2001, 162, 1187–1198. [Google Scholar] [CrossRef]
- Wu, J.E.; Huang, S.; Wang, J.C.; Tong, W.F. Allozyme variation and the genetic structure of populations of Trochodendron aralioides, a monotypic and narrow geographic genus. J. Plant Res. 2001, 114, 45–57. [Google Scholar] [CrossRef]
- Pigg, K.B.; Dillhoff, R.M.; DeVore, M.L.; Wehr, W.C. New diversity among the Trochodendraceae from the early/middle eocene Okanogan highlands of British Columbia, Canada, and northeastern Washington State, United States. Int. J. Plant Sci. 2007, 168, 521–532. [Google Scholar] [CrossRef]
- Yahara, T. Phytogeographical problems in the temperate flora in Japan. In Current Aspects of Biogeography in West Pacific and East Asian Regions; Ohba, H., Hayami, I., Mochizuki, K., Eds.; University Museum, University of Tokyo: Tokyo, Japan, 1989; pp. 99–113. [Google Scholar]
- Stevens, P.F. Angiosperm Phylogeny and Diversification. In Encyclopedia of Evolutionary Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 78–83. ISBN 978-0-12-800426-5. [Google Scholar]
- Su, H.J. Studies on the climate and vegetation types of the natural forests in Taiwan (II): Altitudinal vegetation zones in relation to temperature gradient. Q. J. Chin. For. 1984, 17, 57–73. [Google Scholar]
- Li, H.L.; Chaw, S.M. Trochodendraceae. In Flora of Taiwan, 2nd ed.; Huang, T.C., Editorial Committee of the Flora of Taiwan, Eds.; Department of Botany, Nation Taiwan University: Taipei, Taiwan, 1996; Volume 2, pp. 504–505. [Google Scholar]
- Chiu, C.A.; Chen, W.C.; Wang, C.C.; Chang, K.C.; Liao, M.C.; Hsu, H.S.; Tsai, C.Y. Is it true for “northern descent” phenomenon of Trochodendron aralioides spatial distribution? Q. J. For. Res. 2017, 39, 85–95, (In Chinese with English Abstract). [Google Scholar]
- Huang, S.F.; Hwang, S.Y.; Wang, J.C.; Lin, T.P. Phylogeography of Trochodendron aralioides (Trochodendraceae) in Taiwan and its adjacent areas. J. Biogeogr. 2004, 31, 1251–1259. [Google Scholar] [CrossRef]
- Ponce-Reyes, R.; Nicholson, E.; Baxter, P.W.J.; Fuller, R.A.; Possingham, H. Extinction risk in cloud forest fragments under climate change and habitat loss. Divers. Distrib. 2013, 19, 518–529. [Google Scholar] [CrossRef]
- Nix, H.A. A. A biogeogaphic analysis of Australian Elapid snakes. In Atlas of Elapid Snakes of Australia: Australian Flora and Fauns Series 7; Longmore, R., Ed.; Bureau of Flora and Fauna: Canberra, Australia, 1986; pp. 4–15. [Google Scholar]
- Booth, T.H.; Nix, H.A.; Busby, J.R.; Hutchinson, M.F. BIOCLIM: The first species distribution modelling package, its early applications and relevance to most current MAXENT studies. Divers. Distrib. 2014, 20, 1–9. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Le Maitre, D.C.; Thuiller, W.; Schonegevel, L. Developing an approach to defining the potential distributions of invasive plant species: A case study of Hakea species in South Africa. Glob. Ecol. Biogeogr. 2008, 17, 569–584. [Google Scholar] [CrossRef]
- Gallien, L.; Douzet, R.; Pratte, S.; Zimmermann, N.E.; Thuiller, W. Invasive species distribution models—How violating the equilibrium assumption can create new insights. Glob. Ecol. Biogeogr. 2012, 21, 1126–1136. [Google Scholar] [CrossRef]
- Peterson, A.T.; Soberón, J.; Pearson, R.G.; Anderson, R.P.; Martínez-Meyer, E.; Nakamura, M.; Araújo, M.B. (Eds.) Ecological Niches and Geographic Distributions; Princeton University Press: Princeton, NJ, USA, 2011; ISBN 978-0-691-13686-8. [Google Scholar]
- Guisan, A.; Tingley, R.; Baumgartner, J.B.; Naujokaitis-Lewis, I.; Sutcliffe, P.R.; Tulloch, A.I.T.; Regan, T.J.; Brotons, L.; McDonald-Madden, E.; Mantyka-Pringle, C.; et al. Predicting species distributions for conservation decisions. Ecol. Lett. 2013, 16, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Franklin, J. Moving beyond static species distribution models in support of conservation biogeography: Moving beyond static species distribution models. Divers. Distrib. 2010, 16, 321–330. [Google Scholar] [CrossRef]
- Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J.; Gordon, A.; Kujala, H.; Lentini, P.E.; McCarthy, M.A.; Tingley, R.; Wintle, B.A. Is my species distribution model fit for purpose? Matching data and models to applications. Glob. Ecol. Biogeogr. 2015, 24, 276–292. [Google Scholar] [CrossRef]
- Booth, T.H. Species distribution modelling tools and databases to assist managing forests under climate change. For. Ecol. Manag. 2018, 430, 196–203. [Google Scholar] [CrossRef]
- Nakao, K.; Higa, M.; Tsuyama, I.; Matsui, T.; Horikawa, M.; Tanaka, N. Spatial conservation planning under climate change: Using species distribution modeling to assess priority for adaptive management of Fagus crenata in Japan. J. Nat. Conserv. 2013, 21, 406–413. [Google Scholar] [CrossRef]
- Hällfors, M.H.; Liao, J.; Dzurisin, J.; Grundel, R.; Hyvärinen, M.; Towle, K.; Wu, G.C.; Hellmann, J.J. Addressing potential local adaptation in species distribution models: Implications for conservation under climate change. Ecol. Appl. 2016, 26, 1154–1169. [Google Scholar] [CrossRef]
- Dyderski, M.K.; Paź, S.; Frelich, L.E.; Jagodziński, A.M. How much does climate change threaten European forest tree species distributions? Glob. Chang. Biol. 2018, 24, 1150–1163. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Chiu, C.-A.; Lin, P.-H.; Lu, K.-C. GIS-based tests for quality control of meteorological data and spatial interpolation of climate data: A case study in mountainous Taiwan. Mt. Res. Dev. 2009, 29, 339–349. [Google Scholar] [CrossRef]
- Kimura, M. Quaternary paleogeography of the Ryukyu Arc. J. Geogr. 1996, 105, 259–285. [Google Scholar] [CrossRef]
- Huang, T.C.; Editorial Committee of the Flora of Taiwan. Flora of Taiwan, 2nd ed.; Department of Botany, Nation Taiwan University: Taipei, Taiwan, 2003; Volume 6. [Google Scholar]
- Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction; Cambridge University Press: Cambridge, UK, 2009; ISBN 978-0-521-70002-3. [Google Scholar]
- Martínez-Minaya, J.; Cameletti, M.; Conesa, D.; Pennino, M.G. Species distribution modeling: A statistical review with focus in spatio-temporal issues. Stoch. Environ. Res. Risk Assess. 2018, 32, 3227–3244. [Google Scholar] [CrossRef]
- Chiou, C.R.; Hsieh, C.F.; Wang, J.C.; Chen, M.Y.; Liu, H.Y.; Yeh, C.L.; Yang, S.Z.; Chen, T.Y.; Hsia, Y.J.; Song, G.Z.M. The first national vegetation inventory in Taiwan. Taiwan J. For. Res. 2009, 24, 295–302. [Google Scholar] [CrossRef]
- Araújo, M.B.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Yilmaz, H.; Yilmaz, O.Y.; Akyüz, Y.F. Determining the factors affecting the distribution of Muscari latifolium, an endemic plant of Turkey, and a mapping species distribution model. Ecol. Evol. 2017, 7, 1112–1124. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.P.; Van Niel, K.P. Improving species distribution models for climate change studies: Variable selection and scale: Species distribution models for climate change studies. J. Biogeogr. 2011, 38, 1–8. [Google Scholar] [CrossRef]
- Williams, K.J.; Belbin, L.; Austin, M.P.; Stein, J.L.; Ferrier, S. Which environmental variables should I use in my biodiversity model? Int. J. Geogr. Inf. Sci. 2012, 26, 2009–2047. [Google Scholar] [CrossRef]
- Marino, J.; Bennett, M.; Cossios, D.; Iriarte, A.; Lucherini, M.; Pliscoff, P.; Sillero-Zubiri, C.; Villalba, L.; Walker, S. Bioclimatic constraints to Andean cat distribution: A modelling application for rare species: Bioclimatic constraints to Andean cat distribution. Divers. Distrib. 2011, 17, 311–322. [Google Scholar] [CrossRef]
- Rupprecht, F.; Oldeland, J.; Finckh, M. Modelling potential distribution of the threatened tree species Juniperus oxycedrus: How to evaluate the predictions of different modelling approaches? J. Veg. Sci. 2011, 22, 647–659. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Thuiller, W.; Lafourcade, B.; Engler, R.; Araújo, M.B. BIOMOD—A platform for ensemble forecasting of species distributions. Ecography 2009, 32, 369–373. [Google Scholar] [CrossRef]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS): Assessing the accuracy of distribution models. J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Liu, C.; White, M.; Newell, G. Selecting thresholds for the prediction of species occurrence with presence-only data. J. Biogeogr. 2013, 40, 778–789. [Google Scholar] [CrossRef]
- Ben Rais Lasram, F.; Guilhaumon, F.; Albouy, C.; Somot, S.; Thuiller, W.; Mouillot, D. The Mediterranean Sea as a ‘cul-de-sac’ for endemic fishes facing climate change. Glob. Chang. Biol. 2010, 16, 3233–3245. [Google Scholar] [CrossRef]
- Araujo, M.; New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Thuiller, W.; Georges, D.; Engler, R.; Breiner, F. Package ‘BIOMOD 2’. 2016. Available online: https://cran.r-project.org/web/packages/biomod2/biomod2.pdf (accessed on 30 January 2017).
- Carvalho, S.B.; Brito, J.C.; Crespo, E.G.; Watts, M.E.; Possingham, H.P. Conservation planning under climate change: Toward accounting for uncertainty in predicted species distributions to increase confidence in conservation investments in space and time. Biol. Conserv. 2011, 144, 2020–2030. [Google Scholar] [CrossRef]
- Araújo, M.B.; Alagador, D.; Cabeza, M.; Nogués-Bravo, D.; Thuiller, W. Climate change threatens European conservation area. Ecol. Lett. 2011, 14, 484–492. [Google Scholar] [CrossRef]
- Yesson, C.; Brewer, P.W.; Sutton, T.; Caithness, N.; Pahwa, J.S.; Burgess, M.; Gray, W.A.; White, R.J.; Jones, A.C.; Bisby, F.A.; et al. How global is the global biodiversity information facility? PLoS ONE 2007, 2, e1124. [Google Scholar] [CrossRef]
- Chiu, C.A.; Lin, P.H.; Tsai, C.Y. Spatio-temporal variation and monsoon effect on the temperature lapse rate of a subtropical island. Terr. Atmos. Ocean. Sci. 2014, 25, 203. [Google Scholar] [CrossRef]
- Guisan, A.; Zimmermann, N.E.; Elith, J.; Graham, C.H.; Phillips, S.; Peterson, A.T. What matters for predicting the occurrences of trees: Techniques, data, or species’ characteristics? Ecol. Monogr. 2007, 77, 615–630. [Google Scholar] [CrossRef]
- Mi, C.; Huettmann, F.; Guo, Y.; Han, X.; Wen, L. Why choose Random Forest to predict rare species distribution with few samples in large undersampled areas? Three Asian crane species models provide supporting evidence. PeerJ 2017, 5, e2849. [Google Scholar] [CrossRef] [PubMed]
- Marchi, M.; Ducci, F. Some refinements on species distribution models using tree-level National Forest Inventories for supporting forest management and marginal forest population detection. iForest 2018, 11, 291–299. [Google Scholar] [CrossRef]
- Oppel, S.; Meirinho, A.; Ramírez, I.; Gardner, B.; O’Connell, A.F.; Miller, P.I.; Louzao, M. Comparison of five modelling techniques to predict the spatial distribution and abundance of seabirds. Biol. Conserv. 2012, 156, 94–104. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Ahmadi, M. A comparison of absolute performance of different correlative and mechanistic species distribution models in an independent area. Ecol. Evol. 2016, 6, 5973–5986. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Su, H.; Hu, J. The identification of A-, B-, C-, and E-class MADS-box genes and implications for perianth evolution in the basal eudicot Trochodendron aralioides (Trochodendraceae). Int. J. Plant Sci. 2007, 168, 775–799. [Google Scholar] [CrossRef]
- Chiu, C.A.; Lu, K.C.; Lin, P.H.; Liao, M.J. Mapping Holdridge’s life zones at Taiwan. J. Nat. Park. 2005, 15, 61–78, (In Chinese with English Abstract). [Google Scholar]
- Parolo, G.; Rossi, G. Upward migration of vascular plants following a climate warming trend in the Alps. Basic Appl. Ecol. 2008, 9, 100–107. [Google Scholar] [CrossRef]
- Chambers, D. Challenges in Modeling the Abundance of 105 Tree Species in Eastern North America for Climate Change Research. Master’s Thesis, McGill University, Montreal, QC, Canada, 2011. [Google Scholar]
- Du, H.; Liu, J.; Li, M.-H.; Büntgen, U.; Yang, Y.; Wang, L.; Wu, Z.; He, H.S. Warming-induced upward migration of the alpine treeline in the Changbai Mountains, northeast China. Glob. Chang. Biol. 2018, 24, 1256–1266. [Google Scholar] [CrossRef]
- Wu, J.E. Study on the Genetic Variation of Trochodendron aralioides and its Phylogenetic Relationship with Allies. Master’s Thesis, National Normal Taiwan University, Taipei, Taiwan, 2000. (In Chinese with English Abstract). [Google Scholar]
- Su, H.J. Studies on the climate and vegetation types of the natural forests in Taiwan (III): A scheme of geographical climatic regions. Q. J. Chin. For. 1985, 18, 33–44. [Google Scholar]
- Halpin, P.N. GIS analysis of the potential impacts of climate change on mountain ecosystems and protected areas. In Mountain Environments and Geographic Information Systems; Price, M., Heywood, D.I., Eds.; Taylor & Francis: London, UK, 1994; pp. 281–381. ISBN 07484-0088-5. [Google Scholar]
- Chala, D.; Brochmann, C.; Psomas, A.; Ehrich, D.; Gizaw, A.; Masao, C.A.; Bakkestuen, V.; Zimmermann, N.E. Good-bye to tropical alpine plant giants under warmer climates? Loss of range and genetic diversity in Lobelia rhynchopetalum. Ecol. Evol. 2016, 6, 8931–8941. [Google Scholar] [CrossRef]


| Elevation Zone (m) | Average Occurrence Probability | ||
|---|---|---|---|
| Current | Future | Future–Current | |
| 0–499 | 0.027 | 0.021 | −0.006 |
| 500–999 | 0.169 | 0.096 | −0.073 |
| 1000–1499 | 0.370 | 0.260 | −0.110 |
| 1500–1999 | 0.693 | 0.607 | −0.087 |
| 2000–2499 | 0.811 | 0.830 | 0.019 |
| 2500–2999 | 0.532 | 0.727 | 0.195 |
| 3000–3499 | 0.181 | 0.368 | 0.187 |
| 3500–3999 | 0.134 | 0.172 | 0.038 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-T.; Chiu, C.-A. The Relic Trochodendron aralioides Siebold & Zucc. (Trochodendraceae) in Taiwan: Ensemble Distribution Modeling and Climate Change Impacts. Forests 2019, 10, 7. https://doi.org/10.3390/f10010007
Lin C-T, Chiu C-A. The Relic Trochodendron aralioides Siebold & Zucc. (Trochodendraceae) in Taiwan: Ensemble Distribution Modeling and Climate Change Impacts. Forests. 2019; 10(1):7. https://doi.org/10.3390/f10010007
Chicago/Turabian StyleLin, Cheng-Tao, and Ching-An Chiu. 2019. "The Relic Trochodendron aralioides Siebold & Zucc. (Trochodendraceae) in Taiwan: Ensemble Distribution Modeling and Climate Change Impacts" Forests 10, no. 1: 7. https://doi.org/10.3390/f10010007
APA StyleLin, C.-T., & Chiu, C.-A. (2019). The Relic Trochodendron aralioides Siebold & Zucc. (Trochodendraceae) in Taiwan: Ensemble Distribution Modeling and Climate Change Impacts. Forests, 10(1), 7. https://doi.org/10.3390/f10010007






