Contrasting Effects of Chinese Fir Plantations of Different Stand Ages on Soil Enzyme Activities and Microbial Communities
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Selection and Soil Sampling
2.2. Soil Physicochemical Analyses
2.3. Standing Litter Analyses
2.4. Soil Enzyme Activities Analyses
2.5. Soil Microbial Communities Analyses
2.6. Statistical Analyses
3. Results
3.1. Soil Physicochemical Properties
3.2. Standing Litter Stocks
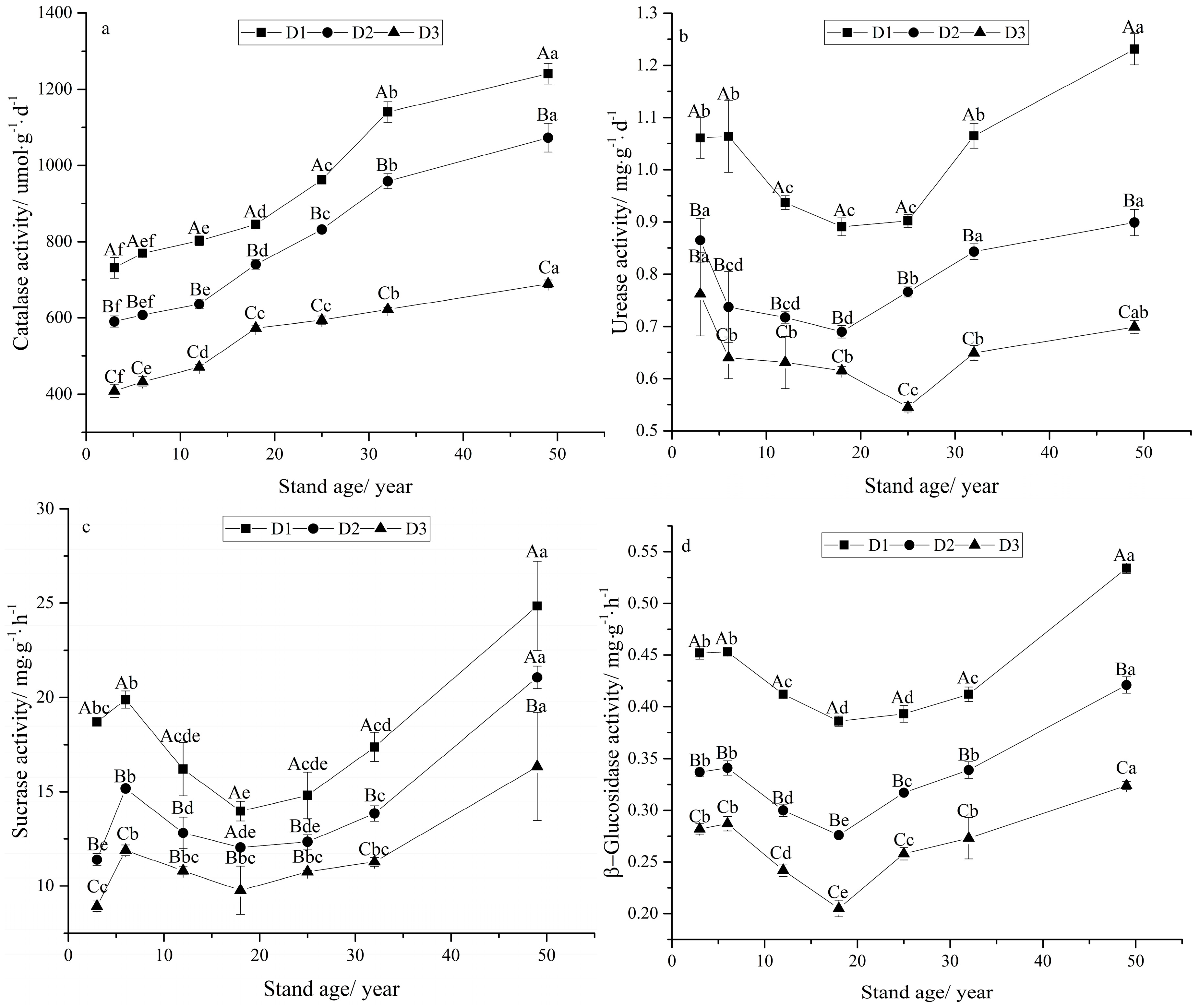
3.3. Soil Enzyme Activities
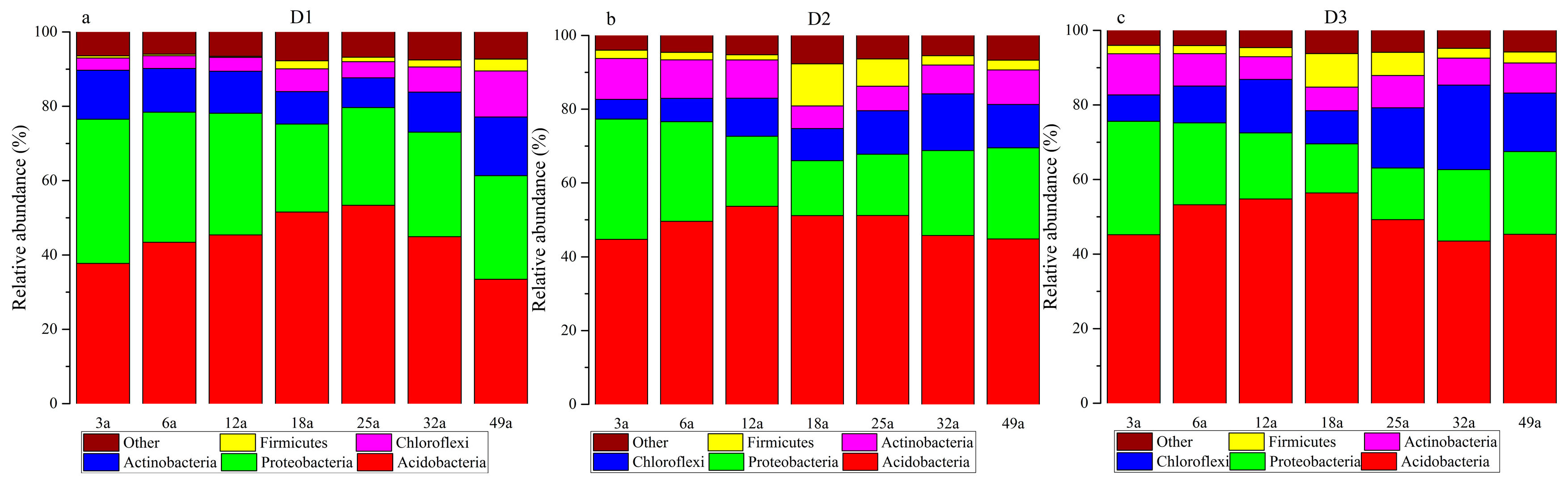
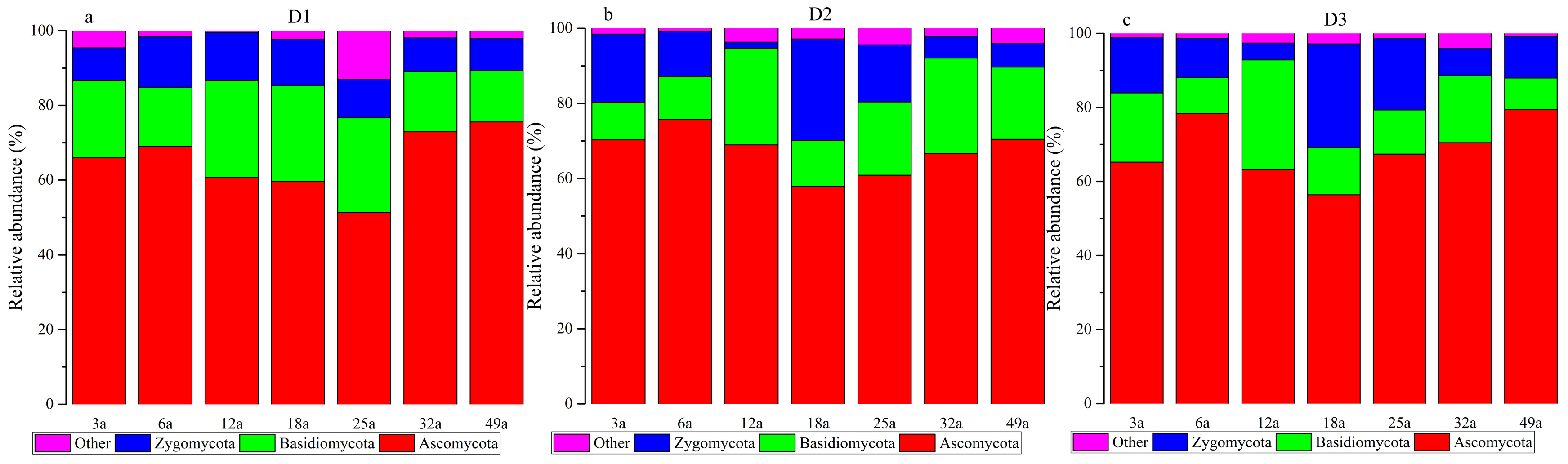
3.4. Microbial Communities
3.5. Links Between Soil Physicochemical Properties, Enzyme Activities and Microbial Diversity Indices
4. Discussion
4.1. Shift in Soil Pyhsicochemical Properties and Standing Litter Stocks
4.2. Shift in Soil Enzyme Activities and Composition and Diversity of Microbial Communities
4.3. Effects of Soil Physicochemical Properties on Enzyme Activities and Microbial Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Bueno de Mesquita, C.P.; Knelman, J.E.; King, A.J.; Farrer, E.C.; Porazinska, D.L.; Schmidt, S.K.; Suding, K.N. Plant colonization of moss-dominated soils in the alpine: Microbial and biogeochemical implications. Soil Biol. Biochem. 2017, 111, 135–142. [Google Scholar] [CrossRef]
- Yuan, X.; Knelman, J.E.; Gasarch, E.; Wang, D.; Nemergut, D.R.; Seastedt, T.R. Plant community and soil chemistry responses to long-term nitrogen inputs drive changes in alpine bacterial communities. Ecology 2016, 97, 1543. [Google Scholar] [CrossRef] [PubMed]
- Li, G.L.; Kim, S.; Han, S.H.; Chang, H.; Du, D.L.; Son, Y.H. Precipitation affects soil microbial and extracellular enzymatic responses to warming. Soil Biol. Biochem. 2018, 120, 212–221. [Google Scholar] [CrossRef]
- Ma, Y.J.; Xu, F.L.; Wang, W.L.; Chen, Q.C.; Zhao, H.Y.; Zhao, Y.F. Increase of soil nutrients and enzymatic activity by adding nitrogen and phosphorus to Larix principis-rupprechtii plantation. J. Plant Nut. Fert. 2015, 21, 664–674. [Google Scholar]
- Yang, J.J.; An, S.S.; Zhang, H.; Chen, Y.N.; Dang, T.H.; Jiao, J.Y. Effects of erosion on soil microbial biomass and enzyme activity in the loess hills. Acta Ecol. Sin. 2015, 35, 5666–5674. [Google Scholar]
- Shu, Y.Y.; Huang, J.S.; Zhao, G.J.; Bao, W.K.; Li, G.Q.; Pang, X.Y. Effects of afforestation with different tree species on soil enzyme activities and nutrient content in eastern Qinghai-Tibetan Plateau, China. Acta Ecol. Sini. 2016, 36, 394–402. [Google Scholar]
- Allison, S.D.; Weintraub, M.N.; Gartner, T.B.; Waldrop, M.P. Evolutionary-economic principles as regulators of soil enzyme production and ecosystem function. In Soil Enzymology; Shukla, G., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 229–243. [Google Scholar]
- Knelman, J.E.; Graham, E.B.; Prevéy, J.S.; Robeson, M.S.; Kelly, P.; Hood, E. Interspecific plant interactions reflected in soil bacterial community structure and nitrogen cycling in primary succession. Front. Microbiol. 2018, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Knelman, J.E.; Graham, E.B.; Ferrenberg, S.; Lecoeuvre, A.; Labrado, A.; Darcy, J.L.; Nemergut, D.R.; Schmidt, S.K. Rapid shifts in soil nutrients and decomposition enzyme activity in early succession following forest fire. Forests 2017, 8, 347. [Google Scholar] [CrossRef]
- Holden, S.R.; Gutierrez, A.; Treseder, K.K. Changes in soil fungal communities, extracellular enzyme activities, and litter decomposition across a fire chronosequence in alaskan boreal forests. Ecosystems 2013, 16, 34–46. [Google Scholar] [CrossRef]
- Smith, A.P.; Marín-Spiotta, E.; Balser, T. Successional and seasonal variations in soil and litter microbial community structure and function during tropical post-agricultural forest regeneration: A multi-year study. Glob. Chang. Biol. 2015, 21, 3532–3547. [Google Scholar] [CrossRef] [PubMed]
- Yesilonis, I.; Szlavecz, K.; Pouyat, R.; Whigham, D.; Xia, L. Historical land use and stand age effects on forest soil properties in the Mid-Atlantic US. For. Ecol. Manag. 2016, 370, 83–92. [Google Scholar] [CrossRef]
- Zhang, J.; Li, M.; Zheng, G. Effect of stand age on soil microbial community structure in wolfberry (Lycium barbarum L.) fields. Acta Ecol. Sin. 2017, 37, 10–17. [Google Scholar] [CrossRef]
- Li, Y.X.; Zhao, Z.; Chen, J.Q.; Liu, E.T. Study on soil microbial diversity of seabuckthorn forest. J. Northwest Agric. For. Univ. 2010, 38, 67–82. [Google Scholar]
- Luo, R.; Yang, M.; Yu, X.; Ma, Z.T.; Hao, R.Q.; Yang, L. Seasonal dynamics of soil microbial community and enzyme activities in Hippophae rhamnoides plantation. Chin. J. Appl. Ecol. 2018, 29, 1163–1169. [Google Scholar]
- Zhang, Y.; Li, H.T.; Wei, Z.C.; Liu, C.H.; Jiang, Z.K.; Ma, X.Q. Soil organic matter characteristics and aggregate stability in different development stages of Chinese fir plantation. Chin. J. Ecol. 2016, 35, 2029–2037. [Google Scholar]
- Selvaraj, S.; Duraisamy, V.; Huang, Z.; Guo, F.; Ma, X. Influence of long-term successive rotations and stand age of chinese fir (Cunninghamia lanceolata) plantations on soil properties. Geoderma 2017, 306, 127–134. [Google Scholar] [CrossRef]
- Wu, Y.L.; Wang, B.; Zhao, C.; Dai, W.; Li, P. Comprehensive evaluation of soil fertility in different developing stages of Chinese fir plantations. Chin. J. Northwest Agric. For. Univ. 2011, 39, 69–75. [Google Scholar]
- Shu, H.L. Soil pyhsicochemical property and enzymatic activity of Chinese fir plantation. Guizhou Agr. Sci. 2010, 38, 81–83. [Google Scholar]
- Wang, D.; Dai, W.; Wang, B.; Li, P.; Deng, Z.C.; Zhao, C. Changes of soil properties at different developmental stages of Chinese Fir plantations. J. Beijing For. Univ. 2010, 32, 59–63. [Google Scholar]
- Liu, L.; Duan, Z.H.; Wang, S.L.; Hu, J.C.; Hu, Z.G.; Zhang, Q.R.; Wang, S.J. Effects of Cunninghamia lanceolata plantations at different development stages on soil microbial community structure. Chin. J. Ecol. 2009, 28, 2417–2423. [Google Scholar]
- Institute of Soil Science, Chinese Academy of Sciences. Soil Physical and Chemical Analysis; Shanghai Scientific & Technical Publishers: Shanghai, China, 1978. [Google Scholar]
- Nguyen, L.T.T.; Osanai, Y.; Lai, K.; Anderson, I.; Bange, M.; Tissue, D. Responses of the soil microbial community to nitrogen fertilizer regimes and historical exposure to extreme weather events: Flooding or prolonged-drought. Soil Biol. Biochem. 2018, 118, 227–236. [Google Scholar] [CrossRef]
- Lefroy, R.D.B.; Blair, G.J.; Strong, W.M. Changes in soil organic matter with cropping as measured by organic carbon fractions and 13C natural isotope abundance. Plant Soil. 1993, 155–156, 399–402. [Google Scholar] [CrossRef]
- Liu, Z.B.; Liu, S.T.; Jiang, T.C.; Song, Y.C.; Wu, T.; Huang, Y.Z.; Luo, Y. Determination of soil total sulfur, total phosphorus and total potassium by inductively coupled plasma atomic emission spectrometry. Soil Fert. Sci. China. 2017, 5, 147–151. [Google Scholar]
- Wang, C.Q.; Xue, L.; Dong, Y.H.; Wei, Y.H.; Jiao, R.Z. Unravelling the functional diversity of the soil microbial community of Chinese fir plantations of different densities. Forests 2018, 9, 532. [Google Scholar] [CrossRef]
- Zanella, A.; Ponge, J.F.; Jabiol, B.; Sartori, G.; Kolb, E.; Le Bayon, R.C.; Gobat, J.M.; Aubert, M.; De Waal, R.; Van Delft, B. Humusica 1, article 5: Terrestrial humus systems and forms—Keys of classification of humus systems and forms. Appl. Soil Ecol. 2017, 122, 75–86. [Google Scholar] [CrossRef]
- DeForest, J.L. The influence of time, storage temperature, and substrate age on potential soil enzyme activity in acidic forest soils using MUB-linked substrates and LDOPA. Soil Biol. Biochem. 2009, 41, 1180–1186. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef]
- Kleivan, E.M. Understanding and analyzing multilevel data from real-time monitoring studies. PsyArXiv. 2017, 1, 1–31. [Google Scholar]
- Nakagawa, S.; Schielzeth, H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 2013, 4, 133–142. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Meng, J.H. On the comparison of soil physical characteristics of Cunninghamia lanceolata plantation amongst different age classes. Hebei J. For. Orchard Res. 2015, 30, 103–107. [Google Scholar]
- Sun, Y.; Wang, Y.Q.; Liu, J.; Xia, F.S.; Wang, J.G.; Li, J.B. Effect of solar greenhouse vegetable cultivation on soil physical quality. Chin. J. Appl. Ecol. 2011, 22, 2054–2060. [Google Scholar]
- Liu, D.Y.; Jiang, Y.Z.; Wang, H.T.; Kong, L.G.; Wang, Y. Effect of continuous cropping on Polar plantation. J. Soil Water Conserv. 2005, 19, 102–105. [Google Scholar]
- Li, H.T.; Zhang, Y.; Wei, Z.C.; Jia, D.D.; Liu, Y.H.; Liu, A.Q. Evaluation on soil fertility of Chinese fir plantations in different development stages. Forest Res. 2017, 30, 322–328. [Google Scholar]
- Springob, G.; Kirchmann, H. Bulk soil c to n ratio as a simple measure of net n mineralization from stabilized soil organic matter in sandy arable soils. Soil Biol. Biochem. 2003, 35, 629–632. [Google Scholar] [CrossRef]
- Paustian, K.; Andrén, O.; Janzen, H.H.; Lal, R.; Smith, P.; Tian, G.; Tiessen, H.; Van Noordwijk, M.; Woomer, P.L. Agricultural soils as a sink to mitigate CO2 emissions. Soil Use Manag. 1997, 13, 230–244. [Google Scholar] [CrossRef]
- Cornelissen, J.H.C.; Sibma, F.; Van Logtestijn, R.S.P.; Broekman, R.A.; Thompson, K. Leaf pH as a plant trait: Species-driven rather than soil-driven variation. Funct. Ecol. 2011, 25, 449–455. [Google Scholar] [CrossRef]
- Henry, H.A.; Juarez, J.D.; Field, C.B.; Vitousek, P.M. Interactive effects of elevated CO2, N deposition and climate change on extracellular enzyme activity and soil density fraction action in a California annual grassland. Glob. Chan. Biol. 2005, 11, 1808–1815. [Google Scholar] [CrossRef]
- Allison, S.D.; Treseder, K.K. Warming and drying suppress microbial activity and carbon cycling in boreal forest soils. Glob. Chan. Biol. 2008, 14, 2898–2909. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, P.; Kong, C.H. Urease, invertase, dehydrogenase and polyphenoloxidase activities in paddy soil influenced by allelopathic rice variety. Eur. J. Soil Biol. 2009, 45, 436–441. [Google Scholar] [CrossRef]
- Geng, Y.Q.; Bai, C.X.; Zhao, T.R.; Wang, S.S.; Chen, J.Q. Soil enzyme activity and its relationship with the soil fertility in Badaling Mountain Area of Beijing. J. Beijing For. Univ. 2006, 28, 7–11. [Google Scholar]
- Wan, Z.M.; Song, C.C. Distribution characteristics of soil enzyme activity in Xiaoyezhang wetland and its relationship with characterization index of active organic carbon. Wetl. Sci. 2008, 6, 249–257. [Google Scholar]
- Wei, Z.C.; Huang, J.; Liu, Y.H.; Jia, D.D.; Li, H.T.; Wu, P.F.; Liu, A.Q. Community characteristics of soil bacteria of Cunninghamia lanceolata plantation at different development stages. J. Southwest For. Univ. 2017, 37, 122–129. [Google Scholar]
- Jesus, E.D.; Susilawati, E.; Smith, S.L.; Wang, Q.; Chai, B.L.; Farris, R.; Rodrigues, J.L.M.; Thelen, K.D.; Tiedje, J.M. Bacterial communities in the rhizosphere of biofuel crops grown on marginal lands as evaluated by 16S rRNA gene pyrosequences. Bioenerg. Res. 2010, 3, 20–27. [Google Scholar] [CrossRef]
- Will, C.; Thurmer, A.; Wollherr, A.; Nacke, H.; Herold, N.; Schrumpf, M.; Gutknecht, J.; Wubet, T.; Buscot, F.; Daniel, R. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. Environ. Microb. 2010, 76, 6751–6759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Penton, C.R.; Xue, C.; Quensen, J.F.; Roley, S.S.; Guo, J. Soil depth and crop determinants of bacterial communities under ten biofuel cropping systems. Soil Biol. Biochem. 2017, 112, 140–152. [Google Scholar] [CrossRef]
- Xiong, W.; Li, R.; Ren, Y.; Liu, C.; Zhao, Q.; Wu, H. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla fusarium, wilt disease. Soil Biol. Biochem. 2017, 107, 198–207. [Google Scholar] [CrossRef]
- Zuppingerdingley, D.; Schmid, B.; Petermann, J.S.; Yadav, V.; De Deyn, G.B.; Flynn, D.F. Selection for niche differentiation in plant communities increases biodiversity effects. Nature 2014, 515, 108–111. [Google Scholar] [CrossRef]
- Dong, L.; Xu, J.; Zhang, L.; Cheng, R.; Wei, G.; Su, H. Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharm. Sin. 2018, 8, 272–282. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M. Three genomes from the phylum acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microb. 2009, 75, 2046–2056. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Hanson, C.A.; Allison, S.D.; Bradford, M.A.; Wallenstein, M.D.; Treseder, K.K. Fungal taxa target different carbon sources in forest soil. Ecosystems 2008, 11, 1157–1167. [Google Scholar] [CrossRef]
- Wang, L.; Luo, X.; Liao, H.; Chen, W.; Wei, D.; Cai, P. Ureolytic microbial community is modulated by fertilization regimes and particle-size fractions in a black soil of northeastern china. Soil Biol. Biochem. 2018, 116, 171–178. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, X.; Tan, Y.; Liu, H.L.; Tian, Z.P.; Zeng, G.P. Dynamics between soil microorganism and soil character actors during Carthamus tinctorius growth periods. Pratacul Sci. 2011, 28, 2084–2091. [Google Scholar]
- Sun, S.; Zhao, H.; Xing, F.; Bai, Z.; Gao, Y.; Dong, Y.; Zhou, J.; Wu, Y.; Yang, Y. Response of soil microbial community structure to increased precipitation and nitrogen addition in a semiarid meadow steppe. Eur. J. Soil Sci. 2017, 68, 524–536. [Google Scholar] [CrossRef]
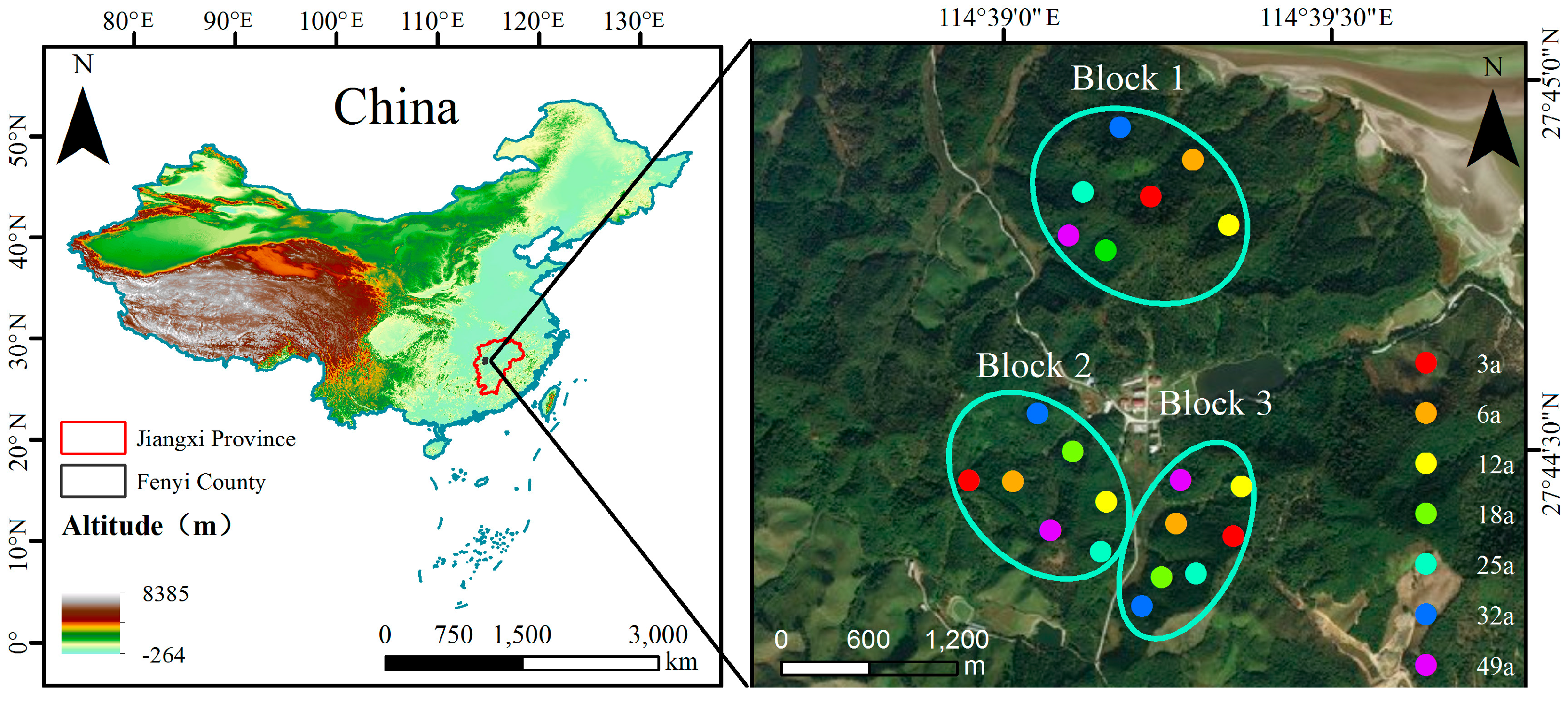


| Stand Age (a) | Mean Elevation (m) | Slope (°) | Aspects | Existing Density (stems·hm−2) | Dominant Undergrowth Vegetation Species |
|---|---|---|---|---|---|
| 3a | 106–113 | 26–27 | southeast | 3333 | Blechnum japonica, Rubus trianthus Foeke., Lophatherum gracile, Ophiopogon japonicus, Maesa japonica (Thunb) Moritzi, Efigeron acris L., Adinandra milletti (Hook. Et Arn.) Benth, Clerodendrum cyrtophyllum Turcz. |
| 6a | 106–116 | 25–27 | southeast, south | 3333 | Blechnum japonica, Maesa japonica (Thunb) Moritzi, Clerodendrum cyrtophyllum Turcz. |
| 12a | 106–107 | 28–32 | southeast | 3294 | Blechnum japonica, Maesa japonica (Thunb) Moritzi |
| 18a | 105–132 | 20–23 | southeast | 3067 | Blechnum japonica, Rubus trianthus Foeke., Maesa japonica (Thunb) Moritzi, Efigeron acris L. |
| 25a | 93–122 | 25–30 | Southeast, south | 2589 | Blechnum japonica, Rubus trianthus Foeke., Lophatherum gracile, Maesa japonica (Thunb) Moritzi, Efigeron acris L., Camellia oleifera Abel. |
| 32a | 103–138 | 28–32 | Southeast, south | 2161 | Blechnum japonica, Rubus trianthus Foeke., Smilax glabra Roxb., Maesa japonica (Thunb) Moritzi, Efigeron acris L., Camellia oleifera Abel. |
| 49a | 93–139 | 25–30 | Southeast, south | 1968 | Blechnum japonica, Rubus trianthus Foeke., Maesa japonica (Thunb) Moritzi, Efigeron acris L., Camellia oleifera Abel., Schima superba Garde. Et Champ. |
| Stand Age (a) | Litter Stocks/ (kg∙hm−2) | Ratio of Litter Stock at Different Layer to Total Litter Stock/% | |||
|---|---|---|---|---|---|
| TOL | BOL | Total | TOL | BOL | |
| 3a | 5864 ± 202 d | 3281 ± 116 e | 9146 ± 225 d | 64.11 | 35.89 |
| 6a | 3155 ± 168 e | 1982 ± 118 f | 5137 ± 122 e | 61.39 | 38.61 |
| 12a | 5261 ± 138 d | 3083 ± 88 e | 8343 ± 162 d | 63.05 | 36.95 |
| 18a | 8590 ± 187 c | 5674 ± 162 d | 14264 ± 250 c | 60.22 | 39.78 |
| 25a | 11073 ± 822 b | 9066 ± 789 c | 20139 ± 808 b | 54.98 | 45.02 |
| 32a | 13201 ± 598 a | 11944 ± 386 b | 25145 ± 774 a | 52.48 | 47.52 |
| 49a | 12495 ± 1367 a | 13108 ± 817 a | 25604 ± 2000 a | 48.71 | 51.29 |
| Stand Age (a) | Bacteria | Fungi | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shannon | Chao1 | Shannon | Chao1 | |||||||||
| D1 | D2 | D3 | D1 | D2 | D3 | D1 | D2 | D3 | D1 | D2 | D3 | |
| 3a | 9.99 ± 0.12 Aab | 9.49 ± 0.19 Bab | 9.24 ± 0.09 Babc | 3864 ± 174 Aab | 3599 ± 207 Bab | 3412 ± 159 Bab | 6.47 ± 0.16 Abc | 5.93 ± 0.03 Bb | 5.85 ± 0.24 Bb | 820 ± 41 Abc | 534 ± 13 Bbc | 517 ± 59 Bb |
| 6a | 10.03 ± 0.11 Aab | 9.53 ± 0.02 Bab | 9.30 ± 0.19 Bab | 4194 ± 104 Aa | 3800 ± 428 ABab | 3475 ± 283 Bab | 7.14 ± 0.24 Aab | 6.00 ± 0.38 Bb | 5.95 ± 0.49 Bb | 856 ± 23 Aabc | 566 ± 12 Bab | 536 ± 91 Bab |
| 12a | 9.97 ± 0.09 Aab | 9.42 ± 0.15 Bab | 9.02 ± 0.31 Bbc | 3997 ± 192 Aab | 3655 ± 98 ABab | 3316 ± 173 Bab | 6.62 ± 0.02 Aabc | 5.89 ± 0.06 Bb | 5.63 ± 0.18 Bbc | 854 ± 38 Abc | 492 ± 46 Bcd | 447 ± 27 Bc |
| 18a | 9.62 ± 0.12 Ab | 9.05 ± 0.11 Bc | 8.99 ± 0.07 Bbc | 3496 ± 336 Aab | 3246 ± 47 Ac | 3155 ± 213 Ab | 6.53 ± 0.10 Aabc | 5.83 ± 0.16 Bb | 5.26 ± 0.14 Bc | 765 ± 10 Ac | 431 ± 27 Bd | 350 ± 27 Bd |
| 25a | 9.70 ± 0.45 Ab | 9.25 ± 0.05 Bbc | 8.94 ± 0.03 Cc | 3219 ± 174 Ab | 3440 ± 19 Abc | 2911 ± 440 Ab | 6.06 ± 0.79 Ac | 5.81 ± 0.40 ABc | 5.32 ± 0.12 Bc | 785 ± 104 Abc | 559 ± 20 Bb | 440 ± 32 Cc |
| 32a | 10.08 ± 0.05 Aab | 9.32 ± 0.08 Bab | 9.29 ± 0.01 Babc | 4202 ± 108 Aa | 3490 ± 264 Bbc | 3472 ± 134 Bab | 7.14 ± 0.23 Aab | 6.33 ± 0.21 Bab | 5.93 ± 0.32 Bb | 887 ± 83 Aab | 587 ± 18 Bab | 548 ± 34 Bab |
| 49a | 10.16 ± 0.15 Aa | 9.55 ± 0.07 Ba | 9.47 ± 0.11 Ba | 4306 ± 450 Aa | 4024 ± 235 Aa | 3507 ± 187 Ba | 7.20 ± 0.01 Aa | 6.70 ± 0.09 Ba | 6.63 ± 0.24 Ba | 966 ± 87 Aa | 609 ± 49 Ba | 594 ± 63 Ba |
| Catalase | Urease | Sucrase | β-glucosidase | Bacteria | Fungi | |||
|---|---|---|---|---|---|---|---|---|
| Shannon | Chao1 | Shannon | Chao1 | |||||
| Estimate (CI) | Estimate (CI) | Estimate (CI) | Estimate (CI) | Estimate (CI) | Estimate (CI) | Estimate (CI) | Estimate (CI) | |
| Fixed Parts | ||||||||
| Intercept | 0.42 (−0.39–1.23) *** | 0.71 (0.37–1.05) *** | 0.76 (−1.22–0.3) *** | 0.14 (0.01–0.27) *** | 1.35 (1.09–1.61) *** | 2.09 (2.05–2.13) *** | 2.96 (1.1–4.82) *** | 1.74 (1.65–1.83) *** |
| SBD | 0.00 (−0.02–0.02) *** | 0.07 (−0.03–0.17) | 0.33 (−0.86–2.18) | 0.05 (0.01–0.09) ** | 0.08 (−0.29–0.45) | 0.64 (0.12–1.16) | 0.78 (0.03–1.53) * | 0.37 (−0.34–1.08) |
| pH | 0.99 (0.98–1.00) | −0.11 (−0.21–0.01) | 0.31 (−0.09–0.71) | −0.01 (−0.05–0.03) | 0.07 (−0.28–0.42) | −0.48 (−1.11–0.15) | −0.45 (−1.02–0.12) * | −0.35 (−1.07–0.37) ** |
| SOM | 0.48 (−0.23–1.19) | 0.49 (−0.21–1.19) | −0.02 (−0.05–0.01) | 0.01 (−0.01–0.03) | 0.00 (−0.02–0.02) | −0.81 (−1.23–0.39) | −0.99 (−1.00–0.98) * | −0.76 (−1.21–0.31) |
| TN | 0.99 (0.98–1.00) | 0.01 (−0.03–0.05) * | 0.43 (−0.93–1.79) * | 0.00 (−0.02–0.02) | 0.22 (0.06–0.38) ** | 0.19 (−0.66–1.04) ** | −0.18 (−0.47–0.11) *** | −0.67 (−0.77–0.57) * |
| TP | −0.58 (−1.14–0.02) | −0.14 (−0.30–0.02) | −0.59 (−0.86–0.32) | −0.03 (−0.09–0.03) | −0.47 (−0.08–0.86) | −0.78 (−1.22–0.34) | −0.12 (−0.28–0.04) | −0.08 (−1.02–0.86) |
| TK | 0.43 (−0.34–1.20) | 0.68 (0.27–1.09) | 0.63 (−0.09–1.35) | 0.00 (−0.01–0.01) | 0.02 (−0.07–0.11) | −0.86 (0-1.25–0.47) | −0.4 (−0.58–0.22) | −0.67 (−1.17–0.17) |
| AN | 0.72 (0.36–1.08) | 0.06 (−0.04–0.16) | 0.00 (−0.02–0.02) | 0.02 (0.00–0.04) | 0.00 (−0.01–0.01) | −0.45 (−0.60–0.30) | 0.75 (0.71–0.79) ** | 0.74 (0.27–1.21) |
| AP | 0.01 (−0.55–0.57) * | 0.00 (−0.09–0.09) *** | 0.66 (0.76–0.56) *** | 0.04 (0.03–0.05) *** | 0.39 (0.29–0.49) *** | 0.62 (0.61–0.63) *** | 0.27 (0.1–0.44) *** | 0.11 (0.07–0.15) *** |
| AK | −0.13 (−0.69–0.43) | 0.88 (0.72–1.04) | 0.01 (−0.05–0.07) | 0.08 (0.07–0.09) | −0.01 (−0.02–0.00) | −0.67 (−1.17–0.17) | 0.00 (−0.01–0.01) | 0.82 (0.40–1.24) |
| C:N | −0.77 (−1.07–0.47) | 0.61 (0.09–1.13) | −0.02 (−0.07–0.03) | 0.02 (−0.01–0.05) | 0.00 (−0.01–0.01) | −0.77 (−1.21–0.33) | −0.01 (−0.02–0.00) | −0.3 (−0.33–0.27) * |
| Random Parts | ||||||||
| σ2 | 0.016 | 0.003 | 0.085 | 0.001 | 0.335 | 0.050 | 0.025 | 0.005 |
| τ00, Location | 0.031 | 0.102 | 0.152 | 0.264 | 0.241 | 0.252 | 0.312 | 0.335 |
| τ00, Layer:Location | 0.085 | 0.142 | 0.114 | 0.251 | 0.284 | 0.302 | 0.216 | 0.284 |
| ICCLocation | 0.235 | 0.413 | 0.433 | 0.512 | 0.280 | 0417 | 0.564 | 0.537 |
| ICCLayer:Location | 0.644 | 0.575 | 0.325 | 0.486 | 0.330 | 0.500 | 0.391 | 0.455 |
| R2c, R2m | 0.085, 0.284 | 0.101, 0.425 | 0.176, 0.378 | 0.129, 0.439 | 0.185, 0.537 | 0.127, 0.485 | 0.238, 0.685 | 0.204, 0.641 |
| Enzymes | Bacteria | Fungi | ||
|---|---|---|---|---|
| Shannon | Chao1 | Shannon | Chao1 | |
| CAT | 0.545 ** | 0.404 ** | 0.642 ** | 0.676 ** |
| UE | 0.793 ** | 0.601 ** | 0.732 ** | 0.833 ** |
| SC | 0.727 ** | 0.570 ** | 0.756 ** | 0.721 ** |
| BG | 0.833 ** | 0.663 ** | 0.798 ** | 0.851 ** |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Xue, L.; Dong, Y.; Hou, L.; Wei, Y.; Chen, J.; Jiao, R. Contrasting Effects of Chinese Fir Plantations of Different Stand Ages on Soil Enzyme Activities and Microbial Communities. Forests 2019, 10, 11. https://doi.org/10.3390/f10010011
Wang C, Xue L, Dong Y, Hou L, Wei Y, Chen J, Jiao R. Contrasting Effects of Chinese Fir Plantations of Different Stand Ages on Soil Enzyme Activities and Microbial Communities. Forests. 2019; 10(1):11. https://doi.org/10.3390/f10010011
Chicago/Turabian StyleWang, Chaoqun, Lin Xue, Yuhong Dong, Lingyu Hou, Yihui Wei, Jiaqi Chen, and Ruzhen Jiao. 2019. "Contrasting Effects of Chinese Fir Plantations of Different Stand Ages on Soil Enzyme Activities and Microbial Communities" Forests 10, no. 1: 11. https://doi.org/10.3390/f10010011
APA StyleWang, C., Xue, L., Dong, Y., Hou, L., Wei, Y., Chen, J., & Jiao, R. (2019). Contrasting Effects of Chinese Fir Plantations of Different Stand Ages on Soil Enzyme Activities and Microbial Communities. Forests, 10(1), 11. https://doi.org/10.3390/f10010011





