Abstract
Enzymes play key roles in the biological functions of living organisms, which serve as catalysts to and regulate biochemical reaction pathways. Recent studies suggest that peptides are promising molecules for modulating enzyme function due to their advantages in large chemical diversity and well-established methods for library synthesis. Experimental approaches to identify protein-binding peptides are time-consuming and costly. Hence, there is a demand to develop a fast and accurate computational approach to tackle this problem. Another challenge in developing a computational approach is the lack of a large and reliable dataset. In this study, we develop a new machine learning approach called PepBind-SVM to predict protein-binding peptides. To build this model, we extract different sequential and physicochemical features from peptides and use a Support Vector Machine (SVM) as the classification technique. We train this model on the dataset that we also introduce in this study. PepBind-SVM achieves 92.1% prediction accuracy, outperforming other classifiers at predicting protein-binding peptides.
1. Introduction
Enzymes are crucial for the biological functions of living organisms. They serve as catalysts to catalyze and regulate biochemical reaction pathways [1,2]. Searching ligands to modulate enzyme functions is one of the central topics for disease diagnosis and therapeutics [3,4,5]. Recent studies suggest that peptides are promising molecules for modulating enzyme function due to their advantages in large chemical diversity, biocompatibility, and well-established methods for library synthesis [6,7,8,9]. For example, peptides and their derivatives can be used to inhibit or activate enzyme catalysis, mediate cell receptor response, and facilitate transmembrane delivery [7,10].
Traditional ligands and drug discovery rely on the synthesis of large chemical libraries. High-throughput screening or selection is often used to identify ligands for binding to a target protein, such as in vitro mRNA display [8], phage display [11], bead-based library screening [12], protein direct evolution [13], aptamer selection [14], peptide microarray [7,10,15], and fragment-based design of small molecules [16]. Unfortunately, these approaches are all very time-consuming and costly. The recent progress of machine learning (ML) and artificial intelligence (AI) have advanced the chemical discovery process [17]. For example, by using a dataset of existing molecular libraries with known functions, ML can be trained to analyze large datasets of ligand structures, sequences, and binding affinities and to predict potential interactions more accurately [18].
During the past two decades, a wide range of ML models has been proposed to predict different types of protein-binding peptides [19,20,21,22,23]. These studies can generally be categorized into two groups. First are studies that introduced new features to extract more discriminatory information for the prediction task [24,25,26]. These studies mainly extract features based on sequential, structural, physicochemical, and evolutionary-based information [27,28].
For instance, Yan and Zhou used chemical interactions and physicochemical properties of amino acids for feature extraction [29]. Later on, Romero-Molina et al. extended the use of physicochemical properties and also included sequential-based features to predict protein-binding peptides [24]. In a different study, Wei et al., employed structural features to detect therapeutic peptides [27]. The structural features are mostly extracted based on the predicted values of the protein local or secondary structure of the proteins using tools such as PSIPRED or SPIDER [30,31,32]. Other studies, such as Akbar et al., used evolutionary information to add to former types of features for better performance [33].
Recent studies have shifted their focus to using Deep Learning (DL) models to automatically extract features from protein sequences instead of different types of properties [33,34,35,36]. However, extracting effective features is still considered a common and effective method to tackle this problem [37].
The second group of studies aims to use effective ML and DL models to predict protein-binding peptides. Among traditional machine learning models, Support Vector Machine (SVM), Random Forest (RF), and Adaptive Boosting have been shown to be very effective for this task [26,37,38,39,40,41,42]. As mentioned earlier, recent studies have shifted their focus to using DL to tackle this problem. However, for smaller datasets, complex DL models cannot be trained properly. Instead, traditional ML methods demonstrate better performance for these cases. In addition, studying the explainability of the complex DL models is more challenging. Therefore, there is a demand for an accurate and explainable model to tackle this problem. Another challenge is to have reliable and large enough datasets to use as a benchmark to train machine learning models to predict common protein-binding peptides. Most of the currently used datasets are either very small or have low resolution.
This study addresses these two challenges by generating a reliable and relatively large peptide-binding protein dataset and proposing an explainable machine learning model. Here, we develop PepBind-SVM to predict protein-binding peptides. To build this model, we extract different sequential and physicochemical features from peptides and use an SVM as the classification technique. We train this model on the dataset that we also introduce in this study. PepBind-SVM outperforms other classifiers in predicting protein-binding peptides. PepBind-SVM, its source code, and our newly generated protein binding peptides are publicly available at https://github.com/MLBC-lab/pepbind-SVM (accessed on 1 September 2024).
2. Dataset Preparation
To generate our own dataset, we first prepared the peptides using an experimental approach. We then filtered the data to generate a training and independent test set to build and validate our machine learning models.
2.1. Microarray Fabrication
Here, we use 10,000 20-mer peptides produced at Arizona State University [7,10,43]. These distinct polypeptide sequences were spotted in duplicate onto a glass slide possessing an amino-silane surface coating. Each polypeptide is 20 residues long, with 17 positions randomly chosen from 19 amino acids (excluding cysteine). The C-terminal three positions of each peptide was a glycine–serine–cysteine (GSC) linker, which was used for conjugating peptides to amino-silane surfaces via a maleimide linker, sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate (Sulfo-SMCC; Pierce, Rockford, IL, USA). The array printing was done using a Telechem Nanoprint60 (Currently Arrayit, Sunnyvale, CA, USA) hot spotted approximately 500 pL of 1 mg/mL peptide per feature on glass slides with 48 Telechem series SMP2-style 946 titanium pins (Currently Arrayit, Sunnyvale, CA, USA).
2.2. Microarray Binding
We labeled β-Gal with Alexa Fluor 647 (Invitrogen) and diluted it to 5 nM in bovine serum albumin (BSA) buffer (1 × PBS with 3% (v/v) BSA and 0.05% Tween 20). The activity of the labeled β-Gal was~75% that of the wild type in solution. Binding of enzymes on the microarray was performed as described in [43,44]. Briefly, a microarray was first pre-washed with surface cleaning solvent (7.33% (v/v) acetonitrile, 37% isopropyl alcohol, and 0.55% trifluoroacetic acid in water), followed by the treatment of a capping buffer (3% (v/v), bovine serum albumin (BSA), 0.02% (v/v) mercaptohexanol, and 0.05% (v/v) Tween 20 in 1 × PBS) to block any active SMCC linker on the array surface. Then, the array was incubated with a solution containing 5 nM Alexa 647-labeled β-Gal or PEP-1–β-Gal complex for two hours, allowing the enzyme to bind with peptides on the array surface. After washing off unbound enzymes, the array was read by a standard array reader (PerkinElmer, Waltham, MA, USA) with color scanning using 647 nm laser lines. The array was then read by a standard array reader (PerkinElmer, Waltham, MA, USA) with dual color scanning using 488 and 647 nm laser lines.
2.3. Generating Training and Testing Data for the Machine Learning Model
After generating the peptides, we prepared training and testing datasets to build our machine learning model in the following manner. We first calculated the affinity of the peptides to interact with proteins and scored each peptide with a number between 0 and 1. After that, we defined a conservative threshold to select protein-binding and non-binding peptides. In this way, we made sure that the protein-binding peptides in our dataset are actual binding and positive samples, while non-binding peptides are actual negative samples. In this way, the model can effectively learn the patterns. Based on the corresponding scores, the number of protein-binding peptides was much smaller than that of non-binding peptides.
The number of non-binding peptides was four times more than the number of binding peptides. We considered this ratio for building our training and testing datasets.
We selected those peptides with over 0.8 (out of 1) affinity of binding to proteins as positive and those peptides with less than 0.2 (out of 1) affinity of binding to proteins as negative samples. In this way, our dataset could be more robust and have clear-cut positive and negative samples. Peptides with binding affinity between 0.2 and 0.8 cannot be accurately categorized as binding or non-binding. Therefore, to make sure that our true positive and true negative samples were accurate, we discarded them. The aim of this study is to accurately identify protein-binding and non-binding peptides. These thresholds were selected experimentally and by studying the similarity of samples with each other. As a result, we generated a training dataset containing 1600 samples. This set was balanced to contain an equal representation of classes, with 800 samples labeled as positives and 800 as negatives. Such a balanced dataset aids in preventing model bias toward the more dominant class. However, to make sure that our model performs well on the actual data, we considered the actual imbalance when we built the testing dataset. For the independent testing, a dataset comprising 1000 samples was prepared. This dataset includes 200 positive samples and 800 negative samples, intentionally skewed to reflect a real-world scenario where negative instances are more prevalent than positive ones.
3. Feature Generation
To encapsulate the complexity and intrinsic characteristics of peptides, we engineered features based on n-gram models, specifically monograms and bigrams, and incorporated the peptides’ physicochemical properties. Initially, several n-lengths were computed recursively. However, no improvement was shown for subsets of length greater than 2. Therefore, this study is restricted to monograms and bigrams as a feature set. The feature generation process is explained as follows.
3.1. Composition and Occurrence (Monogram)
To generate the occurrence, we count the number of each amino acid along the peptide. It consists of 20 features. To generate the composition, we calculate the frequency of the amino acid occurrence. To do this, we simply divide the numbers in the occurrence feature vector by the total number of amino acids in the peptide. It is shown that the performance of occurrence and composition can be different with respect to the other features being used and with respect to the problem at hand [45].
3.2. Bigram
In addition to monogram features, we explored the sequential relationship between amino acids by considering bigram, which is defined as the number of pairs of adjacent amino acids within the sequences. This approach allows the model to capture the local sequence context, which can be a determinant of the peptide’s binding capability. Since there are 20 amino acids, the total number of combinations of pairs is 20 × 20. Hence, the Bigram feature group consists of 400 features. It has been widely used for different protein and peptide-related problems and obtained promising results [34].
3.3. Physicochemical Properties
Here, we incorporate the normalized density of 55 physicochemical properties, which is calculated as follows. For a given amino acid along the peptide sequence, we replace it with the corresponding value of its property. We then sum them all together and divide the summation by the length of the peptide, as was done in [46]. These properties include, but are not limited to, hydrophobicity, charge, and molecular weight. Normalization of these features ensures that each property contributes equally to the model’s learning process, preventing any single feature from dominating due to scale differences. These features were introduced in [46] as the most effective set of physicochemical-based features.
4. Classification Method
In this study, we tried using different classifiers, and, among them, SVM demonstrated the best performance and is used to build PepBind-SVM. The result of this comparison is comprehensively presented in the Results and Discussion Section. SVM has been shown as an effective binary-class classification technique that performs well in similar studies and specifically for protein-binding peptide prediction in the literature [36,37,38]. SVM aims to classify the samples by identifying the maximal marginal hyperplane (MMH) to separate the data [47]. In this study, the Linear Kernel is used for our SVM model. As shown in the Result and Discussion Section, using the linear kernel, we obtained the best results compared to other kernels.
5. Evaluation Methods and Metrics
To evaluate the performance of our model, we used k-fold cross-validation on the training data and an independent testing set. In k-fold cross-validation, the training data are divided into k groups. At every step, K-1 of the groups are used for training purposes while the remaining group is being used for testing purposes. This process is repeated k times until all groups are being used as the testing set. In this way, we can effectively use our data for validation. In this study, we use k = 5. To make sure that our model is general, we also use an independent test set that is not used for training purposes.
For the evaluation of our model’s performance, various metrics, including accuracy (Acc), Sensitivity (Se), Specificity (Sp), the area under the receiver–operating characteristic curve (AUC), Matthews correlation coefficient (MCC), and F1-score are used. These metrics serve as reliable measures to assess the effectiveness and robustness of the model. The calculations for each metric are defined as follows:
where TP represents the count of true positives, TN represents the count of true negatives, FP represents the count of false positives, and FN represents the count of false negatives. These five evaluation metrics were used for results achieved for both testing dataset and five-fold cross-validation.
6. Results and Discussion
In this section, we first present our achieved results and continue with a comprehensive feature analysis. After that, we discuss the explainability of our model using Shapley Additive exPlanations (SHAP).
6.1. Selecting a Base Classifier to Build Our Model
To build our model, we use different classifiers for our extracted features. We studied several classifiers and, among them, chose RF, SVM, and gradient-boosting decision trees (XGBoost). As discussed in the Introduction Section, these three classifiers have been shown to be effective methods in predicting protein-binding peptides [26,37,38,39,40,41,42]. Introduced by Breiman, Random Forest is considered one of the best ensemble classifiers [48]. Based on the Bagging approach, RF promotes randomness to obtain better results. For RF, the employed data are first randomly divided into several groups. Then, for each group, a base decision tree is used for training using a random subset of a total number of features. Therefore, both data and features are randomly used to train each decision tree. After that, the results of all the base learners are combined to present the final classification. On the other hand, XGBoost is based on a boosting method [49]. In this model, a base classifier is trained iteratively on a subset of the data, and based on its performance, the weights for the classified samples are adjusted for the next iteration. In the next iteration, the weights of those samples that are incorrectly classified are increased to make it more costly to misclassify them. This process is being repeated several times. In the end, all those predictors in different iterations are combined to provide the final predictor. For the XGBoost, the employed base learner is gradient descent as its base learner.
We use these three classifiers with different parameters for our three extracted feature groups. We use SVM with linear and Radial Basis function kernels. We also use RF with 50, 100, and 200 decision trees. In this way, we can choose the parameters that perform the best. We used grid search as well as several values manually (those that performed well for similar studies) to tune our classifiers and optimize them. After careful comparison, we used the parameters that are general, and also their results were aligned with the outcome of the grid search to avoid overfitting. The results achieved using these classifiers for Composition, Occurrence, Bigram, and Physicochemical-based feature groups for the independent test set are presented in Table 1, Table 2, Table 3, and Table 4, respectively.

Table 1.
Performance of our employed classifiers using composition feature group on the independent dataset.

Table 2.
Performance of our employed classifiers using occurrence feature group on the independent dataset.

Table 3.
Performance of our employed classifiers using the Physicochemical feature group on the independent dataset.

Table 4.
Performance of our employed classifiers using the Bigram feature group on the independent dataset.
As shown in Table 1, Table 2, Table 3 and Table 4, using SVM, we achieve the best results. After SVM, XGBoost achieves better results than RF. Among different numbers of base learners, using 100, RF achieves its best results. Still, it performs poorer than SVM and XGBoost. As shown in Table 1, using SVM with a linear kernel, we achieved 92.1% accuracy, 86.0% sensitivity, 93.6% specificity, 0.765 MCC, 0.937 AUC, and 0.813 F1 score, which outperforms other combinations. It highlights the importance of the monogram feature group. Also, better results using linear kernel demonstrate that linear transformation of the input data to a higher dimension is better for maintaining the patterns in the data. In other words, the underlying pattern in the data can be expressed in a linear manner.
As shown in Table 4, Bigram’s results are lower than those of other feature groups. This demonstrates that, with respect to the number of generated features (400), Bigram does not provide significant discriminatory information compared to other feature groups. Comparing the results in Table 1 and Table 2 shows that we can achieve slightly better results by using composition. As discussed in [45] and considering the similar nature of composition and occurrence feature groups, we just use the composition feature group for the next step, which is the combination of all feature groups.
6.2. Selecting the Input Feature Groups to Build Our Model
Next, we combine composition, physicochemical, and Bigram feature groups and use our classifiers on the independent test set. The result of this experiment is shown in Table 5.

Table 5.
Performance of our employed classifiers using a combination of the composition, physicochemical, and Bigram feature groups on the independent dataset.
As shown in Table 5, using a combination of different feature groups, we achieve lower results for all the metrics. It shows that we do not add extra discriminatory information, considering the number of additional features when combining these feature groups. Considering the number of added features, Bigram might have the highest weight in reducing the performance. Therefore, in the next step, we just use the combination of composition and physicochemical feature groups. The result of this experiment is shown in Table 6.

Table 6.
Performance of our employed classifiers using a combination of the composition and physicochemical feature groups on the independent dataset.
As shown in Table 6, adding a physicochemical-based feature group reduces the performance compared to using composition or occurrence alone. It again highlights that the discriminatory information presented in the physicochemical-based feature group is already captured in the occurrence and composition feature group. It is actually in line with the fact that the necessary information for understanding the protein sequence is already embedded in its amino acid sequence [50]. To validate and investigate the generality and robustness of achieved results, we use our employed classifiers on the composition and the combination of all three feature groups (composition, Bigram, and physicochemical-based feature groups) using five-fold cross-validation. The results of these experiments are presented in Table 7 and Table 8, respectively.

Table 7.
Performance of our employed classifiers on the composition feature group using five-fold cross-validation.

Table 8.
Performance of our employed classifiers using a combination of the composition and physicochemical feature groups using five-fold cross-validation.
As shown in Table 7 and Table 8, although the results are lower than those reported on the independent test set, the trend is still similar. As shown in these tables, using the SVM classifier and composition as the input feature group, we achieve better performance than using the combination of all the employed feature groups. This highlights the generality of our achieved results. The lower results achieved on the five-fold cross-validation can be associated with using far fewer samples to test our model. Using five-fold cross-validation, we use just 80% of our data to train the model, compared to using 100% of the data when we use an independent test set. As a result, we use an SVM classifier with a linear kernel and composition feature group to build PepBind-SVM. PepBind-SVM, its source code, and our newly generated protein binding peptides are publicly available at https://github.com/MLBC-lab/pepbind-SVM (accessed on 1 September 2024).
Achieving the best results using SVM with linear kernel as the classifier and composition as feature group, which are relatively simple combinations, can be directly related to the employed dataset. We prepared the dataset with several control cases to make sure that it is a very clear representation of protein-binding and non-binding peptides. Achieving promising results using this combination demonstrates that if we use proper data with high resolution and clarity, finding the pattern in the data is much more straightforward. It is important to highlight that despite promising results, we still have a long way to go to solve this problem. Hence, for our future study, we aim at using structural and evolutionary based features to tackle this problem. We also aim to use a complex deep-learning model to solve this problem. One of the limitations of this study is regarding data cleaning. We cleaned up our data using a binding affinity of less than 0.2 for non-binding peptides and over 0.8 for binding peptides. This threshold enables us to make sure that our positive samples and negative samples are correct. In the future, we aim to collect more data and experimentally validate the proper threshold or try other thresholds with more extensive data.
Another limitation of this study is that our model is designed to identify protein-binding and non-binding peptides. However, it is not able to identify peptides with binding affinity between 0.2 and 0.8 (uncertain proteins). Our future direction is to identify a proper threshold, which would be validated experimentally with the help of experts, for uncertain peptides and identify them.
Finally, since the employed dataset is new and was generated by us, there is no previous work with which to compare our model. However, we share this dataset along with this article for future studies to evaluate their model. Our future direction is also to use complex yet explainable models to tackle this problem. Next, we investigate the explainability of the proposed model using SHAP.
6.3. SHAP for Interpreting Model Feature Relation and Black Box Model Explanation
Shapley Additive exPlanations (SHAP) is a widely used method for interpreting machine learning models [51]. Studies have used this method to elucidate the decision-making processes of machine learning models across various types of biological data [52,53]. This technique is created based on the shapely values from cooperative game theory. Each feature in the data plays the role of the cooperative player. SHAP explains the output of machine learning models by assigning each feature an importance value for a specific prediction. Note that we considered top XAI methods such as SHAP, LIME [54], and ELI5 [55], and ultimately chose SHAP since it has been successfully used for similar problems and obtained promising results [56,57]. Shapely values present a fair method for allocating rewards among participants involved in a collaborative game. In such a game, a group of N players collaborates to generate a total payout of v. Shapely value assigns a fair payout to each player based on their contribution. A feature with a higher magnitude of SHAP value indicates a significant contribution of that feature to the prediction. The Shapley value is calculated using the following equation:
where F is the set of all features, and S is a subset of features that does not contain feature i. As shown in this equation, is computed by considering all possible feature subsets S and evaluating how the prediction changes by including or excluding features i. In this study, we investigated both local and global explanations using SHAP. Local explanations are focused on providing a deeper understanding of individual predictions, while global explanations provide insight into the overall behavior of the model [58]. Our goal is to gain a comprehensive understanding of how each feature contributes to the model’s decision-making in peptide-binding site prediction.
We begin by examining local explanations through the computation of SHAP values for our test dataset. We generate a waterfall plot based on all n-gram occurrence and n-gram composition features for n = 1 and n = 2. A waterfall plot is used for local explanation utilizing the SHAP values calculated for a single instance. Figure 1 illustrates waterfall plots for four distinct instances randomly selected from the test dataset.
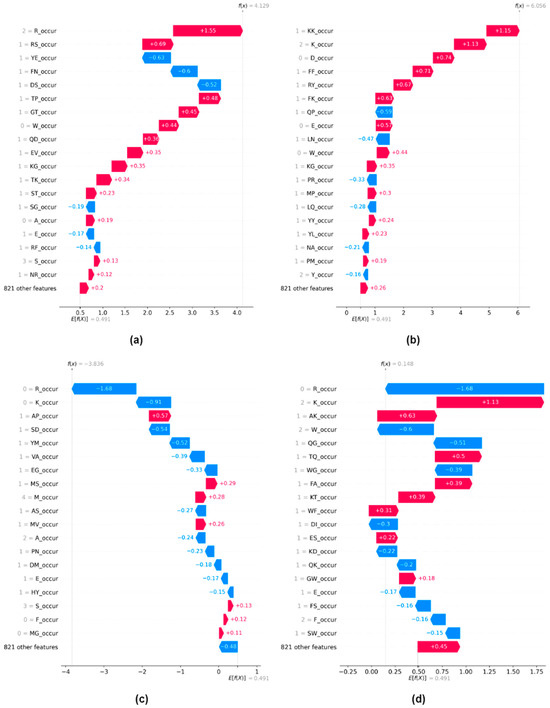
Figure 1.
Local explanations using waterfall plots for n-gram occurrence and n-gram composition features for the values of n = 1 and n = 2. A prediction f(x), for instance, x, and its deviation from E[f(x)] could be measured as the sum of the feature importance values (SHAP values) of individual features. (a) Instance with positive prediction (p = 0.92) (b) Instance with positive prediction (p = 0.97) (c) Instance with negative prediction (p = 0.08) (d) Instance with negative prediction (p = 0.5).
The upper two subfigures (Figure 1a,b) depict instances for which the model shows strong confidence in assigning them to the positive class. The lower two subfigures (Figure 1c,d) show two instances for which the model exhibits strong confidence in assigning them to a negative class. Additionally, the plot provides compelling evidence in favor of utilizing only occurrence-based features in constructing tools for predicting peptide binding sites since among 840 n-gram features, the top 20 contributing features are all generated by calculating the occurrence/composition of amino acids in the sequence. This highlights that n-gram occurrence/composition features are able to provide distinguishable characteristics for peptide-binding site prediction. In addition, amino acids with electrically charged side chains, such as Arginine (R), Lysine (L), Aspartic Acid (D), and Glutamic Acid (E), constantly demonstrate higher values. It shows that charged amino acids are more likely to have an important impact on the binding or non-binding properties of the peptides.
To further investigate the efficacy of amino acid occurrence-based features in our task, we delved into global-level explanations. Leveraging the SHAP library, we computed the global feature importance, which provides a holistic view of each feature’s contribution to the model’s predictions across the entire dataset. We computed the global feature importance by taking the mean absolute SHAP values across all samples. Figure 2 depicts the top 20 features contributing towards model prediction across all samples. In addition, we generated a summary plot using the violine plot type from the SHAP library, which displays the distribution and density of SHAP values for their respective feature. In Figure 3, we present a violine summary plot highlighting the top feature filtered from the pool of 840 n-gram features. Together, these figures strongly indicate the pivotal role of amino acid occurrence-based features in driving the model’s prediction.
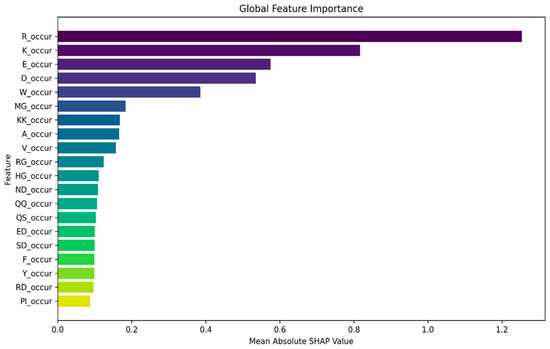
Figure 2.
Global feature importance for n-gram occurrence and n-gram composition features is computed by taking the mean absolute SHAP values across all samples. This plot provides insights into the overall contribution of individual features to the model’s prediction across all samples in the dataset. Only the top 20 contributing features are shown.
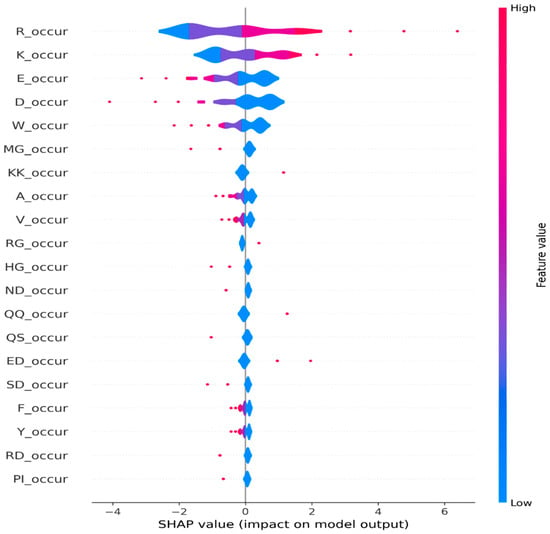
Figure 3.
Global explanations using summary plots from SHAP for n-gram occurrence and n-gram composition features.
Finally, we explore both the local and global explanations for Monogram occurrence features. We observed that using monogram features alone for training our machine learning model yields the best performance in distinguishing protein-binding peptides from non-binding peptides. Figure 4 shows the waterfall plot for four instances from the test dataset. Figure 4a,b depict two samples for which the model predicts positive sites, and Figure 4c,d show samples for which the model is showing strong confidence in assigning them to the negative class.
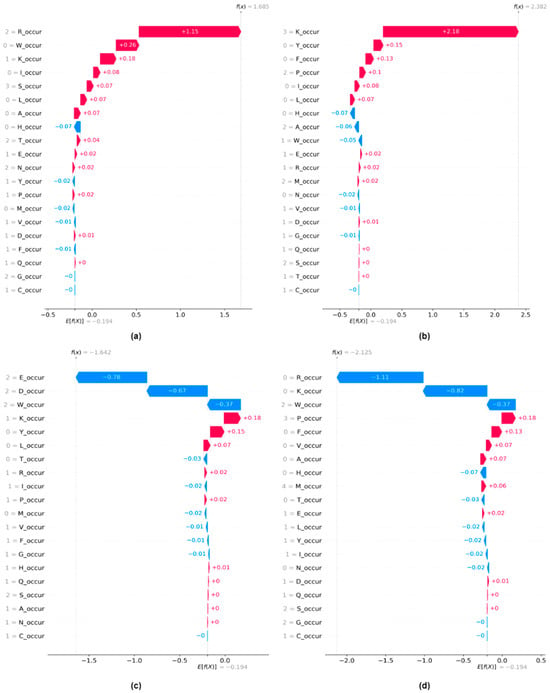
Figure 4.
Local explanations using waterfall plots for monogram occurrence features. A prediction f(x), for instance, x, and its deviation from E[f(x)] could be measured as the sum of the feature importance values (SHAP values) of individual features. (a) Instance with positive prediction (p = 0.91) (b) Instance with positive prediction (p = 0.96) (c) Instance with negative prediction (p = 0.11) (d) Instance with negative prediction (p = 0.06).
Moreover, we generate a global feature importance plot using SHAP values, as depicted in Figure 5. Additionally, we generate a violine summary plot (Figure 6) for monogram features, which provides a compact representation of the distribution and variability of SHAP values for each feature.
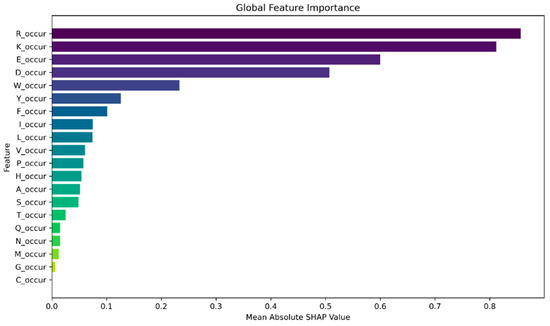
Figure 5.
Global feature importance for monogram occurrence features is computed by taking the mean absolute SHAP values across all samples. Only the top 20 contributing features are shown.
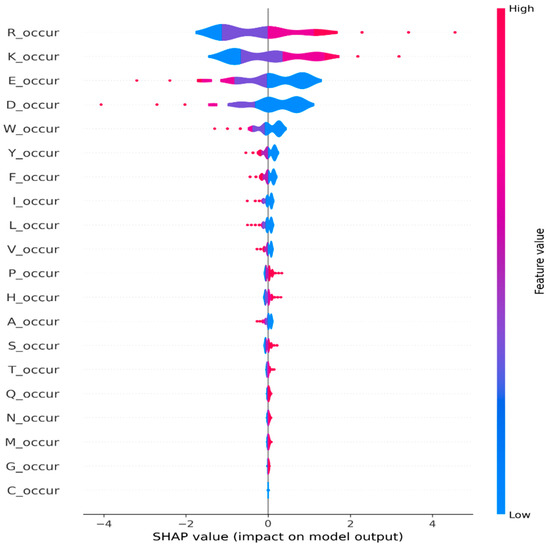
Figure 6.
Global explanations using summary plots from SHAP for monogram occurrence features.
7. Conclusions
Peptides are considered important macromolecules for modulating enzyme function due to their advantages in large chemical diversity, biocompatibility, and well-established methods for library synthesis. Experimental approaches to identify functioning peptides, such as those that are protein binding, are time-consuming and costly. Hence, there is a demand to build a fast and accurate computational approach to tackle this problem. In this study, we proposed PepBind-SVM, a new machine learning-based model to accurately predict protein-binding peptides. To build such a model and make sure that it is able to identify binding patterns in peptides, we generate a new experimentally validated dataset that is released alongside this article.
We then investigate different feature sets and classifiers to identify the most effective combination. Finally, we comprehensively investigate the explainability of our model using SHAP. Our analysis demonstrates the importance of amino acids with electrically charged side chains playing an important role in protein-binding or non-binding properties of the peptides. As mentioned earlier, here we focused on proposing an accurate, explainable machine learning model to tackle this problem. Therefore, our proposed method is relatively simple. However, despite the simplicity and due to the generation of a well-prepared dataset, PepBind-SVM obtained promising results. PepBind-SVM achieves 92.1% prediction accuracy, outperforming other classifiers in predicting protein-binding peptides. For our future study, we aim to investigate other sources of features, such as structural and evolutionary attributes. We also aim to use more sophisticated deep-learning models. PepBind-SVM, its source code, and our newly generated protein-binding peptides are publicly available at https://github.com/MLBC-lab/pepbind-SVM (accessed on 1 September 2024).
Author Contributions
Conceptualization, J.F., I.D. and S.R.I.; methodology, S.M.A., A.B., J.F. and I.D.; software, S.M.A.; validation, S.M.A., I.D., A.B. and S.R.I.; formal analysis, A.B. and S.M.A.; investigation, J.F., I.D., A.B. and S.M.A.; resources, J.F. and I.D.; data curation, J.F.; writing—original draft preparation, I.D., S.M.A., A.B. and S.R.I.; writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
PepBind-SVM, its source code, and our newly generated protein-binding peptides are publicly available at https://github.com/MLBC-lab/pepbind-SVM (accessed on 1 September 2024).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1. [Google Scholar] [CrossRef]
- Medina-Cleghorn, D.; Nomura, D.K. Exploring metabolic pathways and regulation through functional chemoproteomic and metabolomic platforms. Chem. Biol. 2014, 21, 1171–1184. [Google Scholar] [CrossRef][Green Version]
- Drews, J. Drug discovery: A historical perspective. Science 2000, 287, 1960–1964. [Google Scholar] [CrossRef]
- Pathan, S.U.; Kharwar, A.; Ibrahim, M.A.; Singh, S.B.; Bajaj, P. Enzymes as indispensable markers in disease diagnosis. Bioanalysis 2024, 16, 485–497. [Google Scholar] [CrossRef]
- Vegas, A.J.; Fuller, J.H.; Koehler, A.N. Small-molecule microarrays as tools in ligand discovery. Chem. Soc. Rev. 2008, 37, 1385–1394. [Google Scholar] [CrossRef]
- Keppler, A.; Kindermann, M.; Gendreizig, S.; Pick, H.; Vogel, H.; Johnsson, K. Labeling of fusion proteins of O6-alkylguanine-DNA alkyltransferase with small molecules in vivo and in vitro. Methods 2004, 32, 437–444. [Google Scholar] [CrossRef]
- Fu, J.; Cai, K.; Johnston, S.A.; Woodbury, N.W. Exploring peptide space for enzyme modulators. J. Am. Chem. Soc. 2010, 132, 6419–6424. [Google Scholar] [CrossRef]
- Roberts, R.W.; Szostak, J.W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 12297–12302. [Google Scholar] [CrossRef]
- Greving, M.P.; Belcher, P.E.; Diehnelt, C.W.; Gonzalez-Moa, M.J.; Emery, J.; Fu, J.; Johnston, S.A.; Woodbury, N.W. Thermodynamic additivity of sequence variations: An algorithm for creating high affinity peptides without large libraries or structural information. PLoS ONE 2010, 5, e15432. [Google Scholar] [CrossRef]
- Fu, J. Microarray Selection of Cooperative Peptides for Modulating Enzyme Activities. Microarrays 2017, 6, 8. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Qian, Z.; Xiao, Q.; Pei, D. High-throughput screening of one-bead-one-compound libraries: Identification of cyclic peptidyl inhibitors against calcineurin/NFAT interaction. ACS Comb. Sci. 2011, 13, 537–546. [Google Scholar] [CrossRef]
- Guntas, G.; Mansell, T.J.; Kim, J.R.; Ostermeier, M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 11224–11229. [Google Scholar] [CrossRef]
- Blind, M.; Blank, M. Aptamer selection technology and recent advances. Mol. Ther. Nucleic Acids 2015, 4, e223. [Google Scholar] [CrossRef]
- Legutki, J.B.; Zhao, Z.G.; Greving, M.; Woodbury, N.; Johnston, S.A.; Stafford, P. Scalable high-density peptide arrays for comprehensive health monitoring. Nat. Commun. 2014, 5, 4785. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.W.; Rees, D.C. The rise of fragment-based drug discovery. Nat. Chem. 2009, 1, 187–192. [Google Scholar] [CrossRef]
- Dybowski, R. Interpretable machine learning as a tool for scientific discovery in chemistry. New J. Chem. 2020, 44, 20914–20920. [Google Scholar] [CrossRef]
- Goulard Coderc de Lacam, E.; Roux, B.; Chipot, C. Classifying Protein–Protein Binding Affinity with Free-Energy Calculations and Machine Learning Approaches. J. Chem. Inf. Model. 2024, 64, 1081–1091. [Google Scholar] [CrossRef]
- Kozlovskii, I.; Popov, P. Protein–peptide binding site detection using 3D convolutional neural networks. J. Chem. Inf. Model. 2021, 61, 3814–3823. [Google Scholar] [CrossRef]
- Ferdous, S.M.; Mugdha, S.B.S.; Dehzangi, I. New Multi-View Feature Learning Method for Accurate Antifungal Peptide Detection. Algorithms 2024, 17, 247. [Google Scholar] [CrossRef]
- Ahmed, S.; Muhammod, R.; Khan, Z.H.; Adilina, S.; Sharma, A.; Shatabda, S.; Dehzangi, A. ACP-MHCNN: An accurate multi-headed deep-convolutional neural network to predict anticancer peptides. Sci. Rep. 2021, 11, 23676. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Lv, H.; Guo, Y.; Peng, W.; Liu, B. sAMPpred-GAT: Prediction of antimicrobial peptide by graph attention network and predicted peptide structure. Bioinformatics 2023, 39, btac715. [Google Scholar] [CrossRef] [PubMed]
- Brixi, G.; Ye, T.; Hong, L.; Wang, T.; Monticello, C.; Lopez-Barbosa, N.; Vincoff, S.; Yudistyra, V.; Zhao, L.; Haarer, E.; et al. SaLT&PepPr is an interface-predicting language model for designing peptide-guided protein degraders. Commun. Biol. 2023, 6, 1081. [Google Scholar]
- Romero-Molina, S.; Ruiz-Blanco, Y.B.; Mieres-Perez, J.; Harms, M.; Münch, J.; Ehrmann, M.; Sanchez-Garcia, E. PPI-affinity: A web tool for the prediction and optimization of protein–peptide and protein–protein binding affinity. J. Proteome Res. 2022, 21, 1829–1841. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.; Sharma, A.; Dehzangi, I.; Tsunoda, T.; Sattar, A. PepCNN deep learning tool for predicting peptide binding residues in proteins using sequence, structural, and language model features. Sci. Rep. 2023, 13, 20882. [Google Scholar] [CrossRef]
- Azim, S.M.; Sabab, N.H.N.; Noshadi, I.; Alinejad-Rokny, H.; Sharma, A.; Shatabda, S.; Dehzangi, I. Accurately predicting anticancer peptide using an ensemble of heterogeneously trained classifiers. Inform. Med. Unlocked 2023, 42, 101348. [Google Scholar] [CrossRef]
- Wei, L.; Zhou, C.; Su, R.; Zou, Q. PEPred-Suite: Improved and robust prediction of therapeutic peptides using adaptive feature representation learning. Bioinformatics 2019, 35, 4272–4280. [Google Scholar] [CrossRef]
- Kazmirchuk, T.D.D.; Bradbury-Jost, C.; Withey, T.A.; Gessese, T.; Azad, T.; Samanfar, B.; Dehne, F.; Golshani, A. Peptides of a feather: How computation Is taking peptide therapeutics under its wing. Genes 2023, 14, 1194. [Google Scholar] [CrossRef]
- Yan, C.; Zou, X. Predicting peptide binding sites on protein surfaces by clustering chemical interactions. Biophys. J. 2015, 108, 215a. [Google Scholar] [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef]
- Heffernan, R.; Paliwal, K.; Lyons, J.; Dehzangi, A.; Sharma, A.; Wang, J.; Sattar, A.; Yang, Y.; Zhou, Y. Improving prediction of secondary structure, local backbone angles and solvent accessible surface area of proteins by iterative deep learning. Sci. Rep. 2015, 5, 11476. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Heffernan, R.; Paliwal, K.; Lyons, J.; Dehzangi, A.; Sharma, A.; Wang, J.; Sattar, A.; Zhou, Y. SPIDER2: A package to predict secondary structure, accessible surface area, and main-chain torsional angles by deep neural networks. In Prediction of Protein Secondary Structure; Humana Press: New York, NY, USA, 2017; pp. 55–63. [Google Scholar]
- Akbar, S.; Raza, A.; Zou, Q. Deepstacked-AVPs: Predicting antiviral peptides using tri-segment evolutionary profile and word embedding based multi-perspective features with deep stacking model. BMC Bioinform. 2024, 25, 102. [Google Scholar] [CrossRef]
- Wardah, W.; Dehzangi, A.; Taherzadeh, G.; Rashid, M.A.; Khan, M.G.; Tsunoda, T.; Sharma, A. Predicting protein-peptide binding sites with a deep convolutional neural network. J. Theor. Biol. 2020, 496, 110278. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jin, J.; Zou, Q.; Nakai, K.; Wei, L. Predicting protein–peptide binding residues via interpretable deep learning. Bioinformatics 2022, 38, 3351–3360. [Google Scholar] [CrossRef]
- Shanker, S.; Sanner, M.F. Predicting protein–peptide interactions: Benchmarking deep learning techniques and a comparison with focused docking. J. Chem. Inf. Model. 2023, 63, 3158–3170. [Google Scholar] [CrossRef]
- Yin, S.; Mi, X.; Shukla, D. Leveraging machine learning models for peptide–protein interaction prediction. RSC Chem. Biol. 2024, 5, 401–417. [Google Scholar] [CrossRef]
- Devnath, L.; Fan, Z.; Luo, S.; Summons, P.; Wang, D. Detection and visualisation of pneumoconiosis using an ensemble of multi-dimensional deep features learned from Chest X-rays. Int. J. Environ. Res. Public Health 2022, 19, 11193. [Google Scholar] [CrossRef]
- Mishra, R. Support Vector Machines Application for Prediction Binding Elements. In Proceedings of the 2024 IEEE International Conference on Big Data & Machine Learning (ICBDML), Bhopal, India, 24–25 February 2024; pp. 265–269. [Google Scholar]
- Yuan, Q.; Yang, Y. Sequence-based predictions of residues that bind proteins and peptides. In Machine Learning in Bioinformatics of Protein Sequences: Algorithms, Databases and Resources for Modern Protein Bioinformatics; World Scientific Publishing: Singapore, 2023; pp. 237–263. [Google Scholar]
- Ye, J.; Li, A.; Zheng, H.; Yang, B.; Lu, Y. Machine Learning Advances in Predicting Peptide/Protein-Protein Interactions Based on Sequence Information for Lead Peptides Discovery. Adv. Biol. 2023, 7, 2200232. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Fang, G.; Fida, H.; Musleh, S.; Yu, D.J.; Alam, T. iMRSAPred: Improved Prediction of Anti-MRSA Peptides Using Physicochemical and Pairwise Contact-Energy Properties of Amino Acids. ACS Omega 2024, 9, 2874–2883. [Google Scholar] [CrossRef]
- Boltz, K.W.; Gonzalez-Moa, M.J.; Stafford, P.; Johnston, S.A.; Svarovsky, S.A. Peptide microarrays for carbohydrate recognition. Analyst 2009, 134, 650–652. [Google Scholar] [CrossRef]
- Fu, J.; Reinhold, J.; Woodbury, N.W. Peptide-modified surfaces for enzyme immobilization. PLoS ONE 2011, 6, e18692. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, Y.H.; Gromiha, M.M. Application of amino acid occurrence for discriminating different folding types of globular proteins. BMC Bioinform. 2007, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Dehzangi, A.; Sharma, A.; Lyons, J.; Paliwal, K.K.; Sattar, A. A mixture of physicochemical and evolutionary–based feature extraction approaches for protein fold recognition. Int. J. Data Min. Bioinform. 2015, 11, 115–138. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Krissinel, E. On the relationship between sequence and structure similarities in proteomics. Bioinformatics 2007, 23, 717–723. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4765–4774. [Google Scholar]
- Levy, J.J.; Titus, A.J.; Petersen, C.L.; Chen, Y.; Salas, L.A.; Christensen, B.C. MethylNet: An automated and modular deep learning approach for DNA methylation analysis. BMC Bioinform. 2020, 21, 108. [Google Scholar] [CrossRef]
- Yap, M.; Johnston, R.L.; Foley, H.; MacDonald, S.; Kondrashova, O.; Tran, K.A.; Nones, K.; Koufariotis, L.T.; Bean, C.; Pearson, J.V.; et al. Verifying explainability of a deep learning tissue classifier trained on RNA-seq data. Sci. Rep. 2021, 11, 2641. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Fan, A.; Jernite, Y.; Perez, E.; Grangier, D.; Weston, J.; Auli, M. ELI5: Long form question answering. arXiv 2019, arXiv:1907.09190. [Google Scholar]
- Akbar, S.; Ali, F.; Hayat, M.; Ahmad, A.; Khan, S.; Gul, S. Prediction of antiviral peptides using transform evolutionary & SHAP analysis based descriptors by incorporation with ensemble learning strategy. Chemom. Intell. Lab. Syst. 2022, 230, 104682. [Google Scholar]
- Dickinson, Q.; Meyer, J.G. Positional SHAP (PoSHAP) for Interpretation of machine learning models trained from biological sequences. PLoS Comput. Biol. 2022, 18, e1009736. [Google Scholar] [CrossRef]
- Prabhu, H.; Bhosale, H.; Sane, A.; Dhadwal, R.; Ramakrishnan, V.; Valadi, J. Protein feature engineering framework for AMPylation site prediction. Sci. Rep. 2024, 14, 8695. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).