A Visual Mining Approach to Improved Multiple- Instance Learning
Abstract
:1. Introduction
- MILTree—a novel tree layout for multiple-instance data visualization;
- MILTree-Med and MILTree-SI—two new methods for instance prototype selection;
- MILSIPTree—a visual methodology to support multiple-instance data classification.
2. Related Work
3. Background and Related Concepts
3.1. Multiple Instance Learning
3.2. Instance Prototype Selection
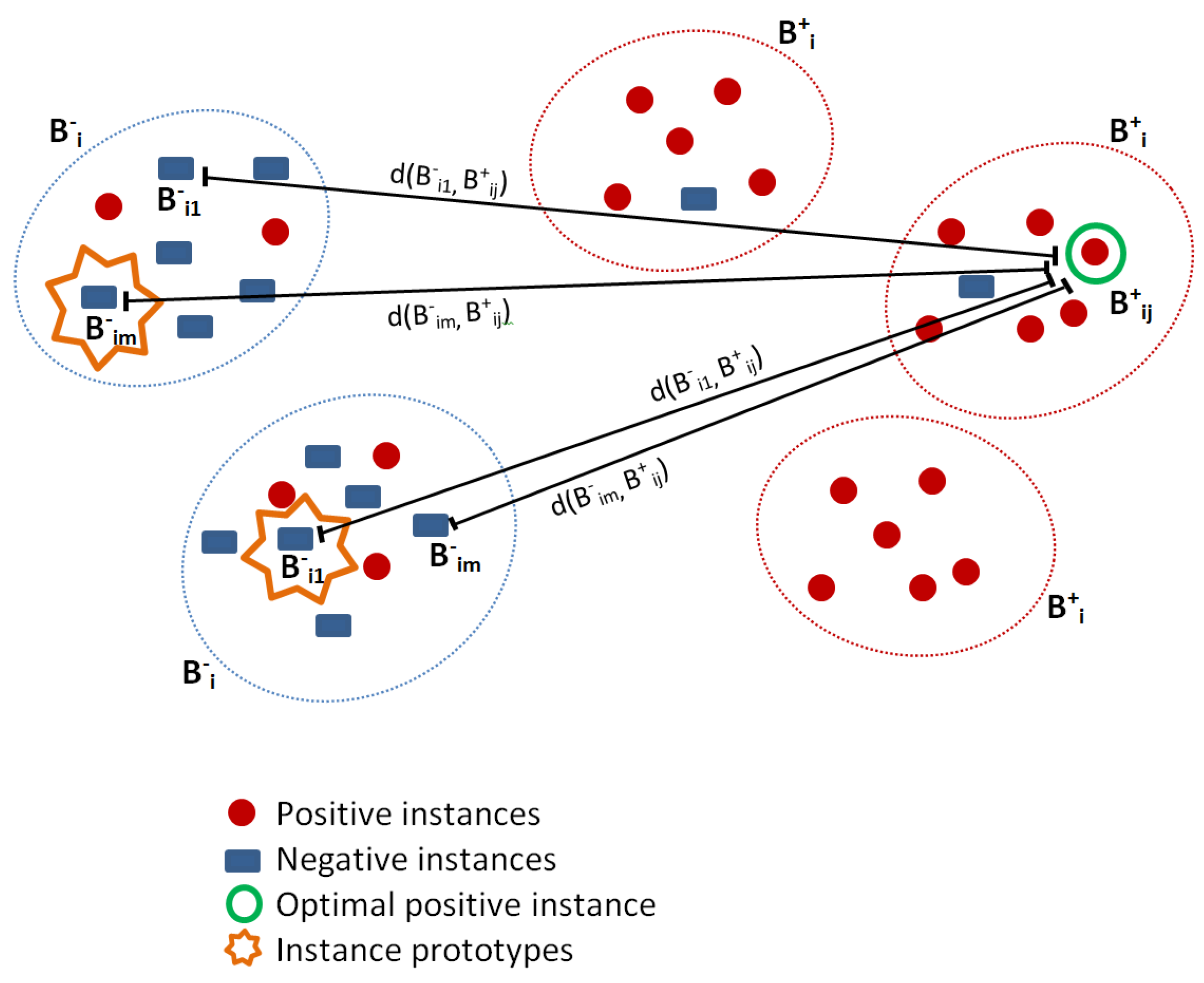
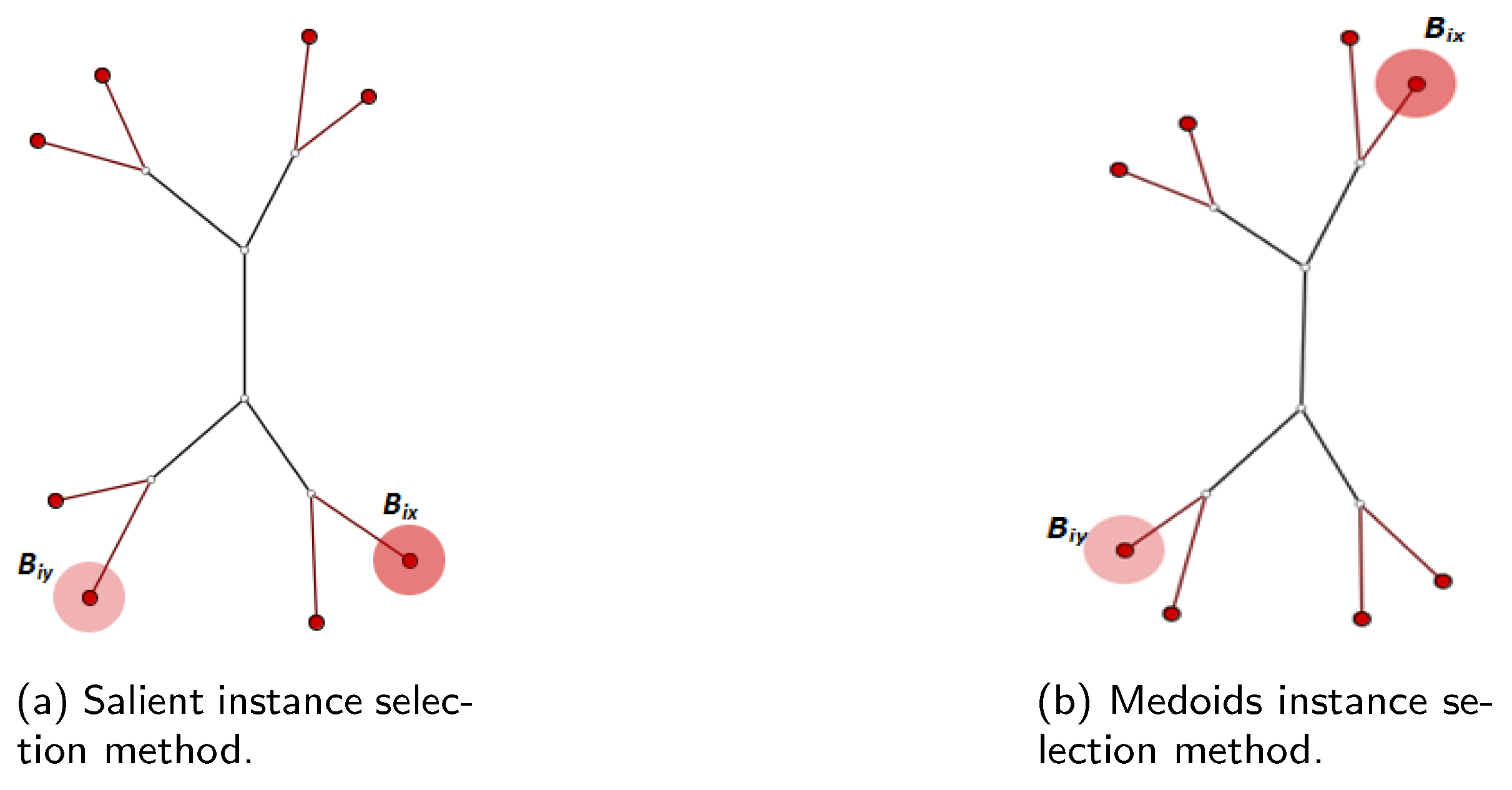
3.2.1. Salient Instance Selection Strategy
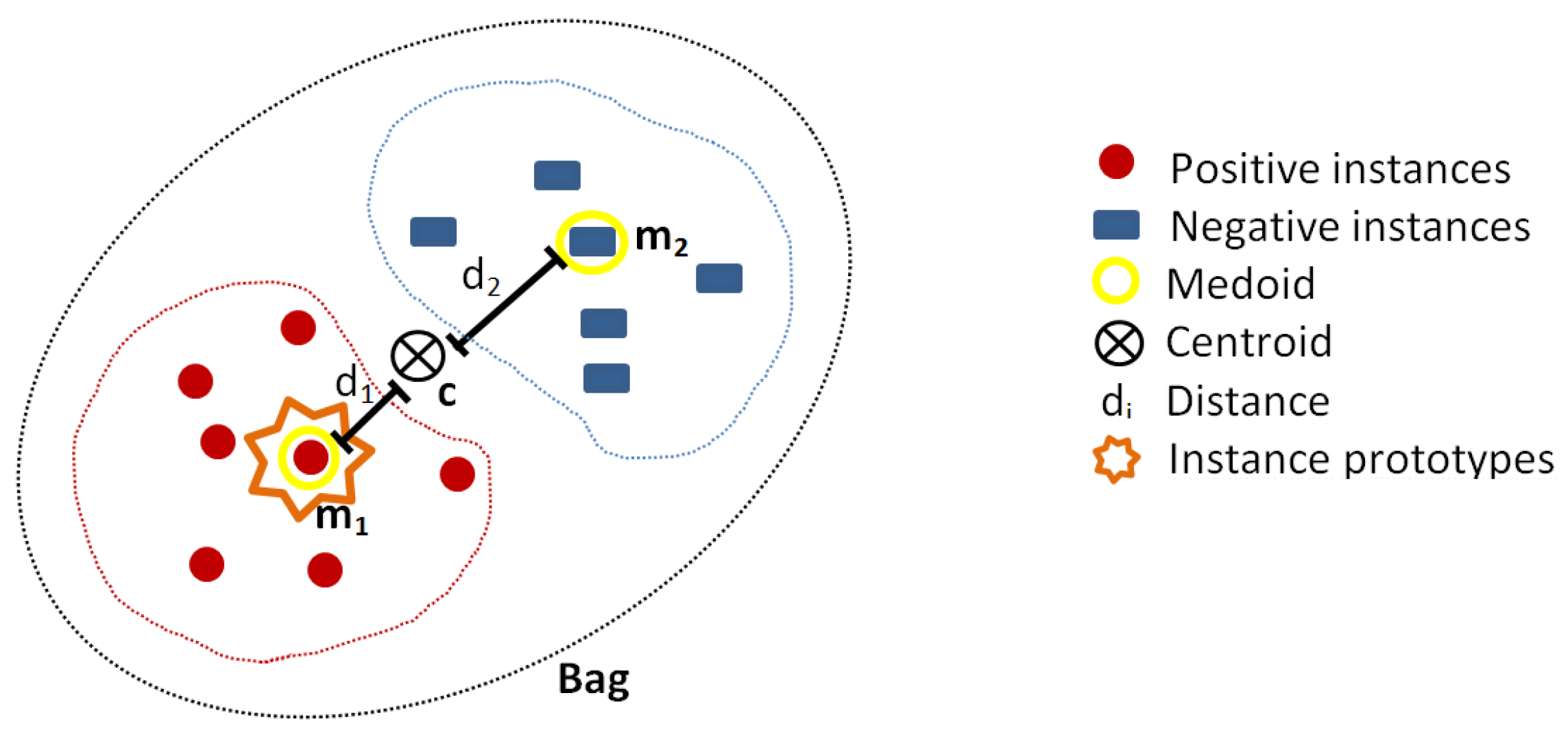
3.2.2. Medoids Instance Selection Strategy
| Algorithm 1: MILSIS: Salience Instance Selection. |
Input: B, SalNum Output: Salient instances (prototypes) T. ; ; //Rough Selection for to do
Compute for each instance in ; //(see Equation (1))Re-sort all instances in in descending order of salience.; Compute and ; //(see Equation (3)) if > and ; then ; ; else if > and then end end end // Fine Selection ; for to do if>then ; // Set of salient instanceselse ; end end Return T; |
4. Visual Multiple-Instance Learning
4.1. Additional Notation
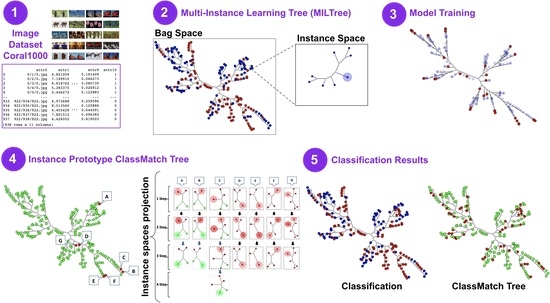
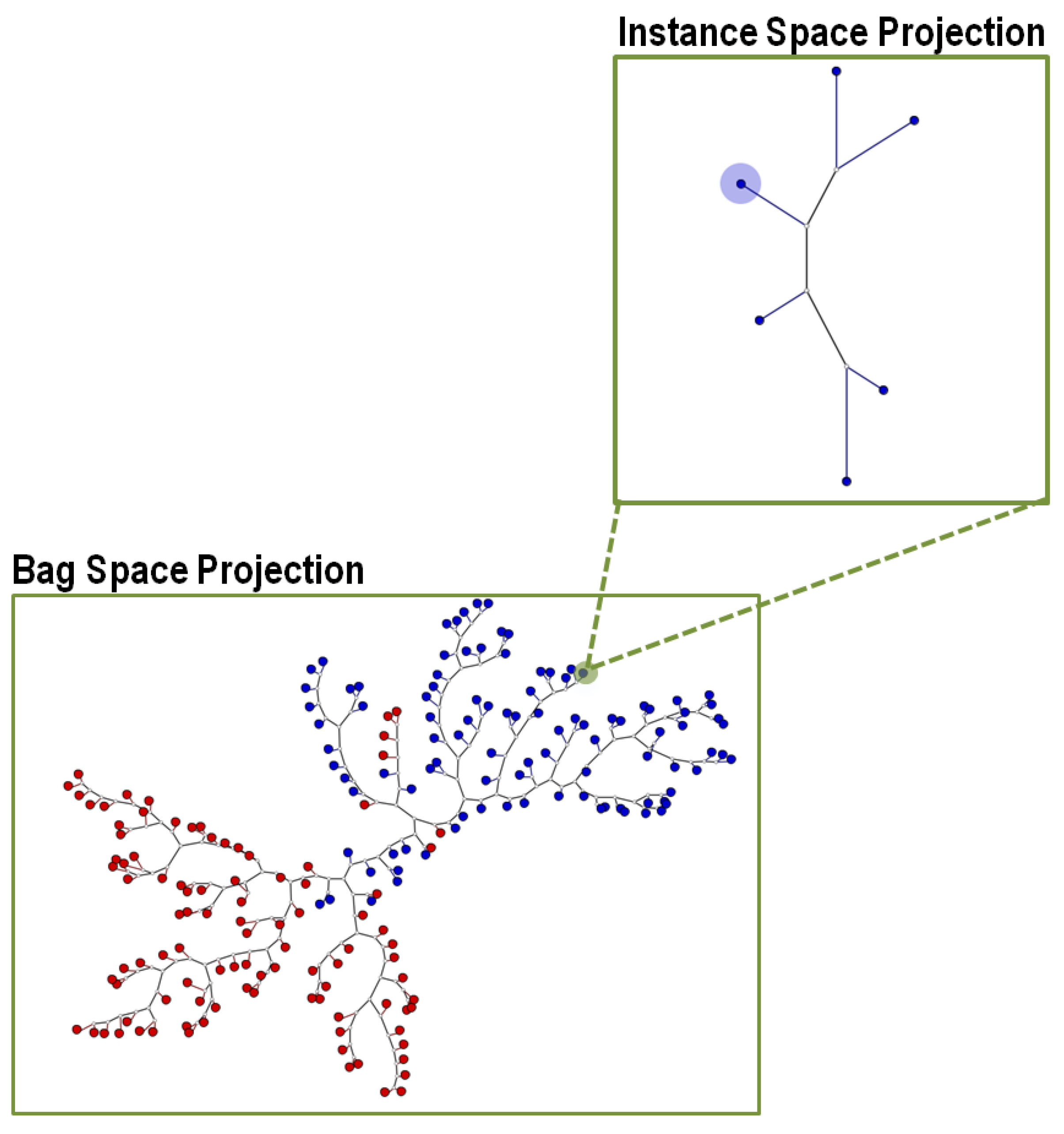
4.2. Creating a Multiple-Instance Tree (MILTree)
| Algorithm 2: NJ tree Algorithm. |
Input: Similarity matrix D. Output: Phylogenetic Neighbor-Joining Tree. ; // Where whiledo
Compute for , and find which is the closest pair of instances ; // (see Equations (4) and (5)). Create a new virtual node u; Compute the lengths of edges and ; //(see Equations (6) and (7)). Replace by u and update the distance matrix D with the new node u ; //(see Equation (8)). Define u as parent of both i and j; ; end |
| Algorithm 3: MILTree Algorithm. |
 |
4.3. Instance Prototype Selection Methods
4.3.1. MILTree-SI
4.3.2. MILTree-Med
4.3.3. Updating Instance Prototypes Using MILTree
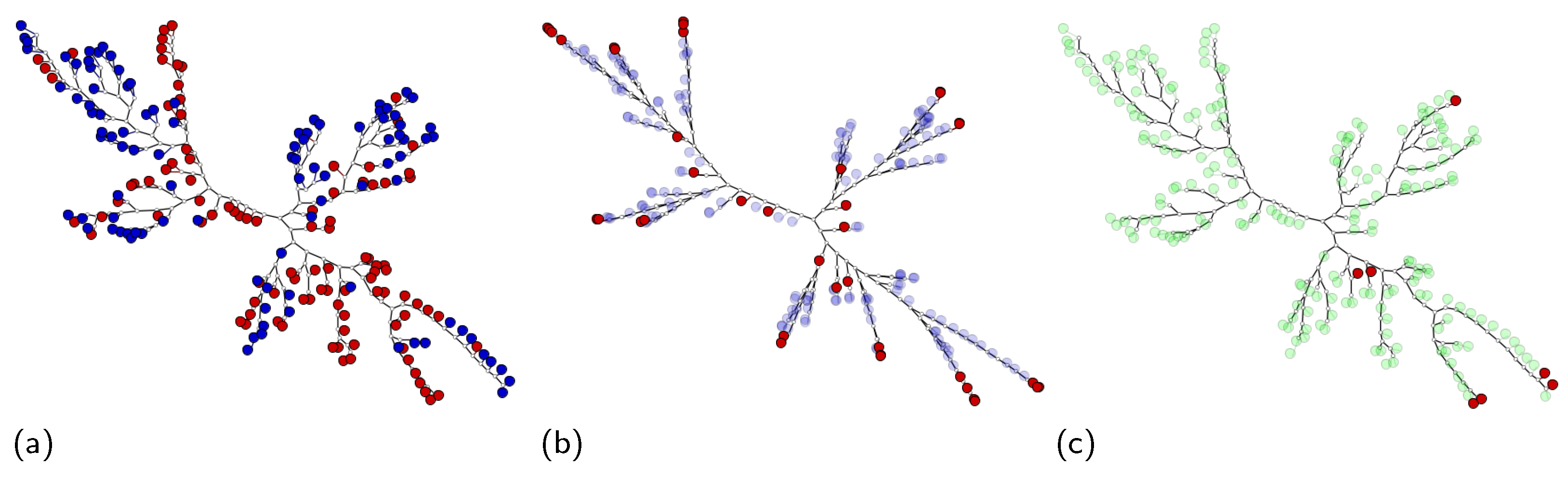
- Prototype highlighting: MILTree highlights the current prototype with a darker color and also , which is the alternative prototype, with a lighter shade. Thus, by inspecting both, the user can validate or update it according to their knowledge by selecting or even another instance in the instance space layout. Figure 4 shows the instance prototypes and projected in the MILTree’s instance space layout. In Figure 4a, the SI selection method is used, and in Figure 4b, the medoids selection is used instead.
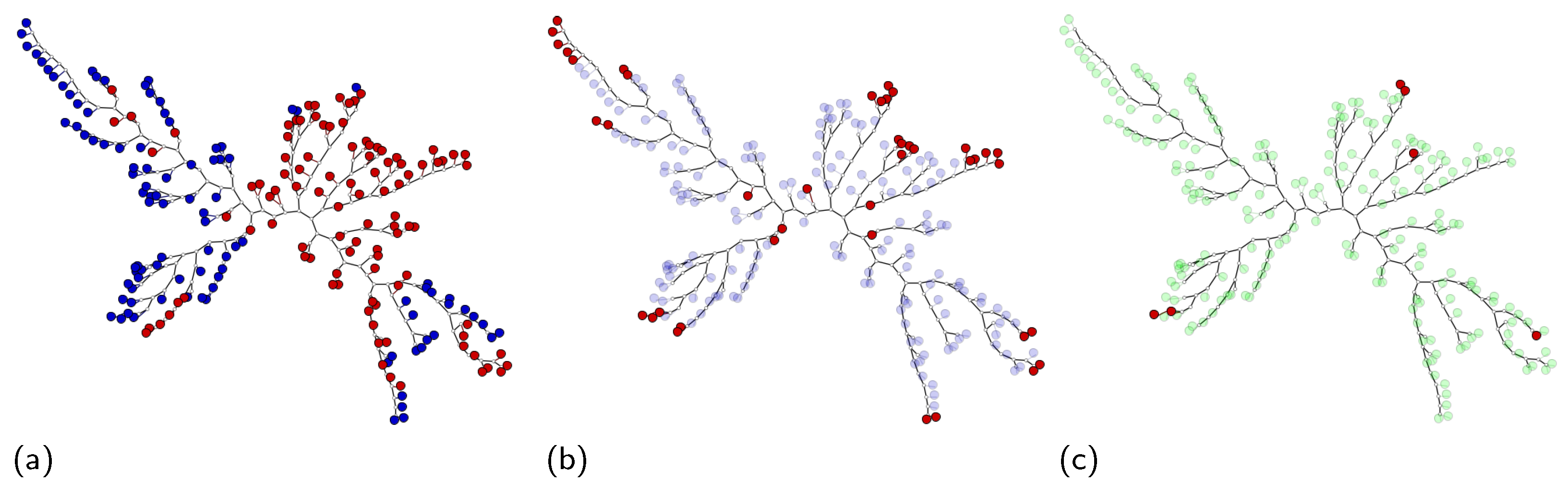
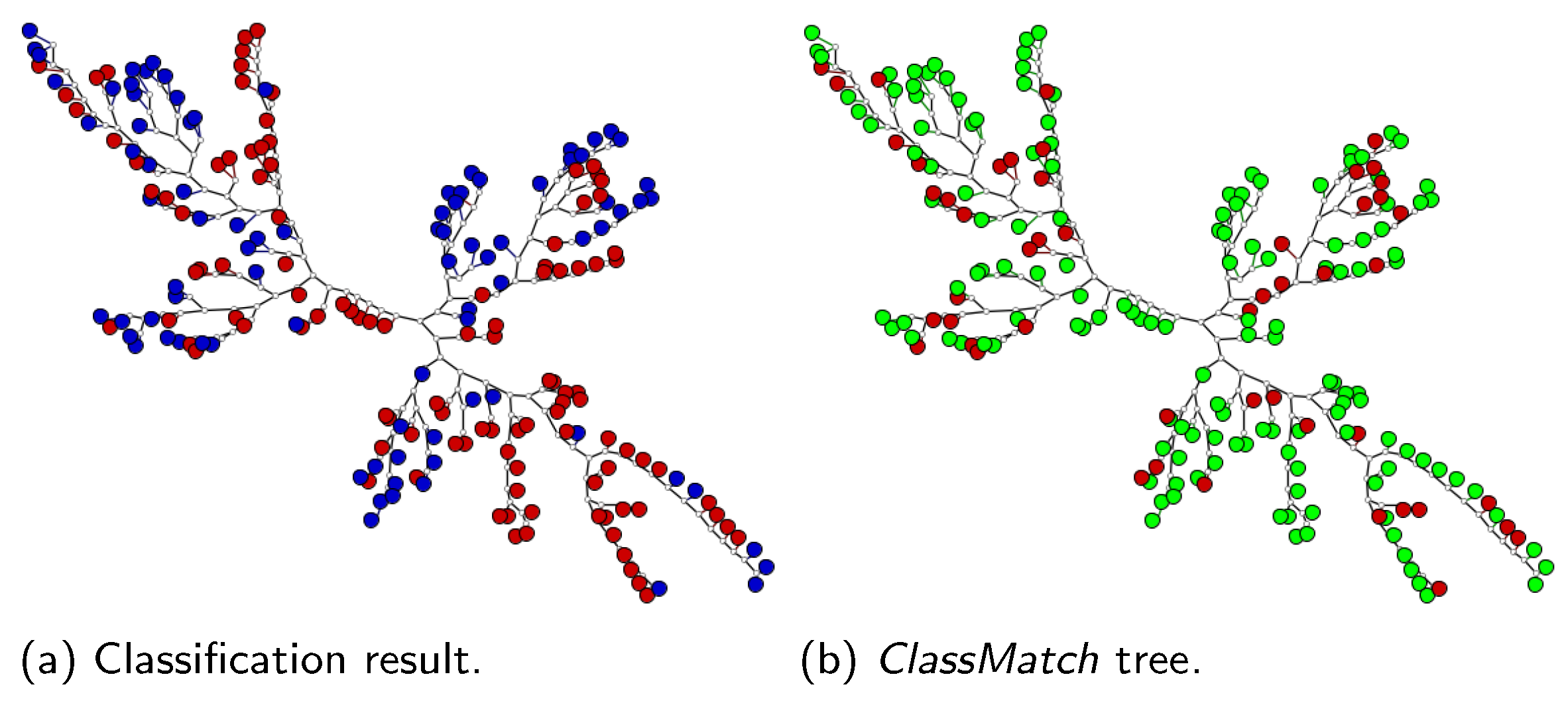
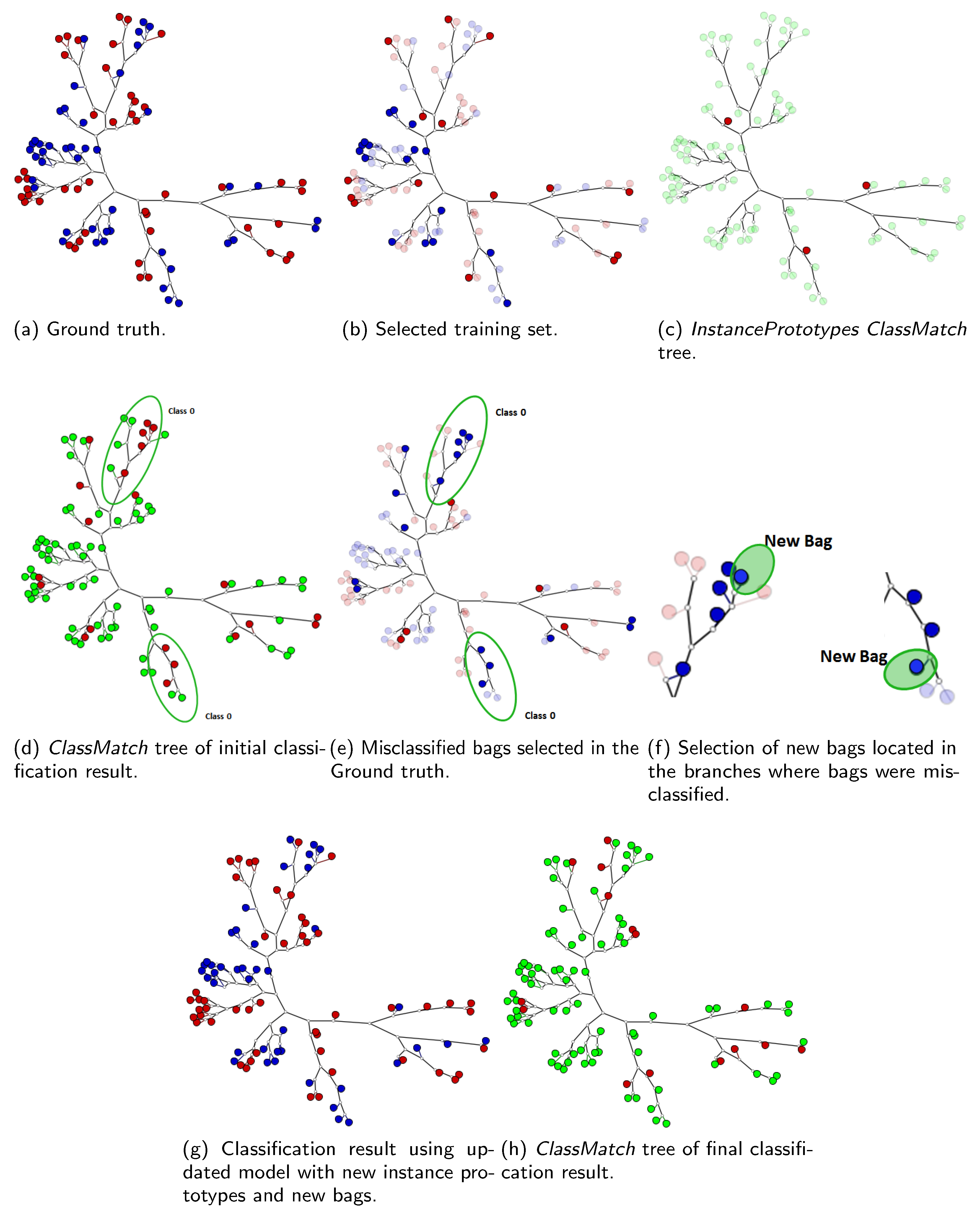
- SVM class match tree: the InstancePrototypes ClassMatch tree uses color to contrast the bags that were misclassified, considering a training or validation set for which the labels are known. A similar approach has been successfully used in [21,33]. In this approach, the instances are used to build an SVM classifier, and the MILTree plots a layout we called InstancePrototypes ClassMatch tree, with colors according to the classification result: pale green for correctly classified and red for misclassified bags. Figure 5a displays the MILTree generated for a subset of the Corel-1000 dataset, where red bags represent positive bags (images of horses) and blue bags represent negative bags (random images from other categories). To find the InstancePrototypes ClassMatch tree for this dataset, we allow users to select a training set to create an SVM classifier. Figure 5b shows the training set that was used to create the classifier; dark red bags are the ones used for training, while the pale blue ones are used as validation/test. Finally, the InstancePrototypes ClassMatch tree shows the classification results (see Figure 5c), where dark red points are misclassified bags, probably with non-representative prototypes. Updating the prototypes will improve the model, as indicated by the results presented later in Section 6.
5. Application of MILTree to Multiple-Instance Learning Scenarios
5.1. Case 1: Instance Space Layout in a Binary Classification Problem
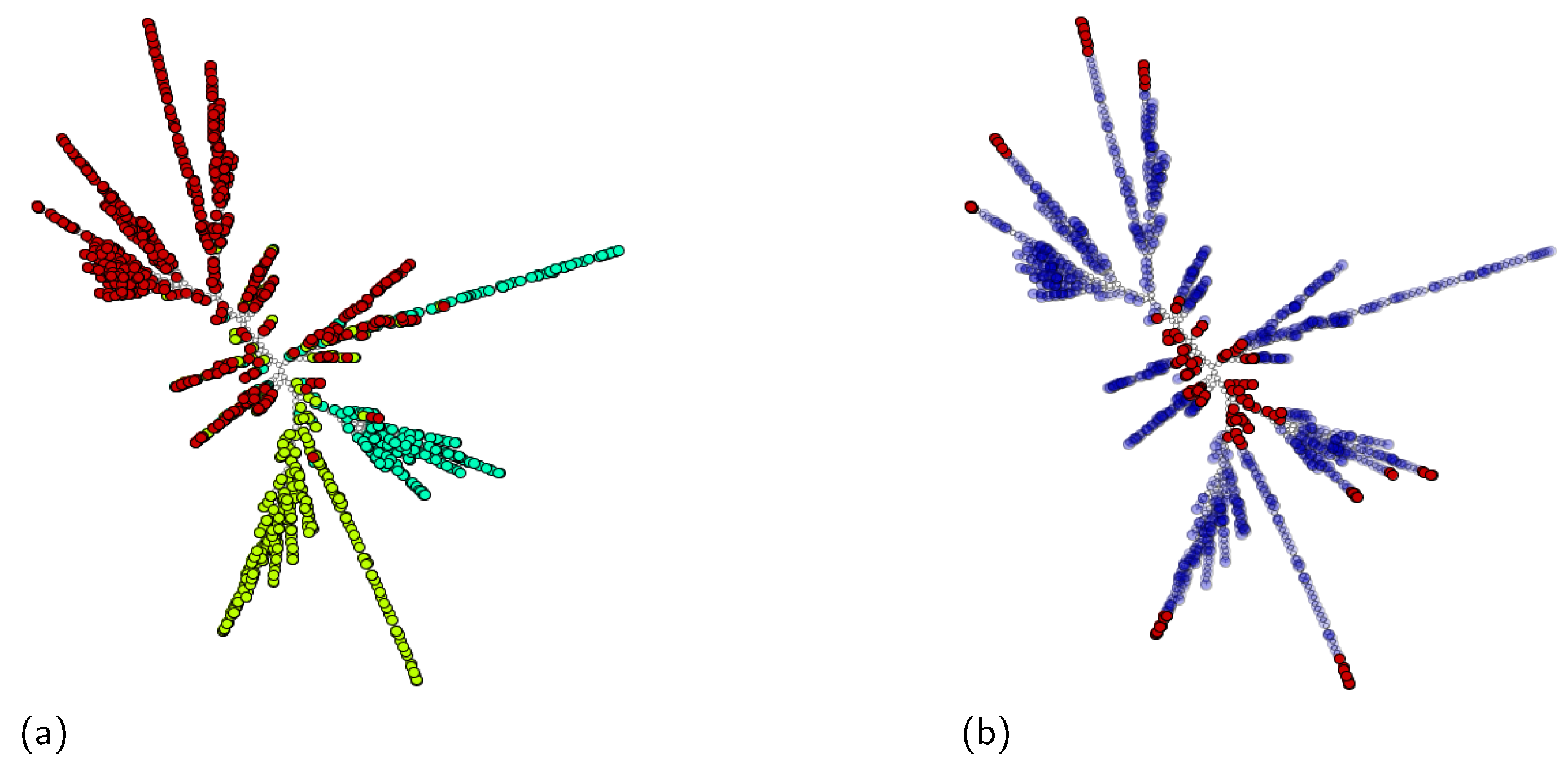
5.2. Case 2: Bag Space Layout and a Multiclass Classification Problem
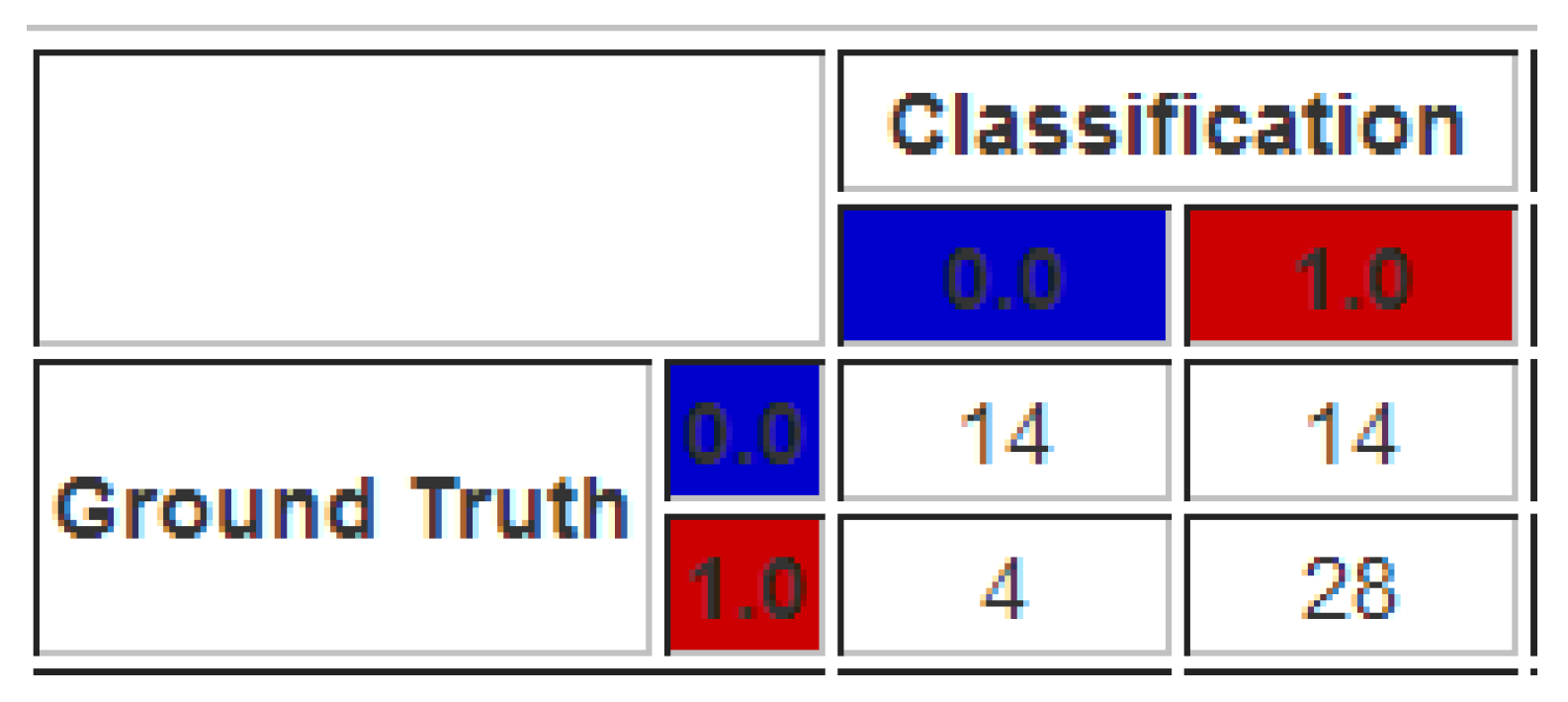
5.3. Case 3: Adding New Bags Using the MILTree Visualization
6. Experiments and Results
6.1. Benchmark Datasets
6.2. Image Classification
6.3. Multiple-Instance Multiclass Datasets
6.4. Scalability Analysis on a MIL Text Classification
6.5. MILTree Layout Bag Positioning
7. User Study
8. Statistical Analysis
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mello, R.F.; Ponti, M.A. Machine Learning: A Practical Approach on the Statistical Learning Theory; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Fu, Z.; Robles-Kelly, A.; Zhou, J. MILIS: Multiple Instance Learning with Instance Selection. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 958–977. [Google Scholar] [CrossRef] [Green Version]
- Dietterich, T.G.; Lathrop, R.H.; Lozano-Perez, T.; Pharmaceutical, A. Solving the Multiple-Instance Problem with Axis-Parallel Rectangles. Artif. Intell. 1997, 89, 31–71. [Google Scholar] [CrossRef] [Green Version]
- Amores, J. MILDE: Multiple instance learning by discriminative embedding. Knowl. Inf. Syst. 2015, 42, 381–407. [Google Scholar] [CrossRef]
- Astorino, A.; Fuduli, A.; Veltri, P.; Vocaturo, E. Melanoma detection by means of multiple instance learning. Interdiscip. Sci. Comput. Life Sci. 2020, 12, 24–31. [Google Scholar] [CrossRef]
- Xiong, D.; Zhang, Z.; Wang, T.; Wang, X. A comparative study of multiple instance learning methods for cancer detection using T-cell receptor sequences. Comput. Struct. Biotechnol. J. 2021, 19, 3255–3268. [Google Scholar] [CrossRef]
- Ray, S.; Craven, M. Supervised versus multiple instance learning: An empirical comparison. In Proceedings of the 22nd International Conference on Machine Learning; ACM: New York, NY, USA, 2005; pp. 697–704. [Google Scholar] [CrossRef]
- Reynolds, D.A.; Quatieri, T.F.; Dunn, R.B. Speaker verification using Adapted Gaussian mixture models. Digit. Signal Process. 2000, 10, 19–41. [Google Scholar] [CrossRef] [Green Version]
- Zafra, A.; Gibaja, E.; Ventura, S. Multiple Instance Learning with MultiObjective Genetic Programming for Web Mining. In Proceedings of the HIS’08, Eighth International Conference on Hybrid Intelligent Systems, Barcelona, Spain, 10–12 September 2008; pp. 513–518. [Google Scholar] [CrossRef]
- Carbonneau, M.A.; Cheplygina, V.; Granger, E.; Gagnon, G. Multiple instance learning: A survey of problem characteristics and applications. Pattern Recognit. 2018, 77, 329–353. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.; Tsochantaridis, I.; Hofmann, T. Support vector machines for multiple-instance learning. In Advances in Neural Information Processing Systems 15; MIT Press: Cambridge, MA, USA, 2003; pp. 561–568. [Google Scholar]
- Fu, Z.; Robles-Kelly, A. An instance selection approach to Multiple instance Learning. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 911–918. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Jiao, J.; Yang, Y.; Wang, B. Multi-instance multi-label learning for automatic tag recommendation. In Proceedings of the 2009 IEEE International Conference on Systems, Man and Cybernetics, San Antonio, TX, USA, 11–14 October 2009; pp. 4910–4914. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, B.; Hao, Z.; Cao, L. A Similarity-Based Classification Framework For Multiple-Instance Learning. IEEE Trans. Cybern. 2014, 44, 500–515. [Google Scholar] [CrossRef]
- Chen, Y.; Bi, J.; Wang, J. MILES: Multiple-Instance Learning via Embedded Instance Selection. Pattern Anal. Mach. Intell. 2006, 28, 1931–1947. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Liu, S.; Huang, Q.; Liu, J.; Tang, X. Salient Instance Selection for Multiple-Instance Learning. In Neural Information Processing; Huang, T., Zeng, Z., Li, C., Leung, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7665, pp. 58–67. [Google Scholar] [CrossRef]
- Ponti, M.A.; da Costa, G.B.P.; Santos, F.P.; Silveira, K.U. Supervised and unsupervised relevance sampling in handcrafted and deep learning features obtained from image collections. Appl. Soft Comput. 2019, 80, 414–424. [Google Scholar] [CrossRef]
- Keim, D.; Kriegel, H.P. Visualization techniques for mining large databases: A comparison. IEEE Trans. Knowl. Data Eng. 1996, 8, 923–938. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Hong, W.; Chen, N.; Li, X.; Liu, W.; Zhang, T. Parallel Filter: A Visual Classifier Based on Parallel Coordinates and Multivariate Data Analysis. In Advanced Intelligent Computing Theories and Applications. With Aspects of Artificial Intelligence; Huang, D.S., Heutte, L., Loog, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4682, pp. 1172–1183. [Google Scholar] [CrossRef]
- Zhang, K.B.; Orgun, M.; Shankaran, R.; Zhang, D. Interactive Visual Classification of Multivariate Data. In Proceedings of the Eleventh International Conference on Machine Learning and Applications (ICMLA 2012), Boca Raton, FL, USA, 12–15 December 2012; Volume 2, pp. 246–251. [Google Scholar] [CrossRef]
- Paiva, J.; Schwartz, W.; Pedrini, H.; Minghim, R. An Approach to Supporting Incremental Visual Data Classification. IEEE Trans. Vis. Comput. Graph. 2015, 21, 4–17. [Google Scholar] [CrossRef]
- Cuadros, A.M.; Paulovich, F.V.; Minghim, R.; Telles, G.P. Point Placement by Phylogenetic Trees and its Application to Visual Analysis of Document Collections. In Proceedings of the 2007 IEEE Symposium on Visual Analytics Science and Technology, Sacramento, CA, USA, 30 October–1 November 2007; pp. 99–106. [Google Scholar]
- Zhang, Q.; Goldman, S.A. EM-DD: An Improved Multiple-Instance Learning Technique. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2001; pp. 1073–1080. [Google Scholar]
- Campello, R.J.; Moulavi, D.; Zimek, A.; Sander, J. Hierarchical density estimates for data clustering, visualization, and outlier detection. ACM Trans. Knowl. Discov. Data 2015, 10, 5. [Google Scholar] [CrossRef]
- Ponti, M.; Nazaré, T.S.; Thumé, G.S. Image quantization as a dimensionality reduction procedure in color and texture feature extraction. Neurocomputing 2016, 173, 385–396. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, Z.; Chen, L.; Guo, B.; Li, W. Featuring, Detecting, and Visualizing Human Sentiment in Chinese Micro-Blog. ACM Trans. Knowl. Discov. Data 2016, 10, 48. [Google Scholar] [CrossRef]
- Tejada, E.; Minghim, R.; Nonato, L.G. On improved projection techniques to support visual exploration of multidimensional data sets. Inf. Vis. 2003, 2, 218–231. [Google Scholar] [CrossRef]
- Ward, M.; Rundensteiner, E. Exploration of Dimensionality Reduction for Text Visualization. In Proceedings of the Coordinated and Multiple Views in Exploratory Visualization (CMV’05), London, UK, 5 July 2005; pp. 63–74. [Google Scholar]
- Jolliffe, I.T. Principal Component Analysis; Springer: New York, NY, USA, 2002. [Google Scholar]
- Cox, T.; Cox, M. Multidimensional Scaling. In Monographs on Statistics and Applied Probability; Chapman & Hall/CRC: Boca Raton, FL, USA, 2001. [Google Scholar]
- Paulovich, F. Mapeamento de dados multi-dimensionais integrando mineração e visualização. PhD Thesis, Universidade de São Paulo, São Paulo, Butanta, 2008. [Google Scholar]
- Joia, P.; Paulovich, F.; Coimbra, D.; Cuminato, J.; Nonato, L. Local Affine Multidimensional Projection. IEEE Trans. Vis. Comput. Graph. 2011, 17, 2563–2571. [Google Scholar] [CrossRef] [PubMed]
- Paiva, J.; Florian, L.; Pedrini, H.; Telles, G.; Minghim, R. Improved Similarity Trees and their Application to Visual Data Classification. IEEE Trans. Vis. Comput. Graph. 2011, 17, 2459–2468. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Zhou, Z.H. Multi-instance clustering with applications to multi-instance prediction. Appl. Intell. 2009, 31, 47–68. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Zhang, M.L. Multi-instance multilabel learning with application to scene classification. In Advances in Neural Information Processing Systems 19; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Lichman, M. UCI Machine Learning Repository. 2013. Available online: https://archive.ics.uci.edu/ml/index.php (accessed on 20 October 2021).
- Chen, Y.; Wang, J.Z. Image Categorization by Learning and Reasoning with Regions. J. Mach. Learn. Res. 2004, 5, 913–939. [Google Scholar]
- Li, W.J.; Yeung, D.Y. MILD: Multiple-Instance Learning via Disambiguation. IEEE Trans. Knowl. Data Eng. 2010, 22, 76–89. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.S.; Wu, J.; Zhou, Z.H. Scalable Multi-instance Learning. In Proceedings of the 2014 IEEE International Conference on Data Mining, Shenzhen, China, 14–17 December 2014; pp. 1037–1042. [Google Scholar] [CrossRef] [Green Version]
- Frank, E.T.; Xu, X. Applying Propositional Learning Algorithms to Multi-Instance Data; Technical Report; University of Waikato: Hamilton, NZ, USA, 2003. [Google Scholar]
- Xu, X. Statistical Learning in Multiple Instance Problems. Master’s Thesis, The University of Waikato, Hamilton, New Zealand, June 2003. [Google Scholar]
- Freund, Y.; Schapire, R.E. Experiments with a New Boosting Algorithm. In Proceedings of the Thirteenth International Conference on Machine Learning (ICML 1996), Bari, Italy, 3–6 July 1996; pp. 148–156. [Google Scholar]
- García, S.; Fernández, A.; Luengo, J.; Herrera, F. Advanced nonparametric tests for multiple comparisons in the design of experiments in computational intelligence and data mining: Experimental analysis of power. Inf. Sci. 2010, 180, 2044–2064. [Google Scholar] [CrossRef]
- Yu, F.X.; Choromanski, K.; Kumar, S.; Jebara, T.; Chang, S.F. On Learning from Label Proportions. arXiv 2014, arXiv:1402.5902. [Google Scholar]
- Stolpe, M.; Morik, K. Learning from label proportions by optimizing cluster model selection. In Machine Learning and Knowledge Discovery in Databases; Springer: Berlin/Heidelberg, Germany, 2011; pp. 349–364. [Google Scholar]


| Bags | Instances | ||||
|---|---|---|---|---|---|
| Dataset | Total | Pos./Neg. | Total | Min/Max | Dim |
| Musk1 | 92 | 47/45 | 476 | 2/40 | 166 |
| Musk2 | 102 | 39/63 | 6598 | 1/1044 | 166 |
| Bags | Instances | ||||
|---|---|---|---|---|---|
| Dataset | Total | Pos./Neg. | Total | Avg. Inst./Bag | Dim |
| Elephant | 200 | 100/100 | 1391 | 6.96 | 230 |
| Fox | 200 | 100/100 | 1220 | 6.10 | 230 |
| Tiger | 200 | 100/100 | 1320 | 6.60 | 230 |
| MILTree-Med | MILTree-SI | |||||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Accur | Prec | Recall | F1 | Accur | Prec | Recall | F1 |
| Musk1 | 83.2 | 83.23 | 81.7 | 0.82 | 83.2 | 82.4 | 81.7 | 0.82 |
| Musk2 | 91.8 | 91.4 | 91.4 | 0.91 | 85.4 | 84.4 | 84.3 | 0.84 |
| Elephant | 83.1 | 81.7 | 81.6 | 0.82 | 81.4 | 79.4 | 79.4 | 0.79 |
| Fox | 72.7 | 68.3 | 68.3 | 0.68 | 72.7 | 68.3 | 68.3 | 0.68 |
| Tiger | 83.0 | 82.0 | 81.4 | 0.82 | 82.9 | 83.4 | 81.4 | 0.82 |
| Method | Musk1 | Musk2 | Elephant | Fox | Tiger | Avg. |
|---|---|---|---|---|---|---|
| MILTree-Med | 83.2 | 91.8 | 83.1 | 72.7 | 83.0 | 82.8 |
| MILTree-SI | 82.3 | 85.4 | 81.4 | 72.7 | 82.9 | 81.1 |
| EM-DD | 84.8 | 84.9 | 78.3 | 56.1 | 72.1 | 75.2 |
| MI-SVM | 77.9 | 84.3 | 73.1 | 58.8 | 66.6 | 72.1 |
| mi-SVM | 87.4 | 83.6 | 80 | 57.9 | 78.9 | 77.6 |
| DD-SVM | 85.8 | 91.3 | 83.5 | 56.6 | 77.2 | 79.0 |
| MILD-B | 88.3 | 86.8 | 82.9 | 55.0 | 75.8 | 77.8 |
| MILIS | 88.6 | 91.1 | - | - | - | - |
| MILES | 86.3 | 87.7 | 84.1 | 63.0 | 80.7 | 80.4 |
| MILSIS | 90.1 | 85.6 | 81.8 | 66.4 | 80.0 | 80.9 |
| MILDE | 87.1 | 91.0 | 85 | 66.5 | 83.0 | 82.5 |
| Measures | |||||||
|---|---|---|---|---|---|---|---|
| Category ID | Inst/Bag | Accur | Prec | Recall | F1 | Proto | AddBags |
| Category0 | 4.84 | 75.96 | 72.68 | 72.67 | 0.73 | 7 | 0 |
| Category1 | 3.54 | 79.07 | 78.74 | 76.74 | 0.78 | 3 | 2 |
| Category2 | 3.1 | 78.97 | 79.16 | 76.67 | 0.78 | 6 | 3 |
| Category3 | 7.59 | 91.25 | 90.89 | 90.85 | 0.91 | 4 | 0 |
| Category4 | 2.00 | 78.4 | 79.68 | 75.95 | 0.78 | 4 | 0 |
| Category5 | 3.02 | 81.91 | 83.21 | 80.26 | 0.82 | 4 | 1 |
| Category6 | 4.46 | 89.09 | 88.68 | 88.49 | 0.88 | 1 | 0 |
| Category7 | 3.89 | 84.23 | 83.62 | 82.91 | 0.83 | 6 | 2 |
| Category8 | 3.38 | 81.19 | 79.46 | 79.25 | 0.79 | 1 | 1 |
| Category9 | 7.24 | 81.26 | 81.26 | 79.39 | 0.8 | 1 | 0 |
| Measures | ||||||||
|---|---|---|---|---|---|---|---|---|
| Category ID | Inst/Bag | Accur | Prec | Recall | F1 | Proto | AddProto | AddBags |
| Category0 | 4.84 | 68.14 | 62.24 | 62.11 | 0.62 | 0 | 11 | 2 |
| Category1 | 3.54 | 75.82 | 74.94 | 72.67 | 0.74 | 3 | 0 | 0 |
| Category2 | 3.1 | 76.47 | 73.33 | 73.33 | 0.73 | 3 | 0 | 2 |
| Category3 | 7.59 | 72.88 | 68.94 | 68.63 | 0.69 | 0 | 12 | 0 |
| Category4 | 2.00 | 83.65 | 84.99 | 82.28 | 0.84 | 4 | 0 | 1 |
| Category5 | 3.02 | 80.86 | 80.84 | 78.95 | 0.8 | 0 | 14 | 0 |
| Category6 | 4.46 | 89.09 | 88.58 | 88.46 | 0.89 | 1 | 0 | 1 |
| Category7 | 3.89 | 80.03 | 78.28 | 77.85 | 0.78 | 0 | 6 | 0 |
| Category8 | 3.38 | 75.21 | 71.7 | 71.7 | 0.72 | 0 | 0 | 2 |
| Category9 | 7.24 | 75.74 | 72.39 | 72.39 | 0.72 | 0 | 6 | 0 |
| Datasets | EMDD | mi-SVM | MI-SVM | DD-SVM | SMILES | MILTree-SI | MILTree-Med |
|---|---|---|---|---|---|---|---|
| Cat0 | 68.7 | 71.1 | 69.6 | 70.9 | 72.4 | 68.1 | 76.0 |
| Cat1 | 56.7 | 58.7 | 56.4 | 58.5 | 62.7 | 75.8 | 79.1 |
| Cat2 | 65.1 | 67.9 | 66.9 | 68.6 | 69.6 | 76.5 | 79.0 |
| Cat3 | 85.1 | 88.6 | 84.9 | 85.2 | 90.1 | 72.9 | 91.3 |
| Cat4 | 96.2 | 94.8 | 95.3 | 96.9 | 96.6 | 83.7 | 78.4 |
| Cat5 | 74.2 | 80.4 | 74.4 | 78.2 | 80.5 | 80.9 | 81.9 |
| Cat6 | 77.9 | 82.5 | 82.7 | 77.9 | 83.3 | 89.1 | 89.1 |
| Cat7 | 91.4 | 93.4 | 92.1 | 94.4 | 94.7 | 80.3 | 84.2 |
| Cat8 | 70.9 | 72.5 | 67.2 | 71.8 | 73.8 | 75.2 | 81.2 |
| Cat9 | 80.2 | 84.6 | 83.4 | 84.7 | 84.9 | 75.8 | 81.3 |
| Method | 1000-Corel | 2000-Corel |
|---|---|---|
| MILTree-Med | 93.1 | 93.9 |
| MILTree-SI | 90.3 | 93.9 |
| MI-SVM | 75.1 | 54.6 |
| mi-SVM | 76.4 | 53.7 |
| DD-SVM | 81.5 | 67.5 |
| MILIS | 83.8 | 70.1 |
| MILES | 82.3 | 68.7 |
| MILDE | - | 74.8 |
| Dataset | Bags | Instances | Dimensions |
|---|---|---|---|
| Components | 423 | 9104 | 200 |
| Functions | 443 | 9387 | 200 |
| Processes | 757 | 25,181 | 200 |
| Total | 1623 | 34,569 | 600 |
| Method | Biocreative |
|---|---|
| MILTree-Med | 99.1 |
| MILTree-SI | 96.3 |
| MI-SVM | 90.9 |
| EM-DD | 91.0 |
| DD | 90.9 |
| MIWrapper | 90.5 |
| TLDSimple | 85.0 |
| MIBoost | 90.5 |
| Cat3 | |||
|---|---|---|---|
| External Bags | Internal Bags | Combined Bags | |
| Matching Bags | 104 (68%) | 134 (73.1%) | 137 (89.5%) |
| Non-Matching Bags | 49 (32%) | 19 (26.9%) | 16 (10.5%) |
| Accuracy | 72.16% | 88.31% | 90.06% |
| Precision | 71.21% | 87.74% | 89.58% |
| Recall | 67.97% | 87.58% | 89.54% |
| Cat6 | |||
| External Bags | Internal Bags | Combined Bags | |
| Matching Bags | 134 (85.9%) | 114 (73.1%) | 139 (89.1%) |
| Non-Matching Bags | 22 (14.1%) | 42 (26.9%) | 17 (10.9%) |
| Accuracy | 86.82% | 76.28% | 89.66% |
| Precision | 85.93% | 73.07% | 89.28% |
| Recall | 85.9% | 73.08% | 89.1% |
| No. | Questions |
|---|---|
| 1 | Is it possible to clearly identify the classes using MILTree? |
| 2 | Does the browsing through MILTree (Bags and Instances projection space) conduce the user to better understand the structure of multiple-instance data? |
| 3 | Is the Prototypes ClassMatch tree useful for identifying misclassified bags? |
| 4 | Is the ClassMatch tree useful for discovering new bags that help to improve or update the model? |
| 5 | Do you feel that MILTree provides useful support in the multiple-instance classification process? |
| Questions | Grade |
|---|---|
| Question 1 | 3.2 |
| Question 2 | 2.8 |
| Question 3 | 3 |
| Question 4 | 2.4 |
| Question 5 | 3 |
| Participants | ||||||
|---|---|---|---|---|---|---|
| Task | User1 | User2 | User3 | User4 | User5 | Average |
| Corel-300 | 85.87 | 87.28 | 85.47 | 87.42 | 84.75 | 86.16 |
| Method | Ranking |
|---|---|
| MILTree-Med | 2.3571 |
| MILDE | 3.0714 |
| MILES | 3.4285 |
| MILTree-SI | 3.5714 |
| DD-SVM | 4.5714 |
| mi-SVM | 5.5714 |
| EMDD | 6.2142 |
| MI-SVM | 7.2142 |
| MT-Med | MT-SI | MI-SVM | mi-SVM | EMDD | DD-SVM | MILES | MILDE | |
|---|---|---|---|---|---|---|---|---|
| MT-Med | 0.000 | 1.112 | 12.94 | 8.693 | 12.05 | 5.450 | 4.530 | 2.825 |
| MT-SI | −1.112 | 0.000 | 11.83 | 7.580 | 10.94 | 4.338 | 3.418 | 1.713 |
| MI-SVM | −12.94 | −11.83 | 0.000 | −4.250 | −0.895 | −7.492 | −8.412 | −10.12 |
| mi-SVM | −8.693 | −7.580 | 4.250 | 0.000 | 3.355 | −3.242 | −4.163 | −5.867 |
| EMDD | −12.05 | −10.94 | 0.895 | −3.355 | 0.000 | −6.597 | −7.518 | −9.222 |
| DD-SVM | −5.450 | −4.338 | 7.492 | 3.242 | 6.597 | 0.000 | −0.920 | −2.625 |
| MILES | −4.530 | −3.418 | 8.412 | 4.163 | 7.518 | 0.920 | 0.000 | −1.705 |
| MILDE | −2.825 | −1.713 | 10.12 | 5.867 | 9.222 | 2.625 | 1.705 | 0.000 |
| i | Method | Holm | Li |
|---|---|---|---|
| 7 | MI-SVM | 0.001 | 0.022 |
| 6 | EMDD | 0.001 | 0.022 |
| 5 | mi-SVM | 0.001 | 0.022 |
| 4 | DD-SVM | 0.010 | 0.022 |
| 3 | MILTree-SI | 0.354 | 0.022 |
| 2 | MILES | 0.413 | 0.022 |
| 1 | MILDE | 0.585 | 0.050 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castelo, S.; Ponti, M.; Minghim, R. A Visual Mining Approach to Improved Multiple- Instance Learning. Algorithms 2021, 14, 344. https://doi.org/10.3390/a14120344
Castelo S, Ponti M, Minghim R. A Visual Mining Approach to Improved Multiple- Instance Learning. Algorithms. 2021; 14(12):344. https://doi.org/10.3390/a14120344
Chicago/Turabian StyleCastelo, Sonia, Moacir Ponti, and Rosane Minghim. 2021. "A Visual Mining Approach to Improved Multiple- Instance Learning" Algorithms 14, no. 12: 344. https://doi.org/10.3390/a14120344
APA StyleCastelo, S., Ponti, M., & Minghim, R. (2021). A Visual Mining Approach to Improved Multiple- Instance Learning. Algorithms, 14(12), 344. https://doi.org/10.3390/a14120344