A Two-Phase Approach for Semi-Supervised Feature Selection
Abstract
1. Introduction
- i.
- To find a subset of features that has maximum relevance and minimum redundancy (abbreviated to MRmr herein) by using the correlation coefficient. For this purpose, an algorithm (Algorithm 1) is presented to maintain a balance between the features with high relevance and the features with minimum redundancy.
- ii.
- To determine a small feature subset that produces high classification accuracy on a supervised classifier to minimize time and complexity of implementing the method.
- iii.
- The proposed method aims to demonstrate the idea that if we have a pair of two clusters that are almost identical or much closer to each other, if we know the class of a cluster of the pair, the same class can be assigned to the other cluster of the pair.
- iv.
- The class or labels of all patterns can be determined using the proposed novel approach, which will save time or cost in collecting patterns for each pattern in the dataset otherwise.
- v.
- The proposed method is a novel concept and can be applied to various real datasets.
2. Existing Feature Selection Techniques
3. Preliminaries of the Methods Used in the Proposed Approach
- P1: Simple PNN method: The inputs fed in the input layer generate PDs in the successive layers [53].
- P2: RCPNN with gradient descent [53]: A reduced and comprehensible polynomial neural network (RCPNN) model generates PDs for the first layer of the basic PNN model, and the outputs of these PDs along with the inputs are fed to the single-layer feed-forward neural network. The network has been trained using gradient descent.
- P3: RCPNN with particle swarm optimization (PSO): This method is the same as the RCPNN except that the network is trained using particle swarm optimization (PSO) [54] instead of the gradient descent technique.
- P4: Condensed PNN with swarm intelligence: In this paper, Dehuri et al. [55] proposed a condensed polynomial neural network using swarm intelligence for the classification task. The model generates PDs for a single layer of the basic PNN model. Discrete PSO (DPSO) selects the optimal set of PDs and input features, which are fed to the hidden layer. Further, the model optimizes the weight vectors using the continuous PSO (CPSO)technique [55].
- P5: All PDs with 50% training used in the proposed scheme of [12].
- P6: All PDs with 80% training used in the proposed scheme of [12].
- P7: Only the best 50% PDs with 50% training used in the proposed scheme of [12].
- P8: Only the best 50% PDs with 80% training used in the proposed scheme of [12].
- P9: Saxena et al. [29] proposed four methods for feature selection in an unsupervised manner by using the GA. The proposed methods also preserve the topology of the dataset despite reducing redundant features.
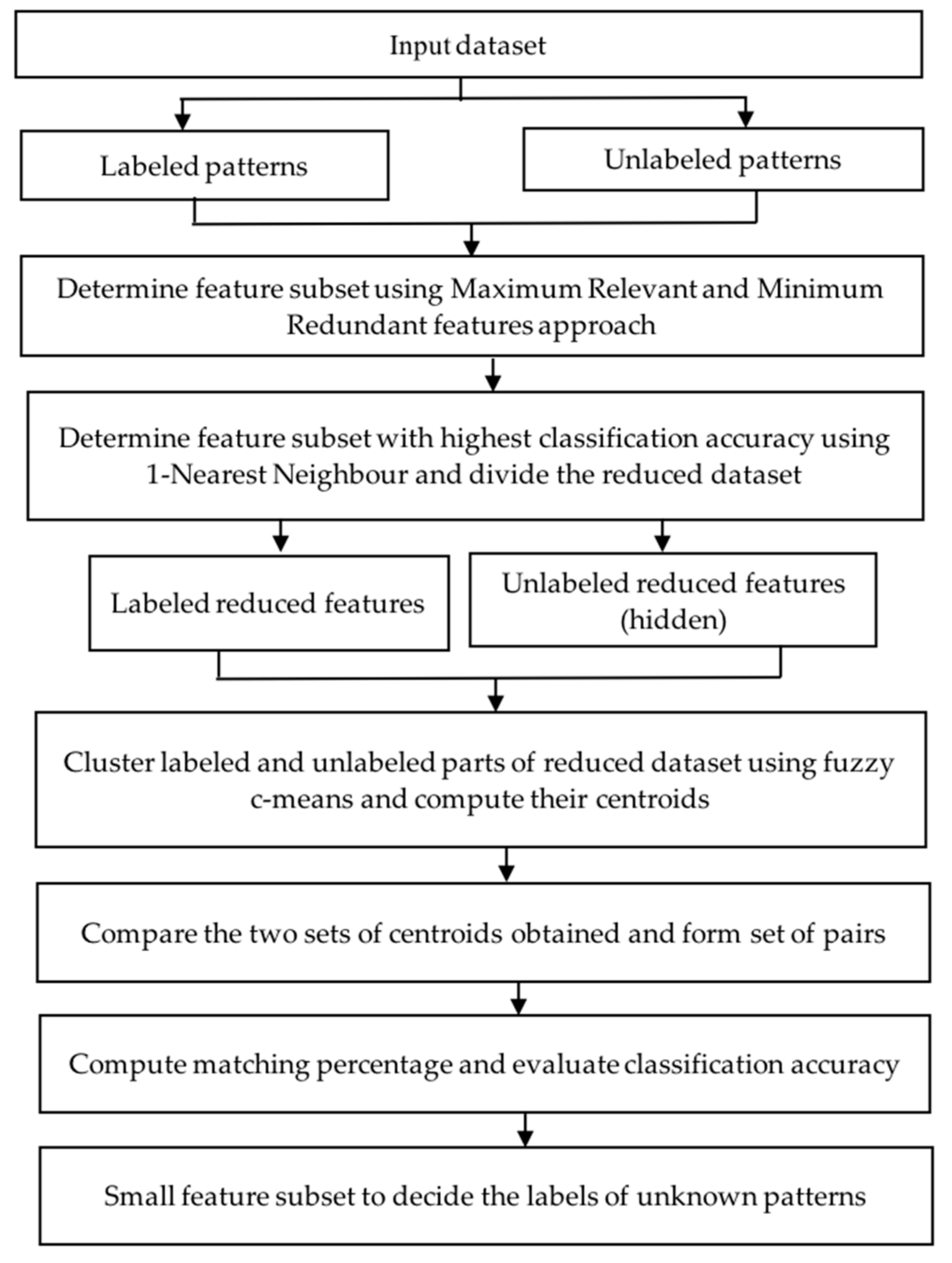
4. Proposed Two-Phase Approach
4.1. Assumptions
4.2. Algorithm of the Two-Phase Approach
| Algorithm 1 The maximum relevant and minimum redundant features |
Input: Dataset with d features; with labels given on some patterns (supervised) and not given on some patterns (unsupervised)
|
| Algorithm 2 The steps of the proposed two-phase approach |
|
| Pseudo-Code: The proposed two-phase approach |
Input DF,P dataset with F features, P number instances and number of classes as C
|
4.3. Complexity of Algorithms
- i.
- Calculating Pearson’s Coefficient O(n)
- ii.
- Calculating Fuzzy C-means clustering O(ndc2i)
- iii.
- Time for calculating KNNO(ndK).
5. Experiments
- *P1–Basic PNN
- *P2–RCPNN with gradient descent
- *P3–RCPNN with PSO
- *P4–Condensed PNN with swarm intelligence
- *P5–All PDs with 50% training in proposed scheme
- *P6–All PDs with 80% training in proposed scheme
- *P7–Only best 50% PDs with 50% training in proposed scheme
- *P8–Only best 50% PDs with 80% training in proposed scheme
- *P9–Unsupervised method using Sammon’s Stress Function
- *P10–Proposed method with 70% known labels
- *P11–Proposed method with 50% known labels
- *P12–Proposed method with 40% known labels
6. Results and Discussion
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification; John Wiley and Sons (Asia): Hoboken, NJ, USA, 2001. [Google Scholar]
- Saxena, A.K.; Dubey, V.K.; Wang, J. Hybrid Feature Selection Methods for High Dimensional Multi–class Datasets. Int. J. Data Min. Model. Manag. 2017, 9, 315–339. [Google Scholar] [CrossRef]
- Saxena, A.; Patre, D.; Dubey, A. An Evolutionary Feature Selection Technique using Polynomial Neural Network. Int. J. Comput. Sci. Issues 2011, 8, 494. [Google Scholar]
- Michalski, R.S.; Karbonell, J.G.; Kubat, M. Machine Learning and Data Mining: Methods and Applications; John Wiley and Sons: New York, NY, USA, 1998. [Google Scholar]
- Kamber, M.; Han, J. Data Mining: Concepts and Techniques, 2nd ed.; Morgan Kaufmann Publisher: San Francisco, CA, USA, 2006. [Google Scholar]
- Kosala, R.; Blockeel, H. Web Mining Research: A Survey. ACM Sig Kdd Explor. Newsl. 2000, 2, 1–5. [Google Scholar] [CrossRef]
- Baldi, P.; Brunak, S. Bioinformatics: The Machine Learning Approach, 2nd ed.; MIT Press: Cambridge, MA, USA, 1998. [Google Scholar]
- Boero, G.; Cavalli, E. Forecasting the Exchangerage: A Comparison between Econometric and Neural Network Models; A FIR Colloquium: Panama City, Panama, 1996; Volume 2, pp. 981–996. [Google Scholar]
- Derrig, R.A.; Ostaszewski, K.M. Fuzzy Techniques of Pattern Recognition in Risk and Claim Classification. J. Risk Insur. 1995, 62, 447. [Google Scholar] [CrossRef]
- Mitchel, T.M. Machine Learning; McGraw Hill: New York, NY, USA, 1997. [Google Scholar]
- Bandyopadhyay, S.; Bhadra, T.; Mitra, P.; Maulik, U. Integration of Densesub Graph Finding with Feature Clustering for Unsupervised Feature Selection. Pattern Recognit. Lett. 2014, 40, 104–112. [Google Scholar] [CrossRef]
- Lin, C.-T.; Prasad, M.; Saxena, A. An Improved Polynomial Neural Network Classifier Using Real-Coded Genetic Algorithm. IEEE Trans. Syst. Man Cybern. Syst. 2015, 45, 1389–1401. [Google Scholar] [CrossRef]
- Campbell, W.M.; Assaleh, K.T.; Broun, C.C. Speaker Recognition with Polynomial Classifiers. IEEE Trans. Speech Audio Process. 2002, 10, 205–212. [Google Scholar] [CrossRef]
- Pal, N.R. Fuzzy Logic Approaches to Structure Preserving Dimensionality Reduction. IEEE Trans. Fuzzy Syst. 2002, 10, 277–286. [Google Scholar] [CrossRef]
- Jain, A.K.; Dubes, R.C. Algorithms for Clustering Data; Prentice–Hall: Upper Saddle River, NJ, USA, 1988. [Google Scholar]
- Sammon, J.W. A Nonlinear Mapping for Data Structure Analysis. IEEE Trans. Comput. 1969, 18, 401–409. [Google Scholar] [CrossRef]
- Schachter, B. A Nonlinear Mapping Algorithm for Large Databases. Comput. Graph. Image Process. 1978, 7, 271–278. [Google Scholar] [CrossRef]
- Pykett, C.E. Improving the Efficiency of Sammon’s Nonlinear Mapping by using Clustering Archetypes. Electron. Lett. 1980, 14, 799–800. [Google Scholar] [CrossRef]
- Pal, N.R. Soft Computing for Feature Analysis. Fuzzy Sets Syst. 1999, 103, 201–221. [Google Scholar] [CrossRef]
- Muni, D.P.; Pal, N.R.; Das, J. A Novel Approach for Designing Classifiers Using Genetic Programming. IEEE Trans. Evol. Comput. 2004, 8, 183–196. [Google Scholar] [CrossRef]
- Mitra, P.; Murthy, C.A.; Pal, S.K. Unsupervised Feature Selection using Feature Similarity. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 301–312. [Google Scholar] [CrossRef]
- Dash, M.; Liu, H. Feature Selection for Clustering. In Proceedings of the Asia Pacific Conference on Knowledge Discovery and Data Mining, Kyoto, Japan, 18–20 April 2000; pp. 110–121. [Google Scholar]
- Dy, J.G.; Brodley, C.E. Feature Subset Selection and Order Identification for Unsupervised Learning; ICML: Vienna, Austria, 2000; pp. 247–254. [Google Scholar]
- Basu, S.; Micchelli, C.A.; Olsen, P. Maximum Entropy and Maximum Likelihood Criteria for Feature Selection from Multivariate Data. In Proceedings of the 2000 IEEE International Symposium on Circuits and Systems (ISCAS), Geneva, Switzerland, 28–31 May 2000; Volume 3, pp. 267–270. [Google Scholar]
- Pal, S.K.; De, R.K.; Basak, J. Unsupervised Feature Evaluation: A Neuro–Fuzzy Approach. IEEE Trans. Neural Netw. 2000, 1, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Muni, D.P.; Pal, N.R.; Das, J. Genetic Programming for Simultaneous Feature Selection and Classifier Design. IEEE Trans. Syst. Man Cyber. 2006, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, R.P. Redundancy in Feature Extraction. IEEE Trans. Comput. 1971, 100, 1051–1054. [Google Scholar] [CrossRef]
- Das, S.K. Feature Selection with a Linear Dependence Measure. IEEE Trans. Comput. 1971, 100, 1106–1109. [Google Scholar] [CrossRef]
- Saxena, A.; Pal, N.R.; Vora, M. Evolutionary Methods for Unsupervised Feature Selection using Sammon’s Stress Function. Fuzzy Inf. Eng. 2010, 2, 229–247. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature Selection based on Mutual Information Criteria of Max–dependency, Max–relevance, and Min–redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Xu, J.; Yang, G.; Man, H.; He, H. L1 Graph base on Sparse Coding for Feature Selection. In Proceedings of the International Symposium on Neural Networks, Dalian, China, 4–6 July 2013; pp. 594–601. [Google Scholar]
- Xu, J.; Yin, Y.; Man, H.; He, H. Feature Selection based on Sparse Imputation. In Proceedings of the 2012 International Joint Conference on Neural Networks (IJCNN), Brisbane, QLD, Australia, 10–15 June 2012; pp. 1–7. [Google Scholar]
- Yang, J.B.; Ong, C.J. Feature Selection using Probabilistic Prediction of Support Vector Regression. IEEE Trans. Neural Netw. Learn. Syst. 2011, 22, 954–962. [Google Scholar] [CrossRef] [PubMed]
- Weston, J.; Mukherjee, S.; Chapelle, O.; Pontil, M.; Poggio, T.; Vapnik, V. Feature Selection for SVMs. In Proceedings of the Advances in Neural Information Processing Systems 13, Cambridge, MA, USA, 27–30 November 2000; pp. 526–532. [Google Scholar]
- Xu, J.; Tang, B.; He, H.; Man, H. Semi–supervised Feature Selection based on Relevance and Redundancy Criteria. IEEE Trans. Neural Netw. Learn. Syst. 2016, 28, 1974–1984. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Elisseeff, A. An Introduction to variable and Feature Selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Hall, M.A. Correlation–based Feature Selection for Discrete and Numeric Class Machine Learning. In Proceedings of the 17th International Conference on Machine Learning (ICML2000), Stanford University, Stanford, CA, USA, 29 June–2 July 2000; pp. 359–366. [Google Scholar]
- He, X.; Caiand, D.; Niyogi, P. Laplacian Score for Feature Selection. In Proceedings of the Advances in Neural Information Processing Systems 18, Vancouver, BC, Canada, 5–8 December 2005; pp. 507–514. [Google Scholar]
- Xu, Z.; King, I.; Lyu, M.R.T.; Jin, R. Discriminative Semi-supervised Feature Selection via Manifold Regularization. IEEE Trans. Neural Netw. 2010, 21, 1033–1047. [Google Scholar]
- Zhao, Z.; Liu, H. Semi–supervised Feature Selection via Spectral Analysis. In Proceedings of the Seventh SIAM International Conference on Data Mining, Minneapolis, MN, USA, 26–28 April 2007; pp. 641–646. [Google Scholar]
- Ren, J.; Qiu, Z.; Fan, W.; Cheng, H.; Yu, P.S. Forward Semi–supervised Feature Selection. In Proceedings of the 12th Pacific-Asia Conference, PAKDD 2008, Osaka, Japan, 20–23 May 2008; pp. 970–976. [Google Scholar]
- Sheikhpour, R.; Sarram, M.A.; Gharaghani, S.; Chahooki, M.A.Z. A Survey on Semi–supervised Feature Selection Methods. Pattern Recognit. 2017, 64, 141–158. [Google Scholar] [CrossRef]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 27 December 1965–7 January 1966; Volume 1, pp. 281–297. [Google Scholar]
- Duan, Y.; Liu, Q.; Xia, S. An Improved Initialization Center k–means Clustering Algorithm based on Distance and Density. AIP Conf. Proc. 2018, 1955, 40046. [Google Scholar]
- Dunn, C. A Fuzzy Relative of the ISO DATA Process and Its Use in Detecting Compact Well–Separated Clusters. J. Cybern. 1973, 3, 32–57. [Google Scholar] [CrossRef]
- Bezdek, J.C. Pattern Recognition with Fuzzy Objective Function Algorithms; Kluwer Academic Publishers: Norwell, MA, USA, 1981. [Google Scholar]
- Saxena, A.; Prasad, M.; Gupta, A.; Bharill, N.; Patel, O.P.; Tiwari, A.; Er, M.J.; Ding, W.; Lin, C.T. A Review of Clustering Techniques and Developments. Neurocomputing 2017, 267, 664–681. [Google Scholar] [CrossRef]
- Thong, P.H. An Overview of Semi–supervised Fuzzy Clustering Algorithms. Int. J. Eng. Technol. 2016, 8, 301. [Google Scholar] [CrossRef]
- Li, L.; Garibaldi, J.M.; He, D.; Wang, M. Semi–supervised Fuzzy Clustering with Feature Discrimination. PLoS ONE 2015, 10, e0131160. [Google Scholar] [CrossRef]
- Haykin, S. A Comprehensive Foundation. Neural Netw. 2004, 2, 20004. [Google Scholar]
- Ivakhnenko, A.G. Polynomial Theory of Complex Systems. IEEE Trans. Syst. Man Cybern. 1971, 4, 364–378. [Google Scholar] [CrossRef]
- Madala, H.R. Inductive Learning Algorithms for Complex Systems Modeling; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Misra, B.B.; Dehuri, S.; Dash, P.K.; Panda, G. A Reduced and Comprehensible Polynomial Neural Network for Classification. Pattern Recognit. Lett. 2008, 29, 1705–1712. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R. Particle Swarm Optimization. In Proceedings of the ICNN’95–International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar]
- Dehuri, S.; Misra, B.B.; Ghosh, A.; Cho, S.B. A Condensed Polynomial Neural Network for Classification using Swarm Intelligence. Appl. Soft Comput. 2011, 11, 3106–3113. [Google Scholar] [CrossRef]
- Dheeru, D.; Taniskidou, E.K. UCI Machine Learning Repository. 2017. Available online: http://archive.ics.uci.edu/ml (accessed on 30 August 2020).
- Rossi, R.A.; Ahmad, N.K. The Network Data Repository with Interactive Graph Analytics and Visualization. 2015. Available online: http://networkrepository.com (accessed on 30 August 2020).
| Dataset | Total Patterns | Attributes | Classes | Patterns in Class1 | Patterns in Class2 | Patterns in Class3 |
|---|---|---|---|---|---|---|
| Iris | 150 | 4 | 3 | 50 | 50 | 50 |
| Wine | 178 | 13 | 3 | 59 | 71 | 48 |
| Pima | 768 | 8 | 2 | 268 | 500 | – |
| Liver | 345 | 6 | 2 | 145 | 200 | – |
| WBC | 699 | 9 | 2 | 458 | 241 | – |
| Thyroid | 215 | 5 | 3 | 150 | 35 | 30 |
| Synthetic | 588 | 5 | 2 | 252 | 336 | – |
| Sonar | 208 | 60 | 2 | 97 | 111 | – |
| Ionos | 351 | 34 | 2 | 225 | 126 | – |
| Experiment No. | Patterns with Labels Known in % | Patterns with Labels Unknown in % |
|---|---|---|
| 1 | 70 | 30 |
| 2 | 50 | 50 |
| 3 | 40 | 60 |
| S No. | Maximum Relevance * | Minimum Redundancy ** |
|---|---|---|
| 1 | 50% (rounded), when total features ≤ 20 | 50% of total features ≤ 20 |
| 2 | 10 when total features > 20 | when total features > 20 |
| 0.702 | 0.816 | 0.814 | 0.717 | 0.699 | 0.809 | 0.774 | 0.744 | 0.405 |
| 0.642 | 0.653 | 0.488 | 0.524 | 0.593 | 0.554 | 0.534 | 0.351 | 0.907 | 0.707 |
| 0.754 | 0.692 | 0.756 | 0.719 | 0.461 | 0.686 | 0.722 | 0.714 | 0.735 | 0.718 |
| 0.441 | 0.595 | 0.671 | 0.669 | 0.603 | 0.419 | 0.586 | 0.618 | 0.629 | 0.481 |
| 0.681 | 0.584 | 0.339 | 0.666 | 0.346 | 0.434 | ||||
| Dataset and Data Label (%) | Features Max Relevancy | Features Min Redundancy | Features Taken | CA (1–nn) Known | Centroid Labeled Cluster#: Centroid | Centroid Unlabeled Cluster#: Centroid | Pairs (Unlabeled, Labeled) | Match % | |
|---|---|---|---|---|---|---|---|---|---|
| Synthetic | 70 | 1,2,3 | 4,5 | 1,2 | 100 | 1:5.06,4.80 2:19.60,19.61 | 1:5.00,4.78 2:19.49,19.63 | (1,1) (2,2) | 100 |
| 50 | 1,2,3 | 4,5 | 1,2 | 100 | 1:5.06,4.85 2:19.70,19.76 | 1:19.42,19.46 2:5.02,4.75 | (1,2) (2,1) | 100 | |
| 40 | 1,2,3 | 4,5 | 1,2 | 100 | 1:19.39,19.71 2:5.14,4.73 | 1:4.98,4.83 2:19.69,19.55 | (1,2) (2,1) | 100 | |
| Iris | 70 | 3,4 | 2 | 4 | 97.8 | 1:0.252 2:2.070 3:1.380 | 1:2.164 2:0.217 3:1.273 | (1,2) (2,1) (3,3) | 97.8 |
| 50 | 3,4 | 2 | 3 | 98.66 | 1:5.638 2:4.271 3:1.478 | 1: 5.740 2:1.471 3:4.427 | (1,1) (2,3) (3,2) | 94.67 | |
| 40 | 3,4 | 2 | 4 | 94.4 | 1:1.357 2:0.280 3:2.050 | 1:0.223 2:2.162 3:1.352 | (1,2) (2,3) (3,1) | 94.4 | |
| Wine | 70 | 1,2,3,4, 5,8,10 | 6,7,11, 12,13 | 1,2,3,10 | 92.45 | 1:13.462,3.054, 2.43,9.201 2:12.440,2.043, 2.27,3.102 3:13.51,2.080, 2.388, 5.520 | 1:12.101,1.641, 2.238,2.824 2:13.222,3.532, 2.467,8.631 3:13.488,2.698, 2.540,5.373 | (1,3) (2,1) (3,2) | 86.79 |
| 50 | 1,2,3,4, 5,8,10 | 6,7,11, 12,13 | 1,2,8,10 | 89.89 | 1:13.543,2.250, 0.39, 5.713 2:12.368,2.058, 0.36, 3.175 3:13.439,2.905, 0.31, 8.856 | 1:13.361,3.482, 0.469,9.326 2:13.506,2.195, 0.338,5.294 3:12.263,1.757, 0.370,2.862 | (1,3) (2,1) (3,2) | 82.02 | |
| 40 | 1,2,3,4, 5,8,10 | 6,7,11, 12,13 | 1,2,8,10 | 87.85 | 1:12.350,1.838, 0.23, 3.198 2:13.521,2.486, 0.30, 5.345 3:13.465,3.397, 0.38, 9.004 | 1:13.522,2.111, 0.336,5.526 2:12.288,1.903, 0.399,2.926 3:13.322,2.984, 0.438,9.038 | (1,2) (2,1) (3,3) | 78.50 | |
| Liver | 70 | 3,4,5 | 1,2,6 | 3,5 | 63.11 | 1:25.726,25.479 2: 51.586,134.642 | 1:58.436,87.142 2:25.631,23.924 | (1,2) (2,1) | 58.25 |
| 50 | 4,5,6 | 1,2,3 | 5,6 | 55.81 | 1:118.762,6.119 2:25.330,3.187 | 1:131.657,5.794 2:25.681,2.879 | (1,1) (2,2) | 50.58 | |
| 40 | 5,4,6 | 1,2,3 | 4,6 | 56.04 | 1:36.709,4.441 2:20.917,2.994 | 1:21.227,2.920 2:47.236,6.706 | (1,2) (2,1) | 52.17 | |
| WBC | 70 | 2,3,6,7,8 | 1,4,9 | 3,6,8 | 96.59 | 1:7.024,8.434, 6.807 2:1.542,1.381, 1.316 | 1: 6.670, 8.579, 5.320 2:1.418,1.309, 1.236 | (1,1) (2,2) | 97.56 |
| 50 | 3,2,6,7,8 | 1,4,9 | 2,6,8 | 95.60 | 1:7.032,8.093, 7.109 2:1.428,1.412, 1.298 | 1:1.368,1.322, 1.259 2:6.851,8.714, 5.765 | (1,2) (2,1) | 95.89 | |
| 40 | 3,2,6,7,8 | 1,4,9 | 3,6,7 | 95.61 | 1:1.648,1.381, 2.237 2:7.000,8.710, 6.165 | 1:6.796,8.741, 6.364 2:1.441,1.287, 2.103 | (1,2) (2,1) | 95.85 | |
| Thyroid | 70 | 3,4,5 | 1,2 | 4,5 | 90.63 | 1:1.422,2.106 2:29.910,15.131 3:11.713,43.082 | 1:1.478,2.435 2:10.785,49.347 3:7.711,15.576 | (1,1) (2,3) (3,2) | 78.13 |
| 50 | 1,4,5 | 2,3 | 4,5 | 85.98 | 1:13.004,52.564 2:1.559,2.111 3:31.312,17.065 | 1:9.924,10.542 2:9.526,41.233 3:1.313,2.177 | (1,2) (2,1) (3,3) | 76.64 | |
| 40 | 3,4,5 | 1,2 | 3,4 | 88.37 | 1:1.265,14.708 2:0.659,53.718 3:2.121,1.430 | 1:0.747,19.634 2:1.712,1.389 3:4.672,2.020 | (1,1) (2,3) (3,2) | 85.27 | |
| Pima | 70 | 1,8,2,6 | 4,5 | 2,6 | 67.83 | 1:154.906,34.164 2:101.265,30.381 | 1: 155.805,34.785 2:100.047,31.197 | (1,1) (2,2) | 76.52 |
| 50 | 2,6,7,8 | 1,4,5 | 2,6 | 68.75 | 1:153.745,35.013 2:101.112,30.301 | 1:156.251,33.646 2:100.490,30.985 | (1,1) (2,2) | 75.26 | |
| 40 | 1,2,6,8 | 4,5 | 2,8 | 77.07 | 1:149.770,36.644 2:102.052,30.263 | 1:100.318,29.902 2:158.792,38.136 | (1,2) (2,1) | 76.57 | |
| Sonar | 70 | 11,12,45, 46,10,9, 13,51,52 44 | 16,17,18,19, 20,21,28,29, 30,31 | 10,12, 13,44, 46 | 80.65 | 1:0.160,0.173, 0.199, 0.189,0.129 2:0.288,0.353, 0.373, 0.257,0.202 | 1:0.234,0.326, 0.346, 0.194,0.139 2:0.127,0.164, 0.188, 0.173,0.130 | (1,2) (2,1) | 75.81 |
| 50 | 11,12,45, 46,10,9, 13,51,52, 44 | 16,17,18,19, 20,21,28,29, 30,31 | 9,12,13, 46,48 | 80.77 | 1:0.142,0.173, 0.194, 0.129,0.083 2:0.239,0.365, 0.387, 0.192,0.105 | 1:0.216,0.350, 0.370, 0.170,0.102 2:0.123,0.156, 0.184, 0.133,0.069 | (1,2) (2,1) | 71.15 | |
| 40 | 11,45,49, 9,46,12, 10,48,47,44 | 16,17,18,19, 20,21,28,29, 30,31 | 10,11, 12,46, 49 | 78.40 | 1:0.140,0.159, 0.187, 0.122, 0.039 2:0.291,0.333, 0.348,0.196,0.058 | 1: 0.305,0.347,0.359, 0.179,0.062 2:0.132,0.147,0.157, 0.132,0.045 | (1,2) (2,1) | 68 | |
| Ionos | 70 | 2,22,27, 34,20,30, 32,26,24, 28 | 8,10,11,12, 13,15,17,19, 21 | 26,27, 28,30, 32 | 92.38 | 1:0.268,0.434, 0.312,–0.200, –0.227 2:0.138,0.638,0.199, 0.179,0.226 | 1:0.016,0.599,0.023, 0.020,0.044, 2:–0.418,0.461,–0.469, –0.369,–0.283 | (1,2) (2,1) | 63.81 |
| 50 | 2,22,32, 27,34,17,28,13,26,24 | 8,10,11,12, 15,19,21 | 13,17, 24,27, 34 | 87.43 | 1:0.756,0.743, –0.024,0.739, –0.015 2:–0.176,–0.197, –0.253 0.275,0.093 | 1:0.743,0.735,–0.016, 0.751,–0.004 2:–0.118,–0.177,–0.021, 0.154,–0.018 | (1,1) (2,2) | 67.43 | |
| 40 | 2,27,22, 30,17,34,24,11,13 | 8,10,12,15, 19,21 | 2,11,13, 24,34 | 87.68 | 1:0.000,0.763,0.767, 0.050,0.034, 2:0.000,0.061, -0.095, 0.198, 0.082 | 1:0.000,0.786,0.753, –0.020,–0.039, 2:0.000,–0.062,–0.213, –0.181,0.027 | (1,1) (2,2) | 72.51 | |
| Dataset and Data Label (in %) | Centroid Label Cluster#: Centroid Values | Centroid Unlabeled Cluster#: Centroid Values | Pairs (Unlabeled, Label) | Match % | Min. Distance between Identified Centroids x:...vs. y:... | |
|---|---|---|---|---|---|---|
| Synthetic | 70 | 1: 5.06,4.80 2: 19.60,19.61 | 1: 5.00,4.78 2: 19.49,19.63 | (1,1) (2,2) | 100 | 0.0632 0.1118 |
| 50 | 1: 5.06,4.85 2: 19.70,19.76 | 1: 19.42,19.46 2: 5.02,4.75 | (1,2) (2,1) | 100 | 0.1077 0.4104 | |
| 40 | 1:19.39,19.71 2:5.14,4.73 | 1: 4.98,4.83 2: 19.69,19.55 | (1,2) (2,1) | 100 | 0.3400 0.1887 | |
| Iris | 70 | 1:0.252 2:2.070 3:1.380 | 1: 2.164 2: 0.217 3: 1.273 | (1,2) (2,1) (3,3) | 97.8 | 0.0350 0.0940 0.1070 |
| 50 | 1:5.638 2:4.271 3:1.478 | 1: 5.740 2: 1.471 3: 4.427 | (1,1) (2,3) (3,2) | 94.67 | 0.1020 0.1560 0.0070 | |
| 40 | 1:1.357 2:0.280 3:2.050 | 1: 0.223 2: 2.162 3: 1.352 | (1,2) (2,3) (3,1) | 94.4 | 0.0050 0.0570 0.1120 | |
| Wine | 70 | 1:13.462,3.054,2.433,9.201 2:12.440,2.043,2.287,3.102 3:13.51,2.080,2.388,5.520 | 1:12.101,1.641,2.238,2.824 2:13.222,3.532,2.467,8.631 3:13.488,2.698,2.540,5.373 | (1,3) (2,1) (3,2) | 86.79 | 0.7824 0.5968 0.6537 |
| 50 | 1:13.543,2.250,0.319,5.713 2: 12.368,2.058,0.356,3.175 3:13.439,2.905,0.381, 8.856 | 1:13.361,3.482,0.469,9.326 2:13.506,2.195,0.338,5.294 3:12.263,1.757,0.370,2.862 | (1,3) (2,1) (3,2) | 82.02 | 0.4246 0.4470 0.7534 | |
| 40 | 1:12.350,1.838,0.293,3.198 2: 13.521,2.486,0.320,5.345 3:13.465,3.397,0.398,9.004 | 1:13.522,2.111,0.336,5.526 2:12.288,1.903,0.399,2.926 3:13.322,2.984,0.438,9.038 | (1,2) (2,1) (3,3) | 78.50 | 0.3054 0.4167 0.4402 | |
| Liver | 70 | 1:25.726,25.479 2: 51.586,134.642 | 1: 58.436,87.142 2: 25.631,23.924 | (1,2) (2,1) | 58.25 | 1.5579 47.9914 |
| 50 | 1:118.762,6.119 2:25.330,3.187 | 1: 131.657,5.794 2: 25.681,2.879 | (1,1) (2,2) | 50.58 | 12.8991 0.4670 | |
| 40 | 1:36.709,4.441 2:20.917,2.994 | 1: 21.227,2.920 2: 47.236,6.706 | (1,2) (2,1) | 52.17 | 10.7679 0.318 | |
| WBC | 70 | 1:7.024,8.434,6.807 2:1.542,1.381,1.316 | 1: 6.670,8.579,5.320 2: 1.418,1.309,1.236 | (1,1) (2,2) | 97.56 | 1.5354 0.1642 |
| 50 | 1: 7.032,8.093,7.109 2:1.428,1.412,1.298 | 1: 1.368,1.322,1.259 2: 6.851,8.714,5.765 | (1,2) (2,1) | 95.89 | 1.4916 0.1150 | |
| 40 | 1:1.648,1.381,2.237 2:7.000,8.710,6.165 | 1: 6.796,8.741,6.364 2: 1.441,1.287,2.103 | (1,2) (2,1) | 95.85 | 0.2639 0.2867 | |
| Thyroid | 70 | 1:1.422,2.106 2:29.910,15.131 3:11.713,43.082 | 1: 1.478,2.435 2: 10.785,49.347 3: 7.711,15.576 | (1,1) (2,3) (3,2) | 78.13 | 0.3337 22.2035 6.3334 |
| 50 | 1:13.004,52.564 2:1.559, 2.111 3:31.312,17.065 | 1: 9.924,10.542 2: 9.526,41.233 3: 1.313,2.177 | (1,2) (2,1) (3,3) | 76.64 | 11.8528 0.2547 22.3606 | |
| 40 | 1:1.265,14.708 2:0.659,53.718 3:2.121,1.430 | 1: 0.747,19.634 2: 1.712,1.389 3: 4.672,2.020 | (1,1) (2,3) (3,2) | 85.27 | 4.9532 34.0841 0.4110 | |
| Pima | 70 | 1:154.906, 34.164 2:101.265,30.381 | 1: 155.805,34.785 2: 100.047,31.197 | (1,1) (2,2) | 76.52 | 1.0926 1.4661 |
| 50 | 1:153.745,35.013 2:101.112,30.301 | 1: 156.251,33.646 2: 100.490,30.985 | (1,1) (2,2) | 75.26 | 2.8546 0.9245 | |
| 40 | 1:149.770,36.644 2:102.052,30.263 | 1: 100.318,29.902 2: 158.792,38.136 | (1,2) (2,1) | 76.57 | 9.1445 1.7712 | |
| Sonar | 70 | 1:0.160,0.173,0.199,0.189,0.129 2:0.288,0.353,0.373,0.257,0.202 | 1:0.234,0.326,0.346,0.194,0.139 2:0.127,0.164,0.188,0.173,0.130 | (1,2) (2,1) | 75.81 | 0.2250, 0.0393 0.3288, 0.1110 |
| 50 | 1:0.142,0.173,0.194,0.129,0.083 2:0.239,0.365,0.387,0.192,0.105 | 1:0.216,0.350,0.370,0.170,0.102 2:0.123,0.156,0.184,0.133,0.069 | (1,2) (2,1) | 71.15 | 0.2642, 0.0310 0.0392, 0.3211 | |
| 40 | 1:0.140,0.159,0.187,0.122,0.039 2:0.291,0.333,0.348,0.196,0.058 | 1:0.305,0.347,0.359,0.179,0.062 2:0.132,0.147,0.157,0.132,0.045 | (1,2) (2,1) | 68 | 0.3097, 0.0353 0.0286, 0.3172 | |
| Ionos | 70 | 1:–0.268,0.434,–0.312, –0.200,–0.227 2:0.138,0.638,0.199, 0.179, 0.226 | 1: 0.016,0.599,0.023 ,0.020,0.044, 2: –0.418,0.461,–0.469, –0.369,–0.283 | (1,2) (2,1) | 63.81 | 0.2821 0.3252 |
| 50 | 1:0.756, 0.743,–0.024, 0.739,–0.015 2:–0.176,–0.197,–.253, 0.275, 0.093 | 1: 0.743, 0.735,–0.016, 0.751,–0.004 2: –0.118,–0.177,–0.021, 0.154,–0.018 | (1,1) (2,2) | 67.43 | 0.0237 0.2908 | |
| 40 | 1:0.000, 0.763, 0.767, 0.050, 0.034, 2:0.000, 0.061,–0.095, –0.198, 0.082 | 1:0.000, 0.786, 0.753, –0.020,–0.039, 2:0.000,–0.062,–0.213, –0.181,0.027 | (1,1) (2,2) | 72.51 | 0.1047 0.1799 | |
| Dataset/ Methods | P1* | P2* | P3* | P4* | *P5 | *P6 | *P7 | *P8 | *P9 | *P10 | *P11 | *P12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Iris | 86.22 | 95.56 | 98.67 | 99.33 | 98 | 99.33 | 96 | 98.67 | 92.30 | 97.80 | 94.70 | 94.40 |
| Wine | 84.83 | 95.13 | 90.95 | 99.43 | 66.33 | 98.33 | 94.34 | 99.44 | 72.80 | 86.80 | 82.20 | 78.50 |
| Pima | 69.45 | 73.35 | 76.04 | 76.82 | 78.12 | 80.70 | 78.52 | 79.94 | – | 76.50 | 75.30 | 76.60 |
| Liver | 65.29 | 69.57 | 70.87 | 73.90 | 75.66 | 76.52 | 72.17 | 76.23 | 60 | 58.30 | 50.60 | 52.10 |
| WBC | 95.90 | 97.14 | 97.64 | – | 97.35 | 97.66 | 96.78 | 97.81 | 96 | 97.60 | 95.90 | 95.90 |
| Thyroid | – | – | – | – | 85.11 | 86.05 | 85.58 | 85.58 | 89.80 | 78.10 | 76.60 | 85.30 |
| Sonar | – | – | – | – | 69.23 | 70.21 | 67.38 | 86.98 | 80.70 | 75.80 | 71.10 | 68 |
| Ionos | – | – | – | – | – | – | – | – | 88 | 63.80 | 67.40 | 72.50 |
| Synthetic | – | – | – | – | – | – | – | – | – | 100 | 100 | 100 |
| Dataset/ Methods | P1* | P2* | P3* | P4* | *P5 | *P6 | *P7 | *P8 | *P9 | *P10 | *P11 | *P12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Iris | 4 | 4 | 4 | 4 | 98 | 4 | 4 | 4 | 2 | 1 | 1 | 1 |
| Wine | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 5 | 4 | 4 | 4 |
| Pima | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | – | 2 | 2 | 2 |
| Liver | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 3 | 2 | 2 | 2 |
| WBC | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 5 | 3 | 3 | 3 |
| Thyroid | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 3 | 2 | 2 | 2 |
| Sonar | 60 | 60 | 60 | 60 | 60 | 60 | 60 | 60 | 18 | 5 | 5 | 5 |
| Ionos | 34 | 34 | 34 | 34 | 34 | 34 | 34 | 34 | 17 | 5 | 5 | 5 |
| Synthetic | – | – | – | – | – | – | – | – | – | 2 | 2 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saxena, A.; Pare, S.; Meena, M.S.; Gupta, D.; Gupta, A.; Razzak, I.; Lin, C.-T.; Prasad, M. A Two-Phase Approach for Semi-Supervised Feature Selection. Algorithms 2020, 13, 215. https://doi.org/10.3390/a13090215
Saxena A, Pare S, Meena MS, Gupta D, Gupta A, Razzak I, Lin C-T, Prasad M. A Two-Phase Approach for Semi-Supervised Feature Selection. Algorithms. 2020; 13(9):215. https://doi.org/10.3390/a13090215
Chicago/Turabian StyleSaxena, Amit, Shreya Pare, Mahendra Singh Meena, Deepak Gupta, Akshansh Gupta, Imran Razzak, Chin-Teng Lin, and Mukesh Prasad. 2020. "A Two-Phase Approach for Semi-Supervised Feature Selection" Algorithms 13, no. 9: 215. https://doi.org/10.3390/a13090215
APA StyleSaxena, A., Pare, S., Meena, M. S., Gupta, D., Gupta, A., Razzak, I., Lin, C.-T., & Prasad, M. (2020). A Two-Phase Approach for Semi-Supervised Feature Selection. Algorithms, 13(9), 215. https://doi.org/10.3390/a13090215








