Edge-Nodes Representation Neural Machine for Link Prediction
Abstract
:1. Introduction
- We propose Edge-Nodes Representation Neural Machine (ENRNM), a novel link prediction framework to learn the link’s representation from the given network automatically.
- We propose a new method to represent the link fully by combining the edge’s representation with the two nodes’ representations on the two sides of the edge, so that the neural network can learn abundant, meaningful patterns and link formation mechanism.
2. Related Work
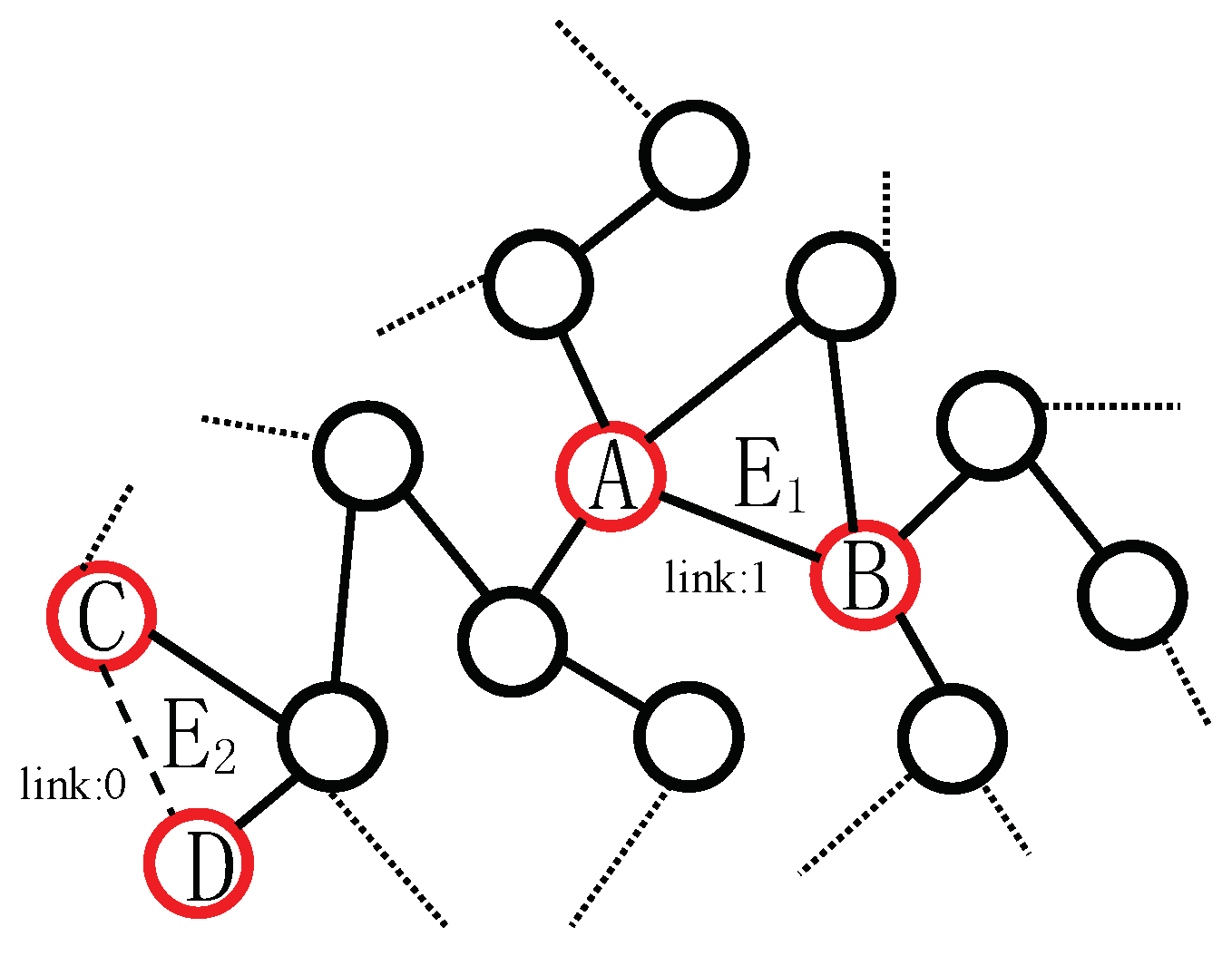
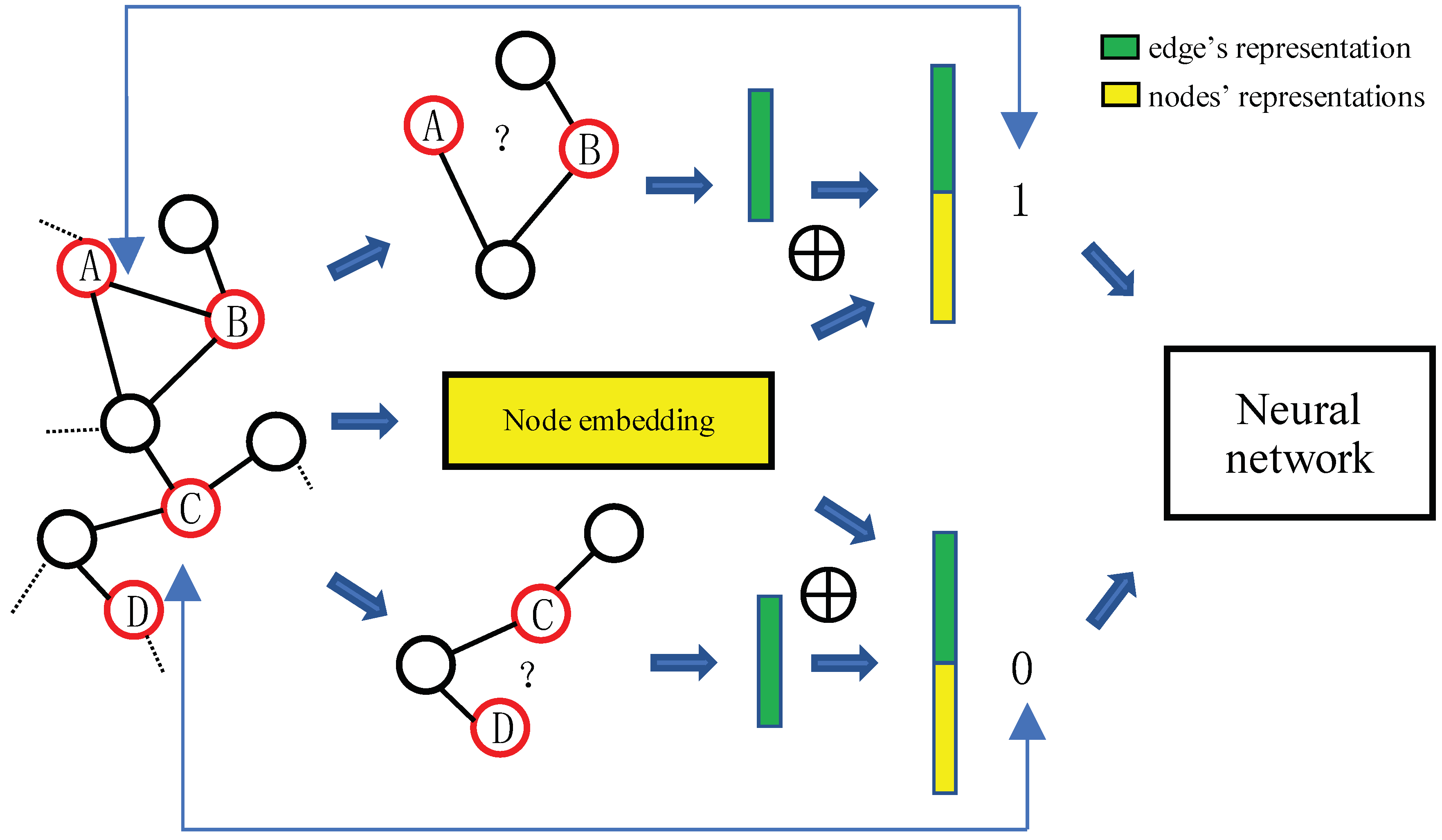
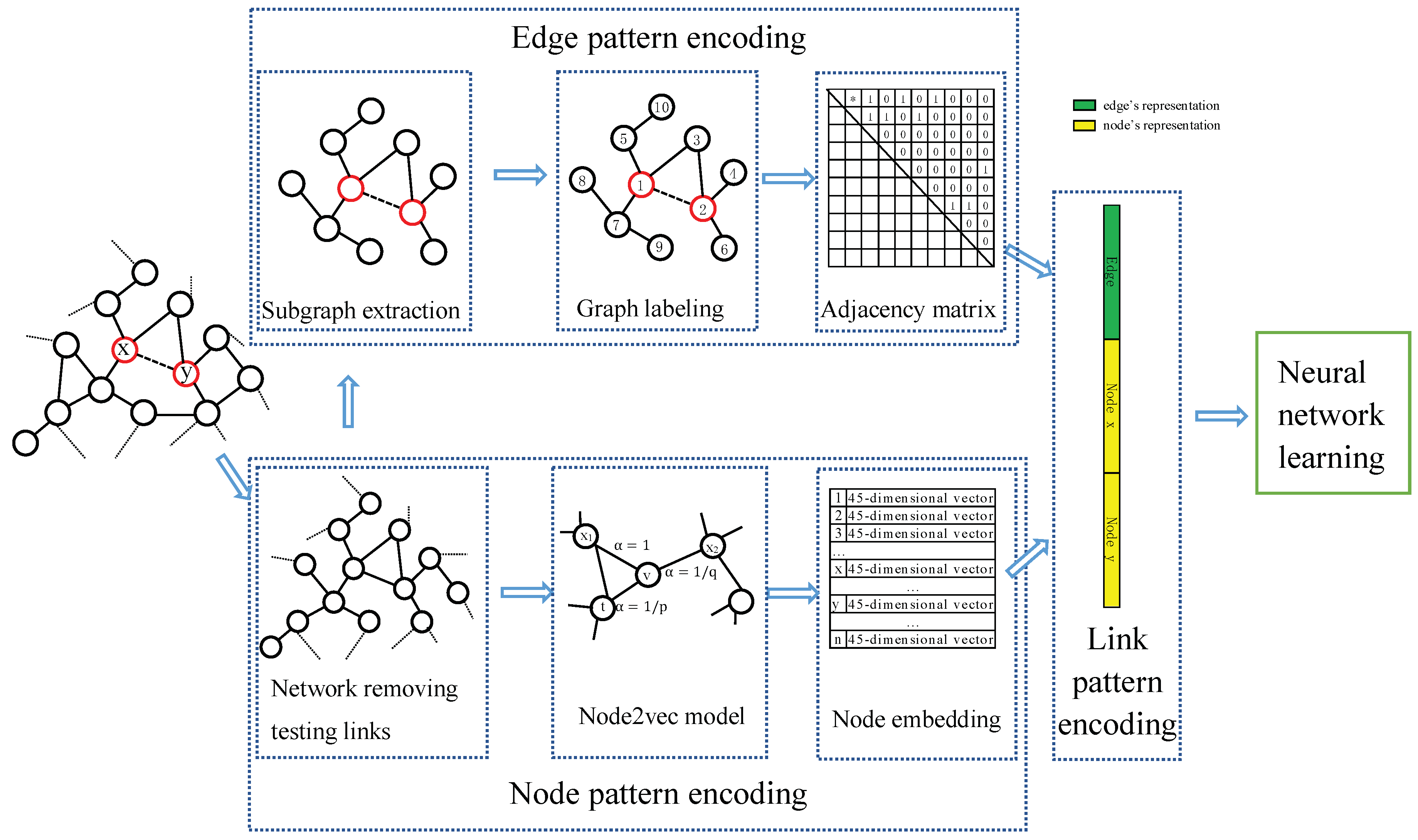
3. Edge-Nodes Representation Neural Machine (ENRNM)
- Edge pattern encoding, which learns edge’s representation by WLNM.
- Node pattern encoding, which learns node’s representation by network embedding model.
- Link pattern encoding, which joints the edge’s representation and two nodes’ on the two sides of the edge as link’s representation for each link.
- Train a fully connected neural network, which learns nonlinear graph topological structures and link’s formation mechanism for link prediction.
| Algorithm 1 ENRNM |
| Input: network G Output: AUC
|
3.1. Edge Pattern Encoding
| Algorithm 2 Edge pattern encoding |
| Input: network G, edge , integer K Output: edge’s representation
|
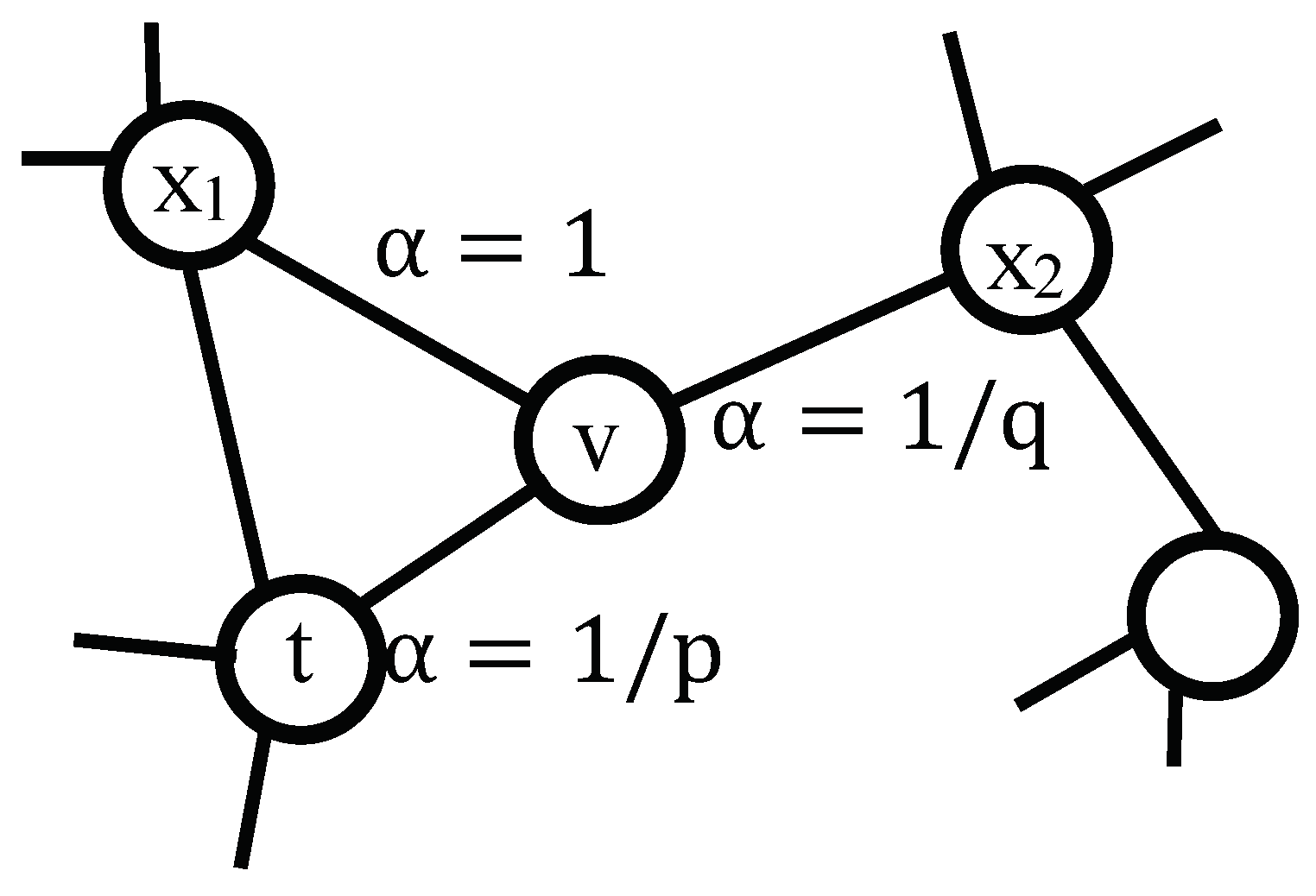
3.2. Node Pattern Encoding
| Algorithm 3 Node pattern encoding |
| Input: network G, dimension d, walks per node r, walk length l, context size k, return p, in-out q Output: every node’s representation
|
3.3. Link Pattern Encoding
3.4. Neural Network Learning
3.4.1. Training
3.4.2. Testing (Link Prediction)
4. Experiments and Results
4.1. Datasets
4.2. Baselines and Experimental Setting
4.3. Time and Computational Complexity
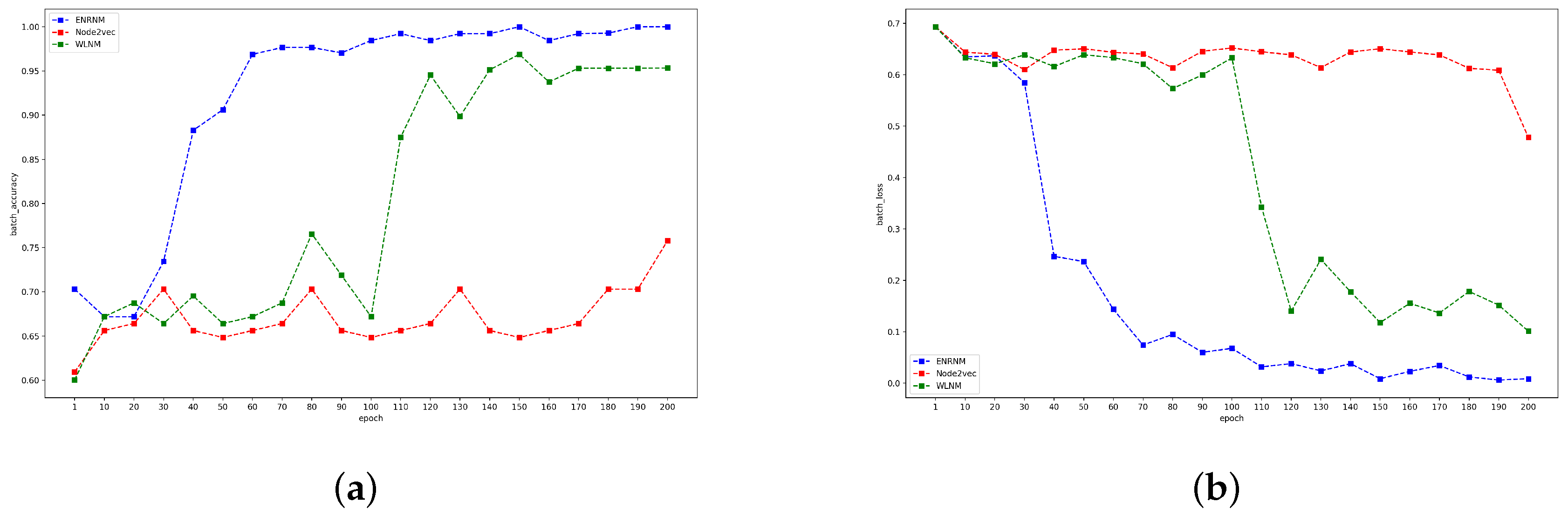
4.4. Results
5. Conclusions and Future Work
5.1. Conclusions
5.2. Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Liben-Nowell, D.; Kleinberg, J. The link-prediction problem for social networks. J. Am. Soc. Inf. Sci. Technol. 2007, 58, 1019–1031. [Google Scholar] [CrossRef] [Green Version]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv, 2016; arXiv:1609.02907. [Google Scholar]
- Von Mering, C.; Krause, R.; Snel, B.; Cornell, M.; Oliver, S.G.; Fields, S.; Bork, P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature 2002, 417, 399. [Google Scholar] [CrossRef]
- Adamic, L.A.; Adar, E. Friends and neighbors on the Web. Soc. Netw. 2003, 25, 211–230. [Google Scholar] [CrossRef] [Green Version]
- Koren, Y.; Bell, R.; Volinsky, C. Matrix factorization techniques for recommender systems. Computer 2009, 42, 30–37. [Google Scholar] [CrossRef]
- Nickel, M.; Murphy, K.; Tresp, V.; Gabrilovich, E. A review of relational machine learning for knowledge graphs. Proc. IEEE 2016, 104, 11–33. [Google Scholar] [CrossRef]
- Oyetunde, T.; Zhang, M.; Chen, Y.; Tang, Y.; Lo, C. BoostGAPFILL: Improving the fidelity of metabolic network reconstructions through integrated constraint and pattern-based methods. Bioinformatics 2016, 33, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Salakhutdinov, R.; Mnih, A. Bayesian probabilistic matrix factorization using Markov chain Monte Carlo. In Proceedings of the 25th International Conference on Machine Learning, Helsinki, Finland, 5–9 July 2008; ACM: New York, NY, USA, 2008; pp. 880–887. [Google Scholar] [Green Version]
- Airoldi, E.M.; Blei, D.M.; Fienberg, S.E.; Xing, E.P. Mixed membership stochastic blockmodels. J. Mach. Learn. Res. 2008, 9, 1981–2014. [Google Scholar] [PubMed]
- Barabási, A.L.; Albert, R. Emergence of scaling in random networks. Science 1999, 286, 509–512. [Google Scholar]
- Zhou, T.; Lü, L.; Zhang, Y.C. Predicting missing links via local information. Eur. Phys. J. B 2009, 71, 623–630. [Google Scholar] [CrossRef] [Green Version]
- Katz, L. A new status index derived from sociometric analysis. Psychometrika 1953, 18, 39–43. [Google Scholar] [CrossRef]
- Jeh, G.; Widom, J. SimRank: A measure of structural-context similarity. In Proceedings of the eighth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Edmonton, AB, Canada, 23–26 July 2002; ACM: New York, NY, USA, 2002; pp. 538–543. [Google Scholar]
- Klein, D.J.; Randić, M. Resistance distance. J. Math. Chem. 1993, 12, 81–95. [Google Scholar] [CrossRef]
- Lü, L.; Zhou, T. Link prediction in complex networks: A survey. Phys. A Stat. Mech. Appl. 2011, 390, 1150–1170. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Chen, Y. Weisfeiler–Lehman neural machine for link prediction. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Halifax, NS, Canada, 13–17 August 2017; ACM: New York, NY, USA, 2017; pp. 575–583. [Google Scholar]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. DeepWalk: Online Learning of Social Representations. arXiv, 2014; arXiv:1403.6652. [Google Scholar]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 855–864. [Google Scholar]
- Tang, J.; Qu, M.; Wang, M.; Zhang, M.; Yan, J.; Mei, Q. LINE: Large-scale Information Network Embedding. arXiv, 2015; arXiv:1503.03578. [Google Scholar]
- Cai, H.; Zheng, V.W.; Chang, K.C. A Comprehensive Survey of Graph Embedding: Problems, Techniques and Applications. arXiv, 2017; arXiv:1709.07604. [Google Scholar]
- Jaccard, P. Etude de la distribution florale dans une portion des Alpes et du Jura. Bulletin De La Societe Vaudoise Des Sciences Naturelles 1901, 37, 547–579. [Google Scholar]
- Cukierski, W.; Hamner, B.; Yang, B. Graph-based features for supervised link prediction. In Proceedings of the 2011 IEEE International Joint Conference on Neural Networks (IJCNN), San Jose, CA, USA, 31 July–5 August 2011; pp. 1237–1244. [Google Scholar]
- Miller, K.; Jordan, M.I.; Griffiths, T.L. Nonparametric latent feature models for link prediction. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 7–10 December 2009; pp. 1276–1284. [Google Scholar]
- Zhang, M.; Chen, Y. Link Prediction Based on Graph Neural Networks. arXiv, 2018; arXiv:1802.09691. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.; Dean, J. Distributed Representations of Words and Phrases and their Compositionality. arXiv, 2013; arXiv:1310.4546. [Google Scholar]
- Weisfeiler, B.; Lehman, A. A reduction of a graph to a canonical form and an algebra arising during this reduction. Nauchno-Technicheskaya Informatsia 1968, 2, 12–16. [Google Scholar]
- USAir Dataset. Available online: http://web.mit.edu/airlinedata/www/default.html (accessed on 2 January 2019).
- Ackland, R. Mapping the Us Political Blogosphere: Are Conservative Bloggers More Prominent? BlogTalk Downunder 2005 Conference, Sydney. Available online: https://core.ac.uk/download/pdf/156616040.pdf (accessed on 2 January 2019).
- Newman, M.E. Finding community structure in networks using the eigenvectors of matrices. Phys. Rev. E 2006, 74, 036104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Cui, Z.; Oyetunde, T.; Tang, Y.; Chen, Y. Recovering Metabolic Networks using A Novel Hyperlink Prediction Method. arXiv, 2016; arXiv:1610.06941. [Google Scholar]
- Spring, N.; Mahajan, R.; Wetherall, D. Measuring ISP topologies with Rocketfuel. ACM SIGCOMM Comput. Commun. Rev. 2002, 32, 133–145. [Google Scholar] [CrossRef]
- Peduzzi, P.; Concato, J.; Kemper, E.; Holford, T.R.; Feinstein, A.R. A simulation study of the number of events per variable in logistic regression analysis. J. Clin. Epidemiol. 1996, 49, 1373–1379. [Google Scholar] [CrossRef]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Freund, Y.; Schapire, R. A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 1999, 14, 1612. [Google Scholar]
- Bengio, Y.; Louradour, J.; Collobert, R.; Weston, J. Curriculum learning. In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; ACM: New York, NY, USA, 2009; pp. 41–48. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Chen, Y.Y.; Gan, Q.; Suel, T. Local methods for estimating pagerank values. In Proceedings of the thirteenth ACM International Conference on Information and Knowledge Management, Washington, DC, USA, 8–13 November 2004; ACM: New York, NY, USA, 2004; pp. 381–389. [Google Scholar]
- Rendle, S. Factorization machines with libfm. ACM Trans. Intell. Syst. Technol. (TIST) 2012, 3, 57. [Google Scholar] [CrossRef]




| Method | Formula |
|---|---|
| common neighbors | |
| Jaccard | |
| preferential attachment | |
| Adamic-Adar | |
| resource allocation | |
| resistance distance | |
| PageRank | |
| Katz |
| Datasets | |V| | |E| |
|---|---|---|
| C.ele | 297 | 2148 |
| USAir | 332 | 2126 |
| PB | 1222 | 16,714 |
| NS | 1589 | 2742 |
| E.coli | 1805 | 14,660 |
| Yeast | 2375 | 11,693 |
| Power | 4941 | 6594 |
| Router | 5022 | 6258 |
| Datasets | Logistic Regression | Decision Tree | Random Forest | Adaboost | Neural Network |
|---|---|---|---|---|---|
| C.ele | 0.8429 | 0.7186 | 0.757 | 0.7581 | 0.870 |
| USAir | 0.958 | 0.8697 | 0.8803 | 0.8756 | 0.968 |
| PB | 0.937 | 0.7992 | 0.8663 | 0.8488 | 0.9453 |
| NS | 0.937 | 0.9273 | 0.9345 | 0.9218 | 0.9832 |
| E.coli | 0.9563 | 0.8762 | 0.9212 | 0.889 | 0.9787 |
| Yeast | 0.9378 | 0.8861 | 0.9066 | 0.8878 | 0.9618 |
| Power | 0.7968 | 0.6534 | 0.6924 | 0.6693 | 0.866 |
| Router | 0.9134 | 0.6158 | 0.623 | 0.7768 | 0.921 |
| Features | Heuristics | Node2vec | WLNM | ENRNM |
|---|---|---|---|---|
| Graph structure feature | Yes | Yes | Yes | Yes |
| Node’s information | No | Yes | No | Yes |
| Edge’s information | No | No | Yes | Yes |
| Model | / | NN | NN | NN |
| Methods | Process | Computational Complexity/Sample | Time Needed (s) |
|---|---|---|---|
| WLNM | edge (link) pattern encoding | 44.912 | |
| Node2vec | node pattern encoding link pattren encoding | 10.326 0.500 | |
| ENRNM | node pattern encoding edge pattern encoding link pattren encoding | 10.628 46.646 1.000 |
| Datesets | CN | Jac | AA | RA | PA | Katz | RD | PR | SR | SBM | MF-c | MF-r | ENRNM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C.ele | 0.849 | 0.793 | 0.863 | 0.867 | 0.756 | 0.865 | 0.741 | 0.902 | 0.761 | 0.866 | 0.838 | 0.843 | 0.870 |
| USAir | 0.941 | 0.904 | 0.949 | 0.957 | 0.893 | 0.930 | 0.897 | 0.943 | 0.783 | 0.945 | 0.919 | 0.845 | 0.968 |
| PB | 0.92 | 0.874 | 0.923 | 0.922 | 0.9012 | 0.927 | 0.882 | 0.934 | 0.772 | 0.937 | 0.933 | 0.942 | 0.9453 |
| NS | 0.939 | 0.939 | 0.939 | 0.937 | 0.6823 | 0.941 | 0.583 | 0.941 | 0.941 | 0.921 | 0.638 | 0.721 | 0.9832 |
| E.coli | 0.933 | 0.807 | 0.953 | 0.959 | 0.913 | 0.928 | 0.887 | 0.952 | 0.638 | 0.938 | 0.906 | 0.918 | 0.9787 |
| Yeast | 0.892 | 0.891 | 0.892 | 0.893 | 0.825 | 0.922 | 0.881 | 0.926 | 0.915 | 0.915 | 0.839 | 0.883 | 0.9618 |
| Power | 0.592 | 0.591 | 0.592 | 0.591 | 0.442 | 0.656 | 0.844 | 0.665 | 0.762 | 0.664 | 0.522 | 0.515 | 0.866 |
| Router | 0.561 | 0.562 | 0.561 | 0.562 | 0.472 | 0.379 | 0.925 | 0.381 | 0.368 | 0.852 | 0.775 | 0.782 | 0.921 |
| Datasets | Node2vec | WLNM | ENRNM |
|---|---|---|---|
| C.ele | 0.6604 | 0.859 | 0.870 |
| USAir | 0.8477 | 0.958 | 0.968 |
| PB | 0.9168 | 0.933 | 0.9453 |
| NS | 0.9657 | 0.984 | 0.9832 |
| E.coli | 0.9691 | 0.971 | 0.9787 |
| Yeast | 0.9524 | 0.956 | 0.9618 |
| Power | 0.8567 | 0.848 | 0.866 |
| Router | 0.640 | 0.944 | 0.921 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, G.; Wang, X.; Wang, Y.; Lin, D.; Sun, X.; Fu, K. Edge-Nodes Representation Neural Machine for Link Prediction. Algorithms 2019, 12, 12. https://doi.org/10.3390/a12010012
Xu G, Wang X, Wang Y, Lin D, Sun X, Fu K. Edge-Nodes Representation Neural Machine for Link Prediction. Algorithms. 2019; 12(1):12. https://doi.org/10.3390/a12010012
Chicago/Turabian StyleXu, Guangluan, Xiaoke Wang, Yang Wang, Daoyu Lin, Xian Sun, and Kun Fu. 2019. "Edge-Nodes Representation Neural Machine for Link Prediction" Algorithms 12, no. 1: 12. https://doi.org/10.3390/a12010012
APA StyleXu, G., Wang, X., Wang, Y., Lin, D., Sun, X., & Fu, K. (2019). Edge-Nodes Representation Neural Machine for Link Prediction. Algorithms, 12(1), 12. https://doi.org/10.3390/a12010012





