Effect of Silver or Copper Nanoparticles-Dispersed Silane Coatings on Biofilm Formation in Cooling Water Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimens
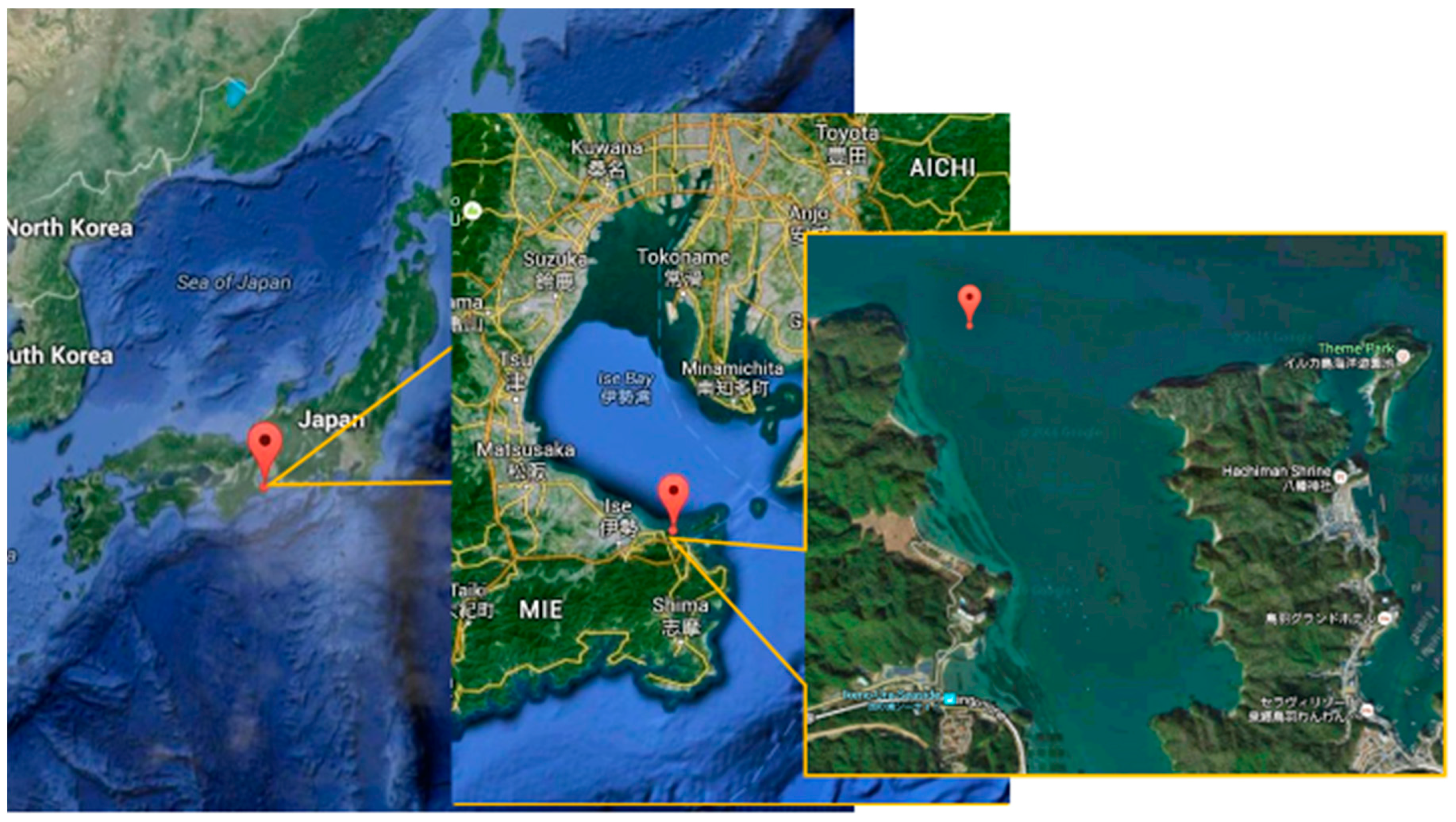
2.2. Sampling Seawater
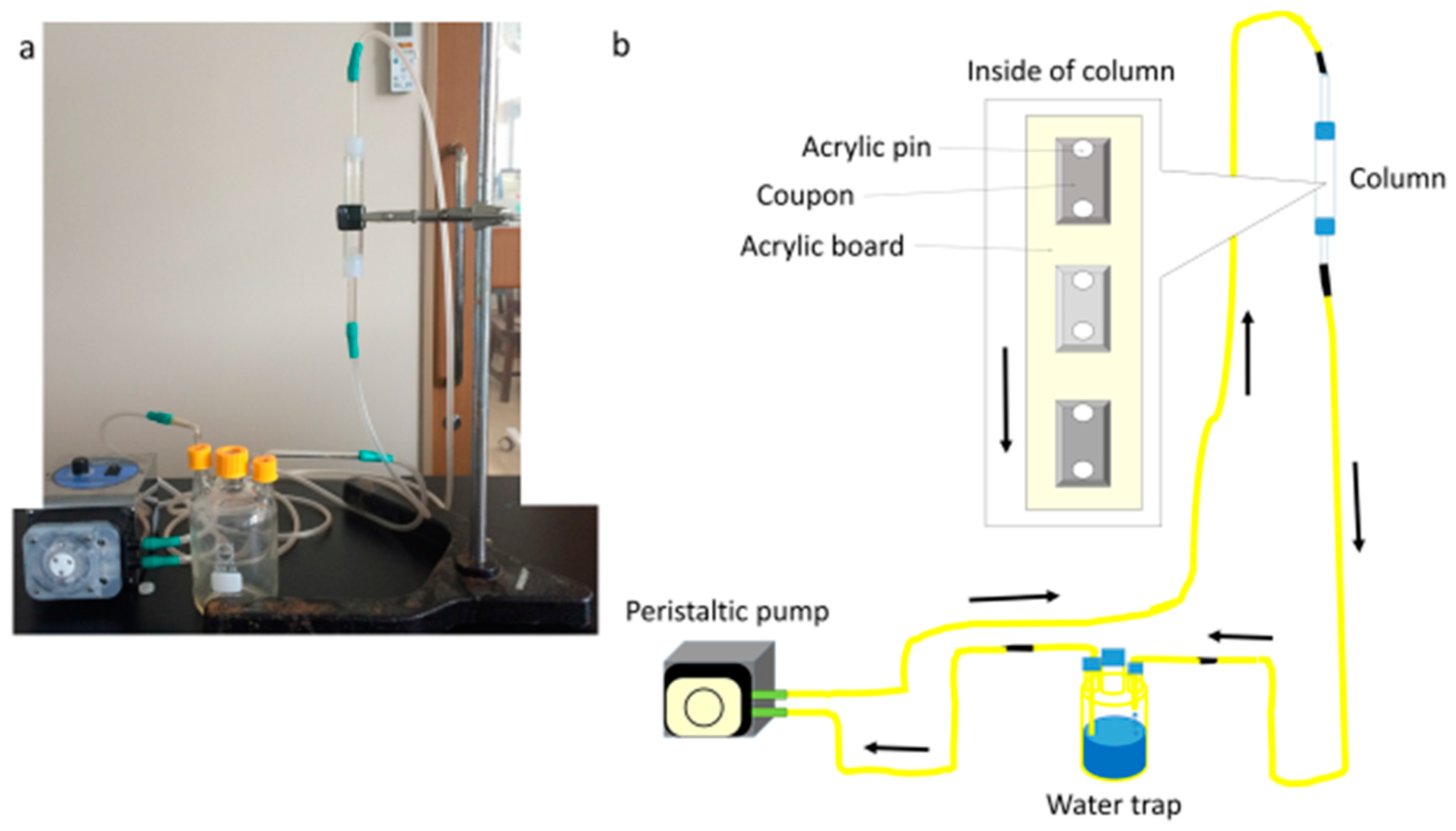
2.3. Biofilm Formation
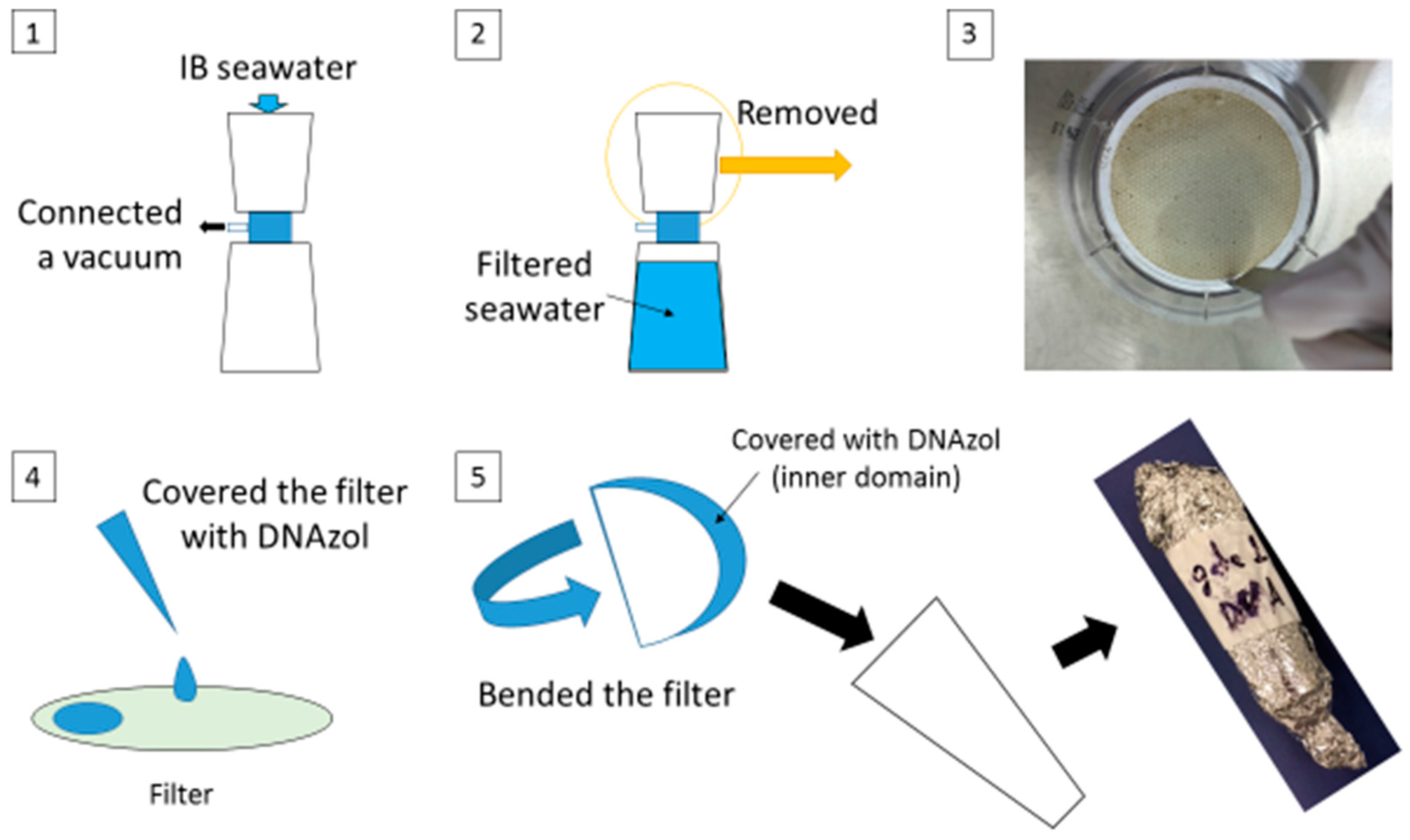
2.4. DNA Extraction from Seawater
2.5. 16S rRNA Gene-Based Bacterial Community Analysis
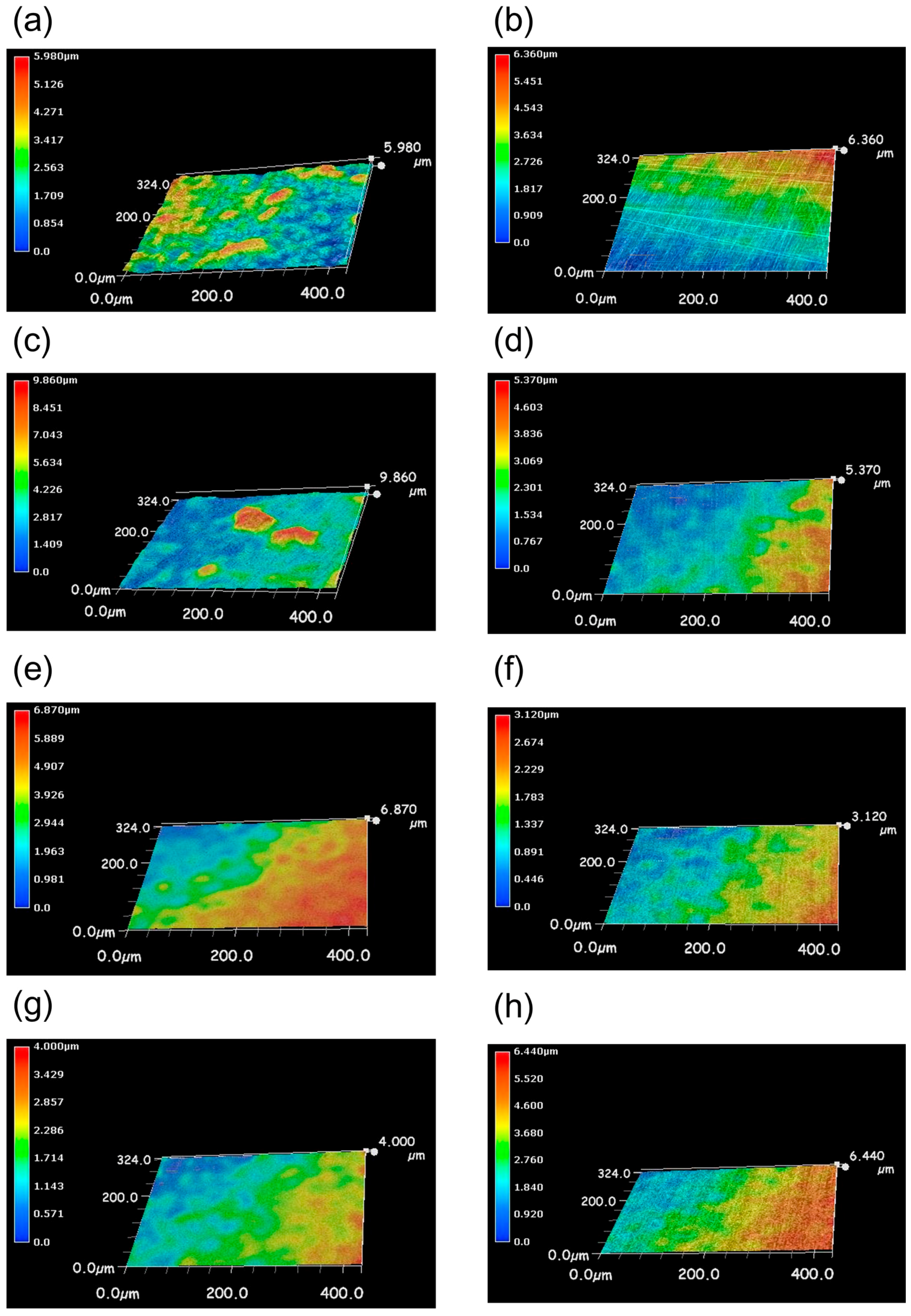
2.6. Three-Dimensional Image of the Surface of Coupons
2.7. Raman Spectroscopy Analysis
3. Results and Discussions
3.1. Biofilm Formation on the Surface of Stainless Steel Using Seawater
3.2. Bacteria Community of IB Seawater
3.3. Comparison of Biofilm Formation among Several Coatings
3.4. The Effect of Silver or Copper Dispersed Silane Coating Stainless Steel on Anti-Biofilm Formation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rondum, K.D.; Cabell, A.C.; Beardwood, E.S. Performance-based cooling water treatment. Power 2012, 156, 46–51. [Google Scholar]
- Sriyutha Murthy, P.; Venkatesan, R. Industrial biofilms and their control. In Marine and Industrial Biofouling; Flemming, H.C., Sriyutha Murthy, P., Venkatesan, R., Coolsey, K.E., Eds.; Springer: Berlin, Germany, 2009; Volume 4, pp. 65–101. [Google Scholar]
- Venugopalan, V.P.; Sanjeevi Rajagopal, S.; Jenner, H.A. Operational and environmental issues relating to industrial cooling water systems. In An Overview in Operational and Environmental Consequences of Large Industrial Cooling Water Systems; Rajagopal, S., Jenner, H.A., Venugopalan, V.P., Eds.; Springer: New York, NY, USA, 2012; pp. 1–12. [Google Scholar]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Ann. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [PubMed]
- Zbigniew Lewandowski, Z.; Beyenal, H. Fundamentals of Biofilm Research, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Benevides, J.M.; Overman, S.A.; Thomas, G.J. Raman, polarized Raman and ultraviolet resonance Raman spectroscopy of nucleic acids and their complexes. J. Raman Spectrosc. 2005, 36, 279–299. [Google Scholar] [CrossRef]
- Bunaciu, A.A.; Aboul-Enein, H.Y.; Hoang, V.D. Raman Spectroscopy for Protein Analysis. Appl. Spectrosc. Rev. 2004, 50, 377–386. [Google Scholar] [CrossRef]
- Baeten, V. Raman spectroscopy in lipid analysis. Lipid Technol. 2010, 22, 36–38. [Google Scholar] [CrossRef]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Sandt, C.; Smith-Palmer, T.; Pink, J.; Brennan, L.; Pink, D. Confocal Raman microspectroscopy as a tool for studying the chemical heterogeneities of biofilms in situ. J. Appl. Microbiol. 2007, 103, 1808–1820. [Google Scholar] [CrossRef] [PubMed]
- Samek, O.; Al-Marashi, J.F.M.; Telle, H.H. The potential of Raman spectroscopy for the identification of biofilm formation by Staphylococcus Epidermidis. Laser Phys. Lett. 2010, 7, 378–383. [Google Scholar] [CrossRef]
- Garima Sharma, G.; Prakash, A. Combined use of Fourier transform infrared and Raman spectroscopy to study planktonic and biofilm cells of Cronobacter sakazakii. J. Microbiol. Biotech. Food Sci. 2014, 3, 310–314. [Google Scholar]
- Samek, O.; Mlynarikova, K.; Bernatova, S.; Jezek, J.; Krzyzanek, V.; Siler, M.; Zemanek, P.; Ruzicka, F.; Hola, V.; Mahelova, M. Candida parapsilosis biofilm identification by Raman spectroscopy. Int. J. Mol. Sci. 2014, 15, 23924–23935. [Google Scholar] [CrossRef] [PubMed]
- Sano, K.; Kanematsu, H.; Kogo, T.; Hirai, N.; Tanaka, T. Corrosion and biofilm for a composite coated iron observed by FTIR-ATR and Raman spectroscopy. Trans. IMF 2016, 94, 139–145. [Google Scholar] [CrossRef]
- Ogawa, A.; Noda, M.; Kanematsu, H.; Sano, K. Application of bacterial16S rRNA gene analysis to a comparison of the degree of biofilm formation on the surface of metal coated glasses. Mater. Technol. 2015, 30, B61–B65. [Google Scholar] [CrossRef]
- Sano, K.; Kanematsu, H.; Hirai, N.; Tanaka, T. Preparation and its anti-biofouling effect observation of organic metal dispersed silane-based composite coating. J. Surf. Finish. Soc. Jpn. 2016, 67, 268–273. (In Japanese) [Google Scholar]
- Faúndez, G.; Troncoso, M.; Navarrete, P.; Figueroa, G. Antimicrobial activity of copper surfaces against suspensions of Salmonella enterica and Campylobacter jejuni. BMC Microbiol. 2004, 4, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehtar, S.; Wiid, I.; Todorov, S.D. The antimicrobial activity of copper and copper alloys against nosocomial pathogens and Mycobacterium tuberculosis isolated from healthcare facilities in the Western Cape: An in vitro study. J. Hosp. Infect. 2008, 68, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Sano, K.; Kanematsu, H.; Tanaka, T. Overview of Silane-Based Polymer Coatings and Their Applications; Hosseini, M., Makhlouf, H.A.S., Eds.; Springer: New York, NY, USA, 2016; pp. 493–509. [Google Scholar]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. drive5. Available online: http://www.drive5.com/ (accessed on 26 July 2016).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Quantitative Insights Into Microbial Ecology. Available online: http://qiime.org/index.html (accessed on 26 July 2016).
- Aronesty, E. ea-utils: “Command-line tools for processing biological sequencing data”. Available online: https://github.com/ExpressionAnalysis/ea-utils/ (accessed on 26 July 2016).
- Aronesty, E. Comparison of sequencing utility programs. Open Bioinform. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Kanematsu, H.; Barry, D.M. Chapter New Evaluation Techniques for Biofilm in Materials Science. In Biofilm and Materials Science; Kanematsu, H., Barry, D.M., Eds.; Springer: Cham, Switzerland, 2015; pp. 187–188. [Google Scholar]
- Frost, R.L.; Xi, Y.; Scholz, R.; López, A.; Belotti, F.M. Vibrational spectroscopic characterization of the phosphate mineral hureaulite—(Mn, Fe)5(PO4)2(HPO4)24(H2O). Vib. Spectrosc. 2013, 66, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Chao, Y.; Zhang, T. Surface-enhanced Raman scattering (SERS) revealing chemical variation during biofilm formation: From initial attachment to mature biofilm. Anal. Bioanal. Chem. 2012, 404, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.W.; Ng, C.Y.; Ren, J.; Connie, Y.M.N.; Ren, J.; Simon, C.L.L.; Qian, P.; Wong, P.K.; Lau, T.C.; Wu, M. Owenweeksia hongkongensis gen. nov., sp. nov., a novel marine bacterium of the phylum ‘Bacteroidetes’. Int. J. Syst. Evol. Microbiol. 2005, 55, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Elifantz, H.; Horn, G.; Ayon, M.; Cohen, Y.; Minz, D. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 2013, 85, 348–357. [Google Scholar] [CrossRef] [PubMed]
- World Register of Marine Species. Available online: http://www.marinespecies.org/index.php (accessed on 26 July 2016).
- Suzuki, M.; Nakagawa, Y.; Harayama, S.; Yamamoto, S. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int. J. System. Evolut. Microbiol. 2001, 51, 1639–1652. [Google Scholar]
- Barry, D.M.; Kanematsu, H. Cooling water. In Biofilm and Materials Science; Kanematsu, H., Barry, D.M., Eds.; Springer: Cham, Switzerland, 2015; pp. 79–83. [Google Scholar]
- Angelos, G.; Kalampounias, A.G. IR and Raman spectroscopic studies of sol-gel derived alkaline-earth silicate glass. Bull. Mater. Sci. 2011, 34, 299–303. [Google Scholar]
- McMillan, P.; Putnis, A.; Carpenter, M.A. A Raman Spectroscopic Study of AI-Si Ordering in Synthetic Magnesium Cordierite. Phys. Chem. Miner. 1984, 10, 256–260. [Google Scholar] [CrossRef]
- Lin-Vien, D.; Colthrup, N.B.; fateley, W.G.; Grasselli, J.G. Alkanes. In The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules, 1st ed.; Academic Press: London, UK, 1991; pp. 9–28. [Google Scholar]
- Colangeli, L.; Mennella, V.; Baratta, G.A.; Bussoletti, E.; Strazzulla, G. Raman and infrared spectra of polycyclic aromatic hydrocarbon molecules of possible astrophysical interest. Astrophys. J. 1992, 396, 369–377. [Google Scholar] [CrossRef]
- Larkin, P. General outline and strategies for IR and Raman spectral interpretation. In Infrared and Raman Spectroscopy Principles and Spectral Interpretation; Elsevier: Burlington, MA, USA, 2011; pp. 117–133. [Google Scholar]
- Kalishwaralal, K.; BarathManiKanth, S.; Pandian, S.R.; Deepak, V.; Gurunathan, S. Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surf. B Biointerfaces 2010, 79, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Kwon, D.N.; Kim, J.H. Enhanced antibacterial and anti-biofilm activities of silver nanoparticles against Gram-negative and Gram-positive bacteria. Nanoscale Res. Lett. 2014, 9, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Ruparelia, J.P.; Chatterjee, A.K.; Duttagupta, S.P.; Mukherji, S. Strain specificity in antimicrobial activity of silver and copper nanoparticles. Acta Biomater. 2008, 4, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Vines, F.; Gomes, J.R.; Illas, F. Understanding the reactivity of metallic nanoparticles: Beyond the extended surface model for catalysis. Chem. Soc. Rev. 2014, 43, 4922–4939. [Google Scholar] [CrossRef] [PubMed]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]


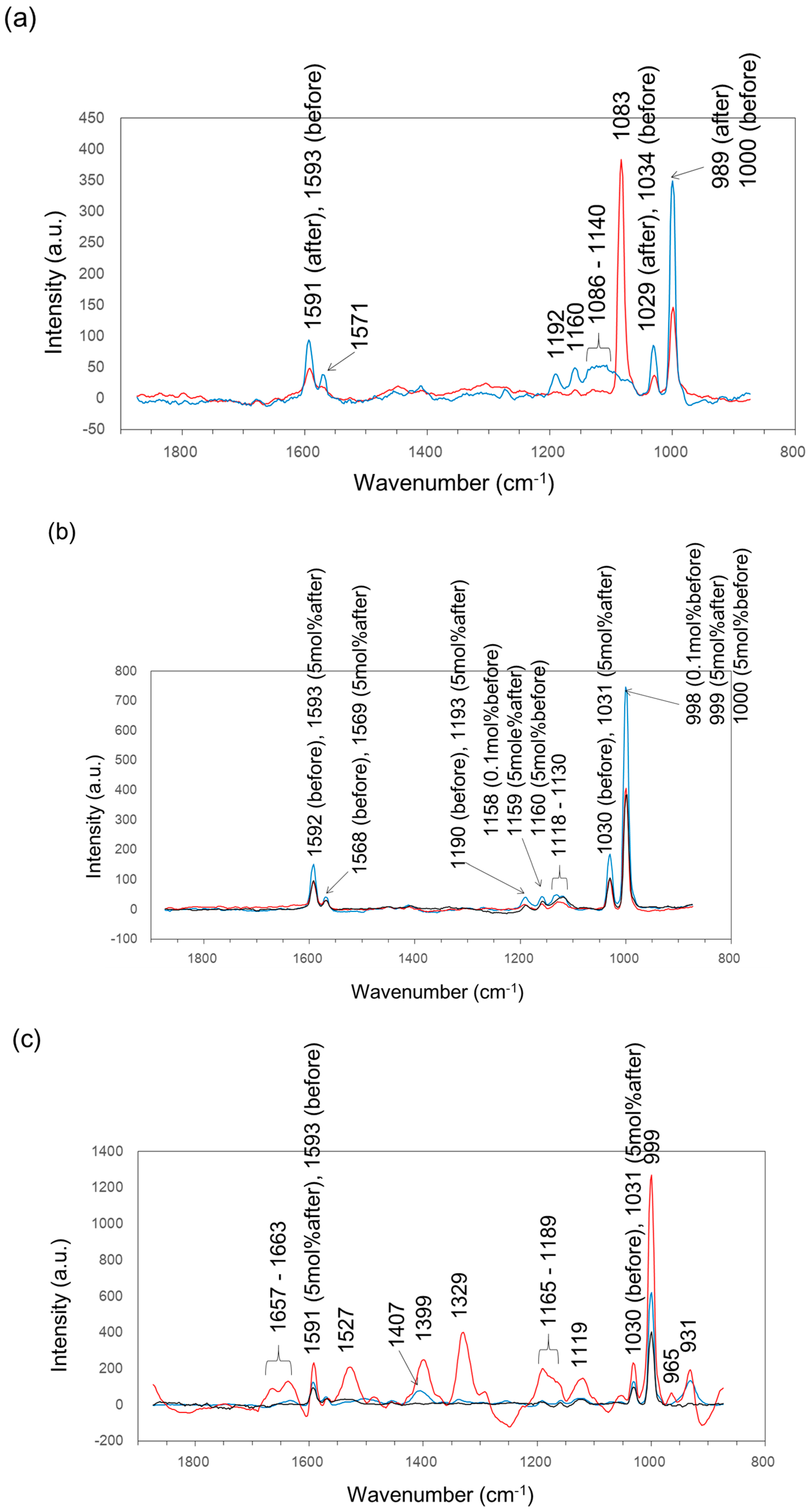
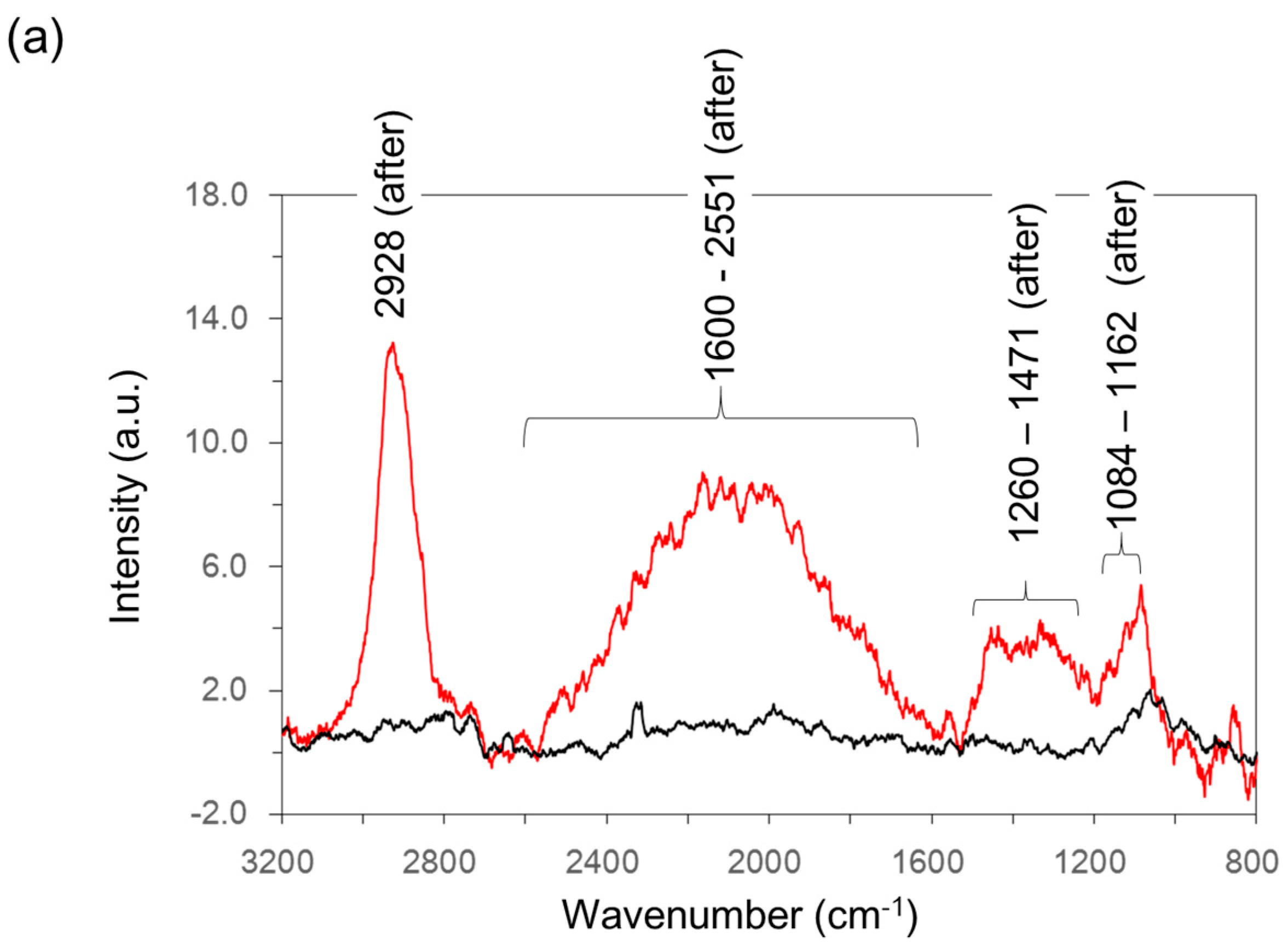
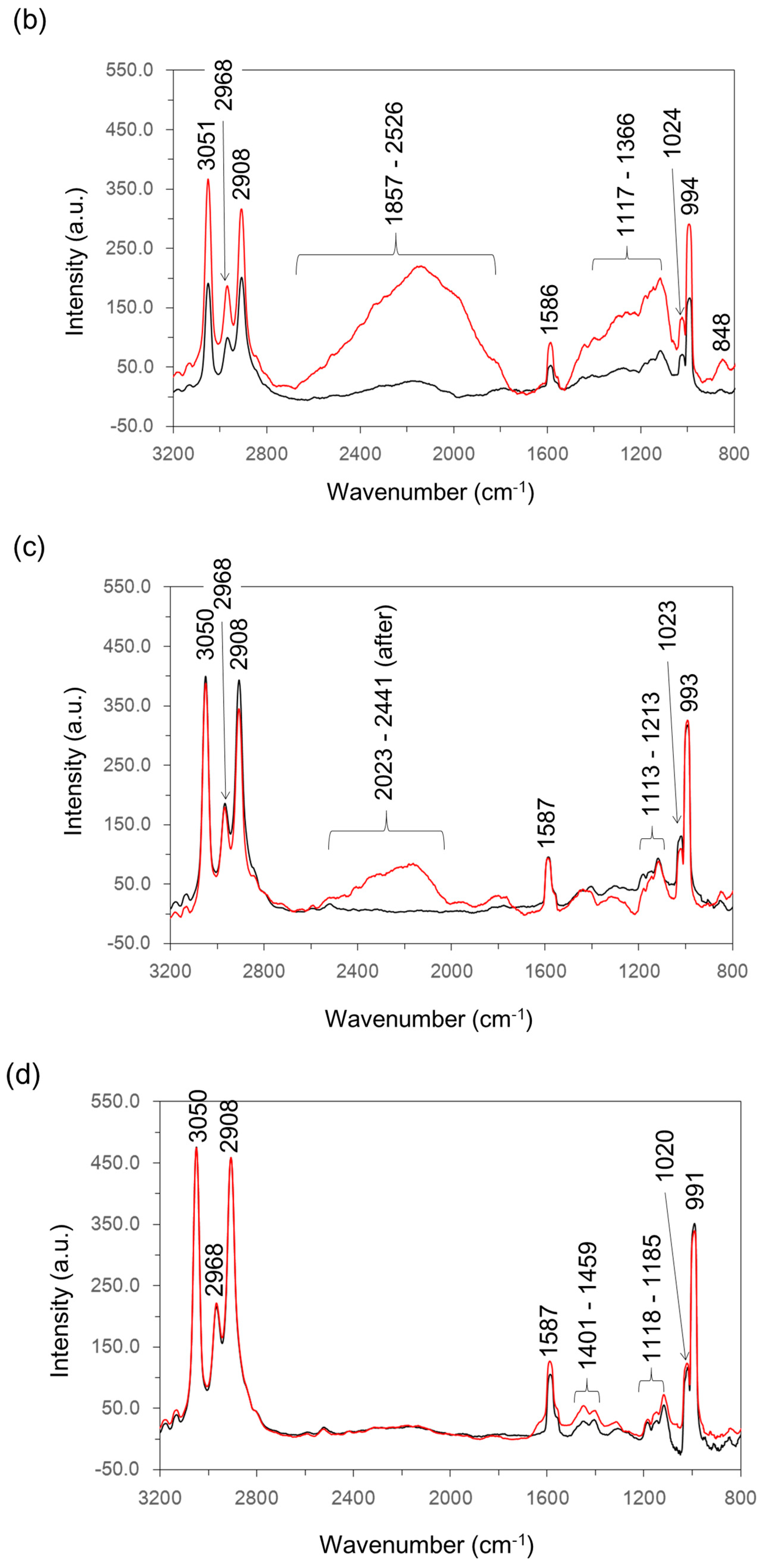
| Wavenumber (cm−1) | Assignment | Reference |
|---|---|---|
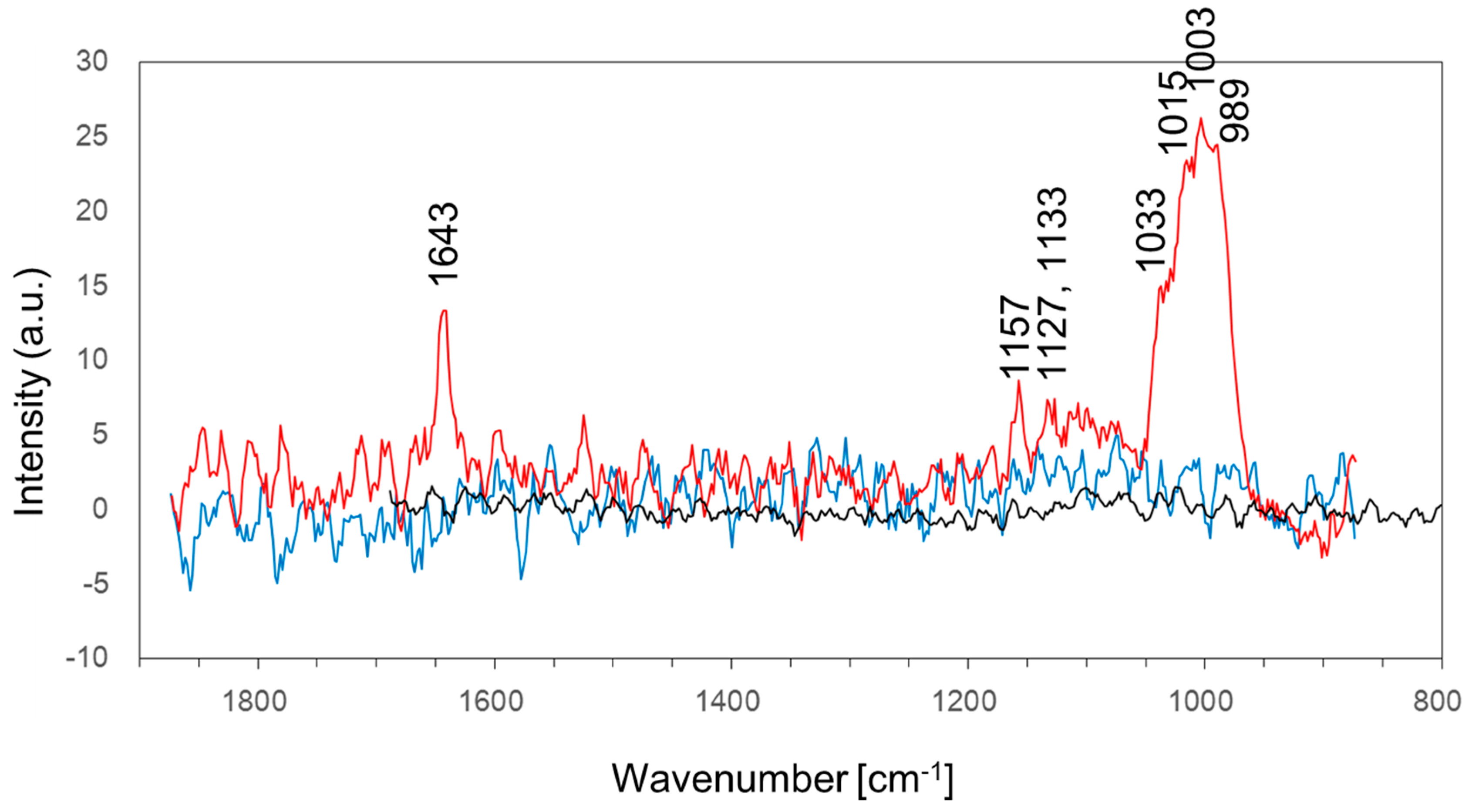
| 989 | phosphate ion stretching vibration | [33] |
| 1003 | phenylalanine | [34] |
| 1015 | carbohydrates peak for solids | [34] |
| 1033 | C–H in-plane phenylalanine of proteins n(CO), n(CC) and n(CCO) of polysaccharides or pectin | [34] |
| 1127 | ν(C–N) | [34] |
| 1133 | palmitic acid and fatty acid | [34] |
| 1153 | C–C bond of lipid | [34] |
| 1643 | amide I bond of the protein | [35] |
| Domain | Phylum | Class | Order | Family | Genus | Abundance (%) |
|---|---|---|---|---|---|---|
| Archaea | Euryarchaeota | Thermoplasmata | Thermoplasmatales | Marine_Group_II | - | 2.0 |
| Bacteria | Actinobacteria | Acidimicrobiia | Acidimicrobiales | OCS155_marine_group | - | 1.8 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Cryomorphaceae | Owenweeksia | 11.9 | |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | Tenacibaculum | 4.7 | |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | NS5 marine group | 4.4 | |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | NS4 marine group | 4.2 | |
| Bacteroidetes | Sphingobacteriia | Sphingobacteriales | NS11-12_marine_group | - | 6.3 | |
| Cyanobacteria | Cyanobacteria | Chloroplast | Chloroplast | Chloroplast | 5.5 | |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Roseobacter clade OCT lineage | 5.4 | |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | - | 5.3 | |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Roseobacter_clade NAC11-7 lineage | 4.8 | |
| Proteobacteria | Alphaproteobacteria | Rhodospirillales | Rhodospirillaceae | AEGEAN-169 marine group | 1.2 | |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | SAR116 clade | - | 1.7 | |
| Proteobacteria | Alphaproteobacteria | SAR11 clade | Surface_1 | - | 3.6 | |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | OM60(NOR5) clade | 4.7 | |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | SAR92_clade | 1.0 | |
| Proteobacteria | Gammaproteobacteria | KI89A clade | - | - | 1.6 | |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Oceanospirillaceae | Pseudospirillum | 1.5 | |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | SAR86 clade | - | 1.1 | |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | ZD0405 | - | 7.5 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogawa, A.; Kanematsu, H.; Sano, K.; Sakai, Y.; Ishida, K.; Beech, I.B.; Suzuki, O.; Tanaka, T. Effect of Silver or Copper Nanoparticles-Dispersed Silane Coatings on Biofilm Formation in Cooling Water Systems. Materials 2016, 9, 632. https://doi.org/10.3390/ma9080632
Ogawa A, Kanematsu H, Sano K, Sakai Y, Ishida K, Beech IB, Suzuki O, Tanaka T. Effect of Silver or Copper Nanoparticles-Dispersed Silane Coatings on Biofilm Formation in Cooling Water Systems. Materials. 2016; 9(8):632. https://doi.org/10.3390/ma9080632
Chicago/Turabian StyleOgawa, Akiko, Hideyuki Kanematsu, Katsuhiko Sano, Yoshiyuki Sakai, Kunimitsu Ishida, Iwona B. Beech, Osamu Suzuki, and Toshihiro Tanaka. 2016. "Effect of Silver or Copper Nanoparticles-Dispersed Silane Coatings on Biofilm Formation in Cooling Water Systems" Materials 9, no. 8: 632. https://doi.org/10.3390/ma9080632
APA StyleOgawa, A., Kanematsu, H., Sano, K., Sakai, Y., Ishida, K., Beech, I. B., Suzuki, O., & Tanaka, T. (2016). Effect of Silver or Copper Nanoparticles-Dispersed Silane Coatings on Biofilm Formation in Cooling Water Systems. Materials, 9(8), 632. https://doi.org/10.3390/ma9080632








