An Image-Analysis-Based Method for the Prediction of Recombinant Protein Fiber Tensile Strength
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Expression, Purification and Spinning of NT2RepCT Fibers
2.2. Native Silk Materials
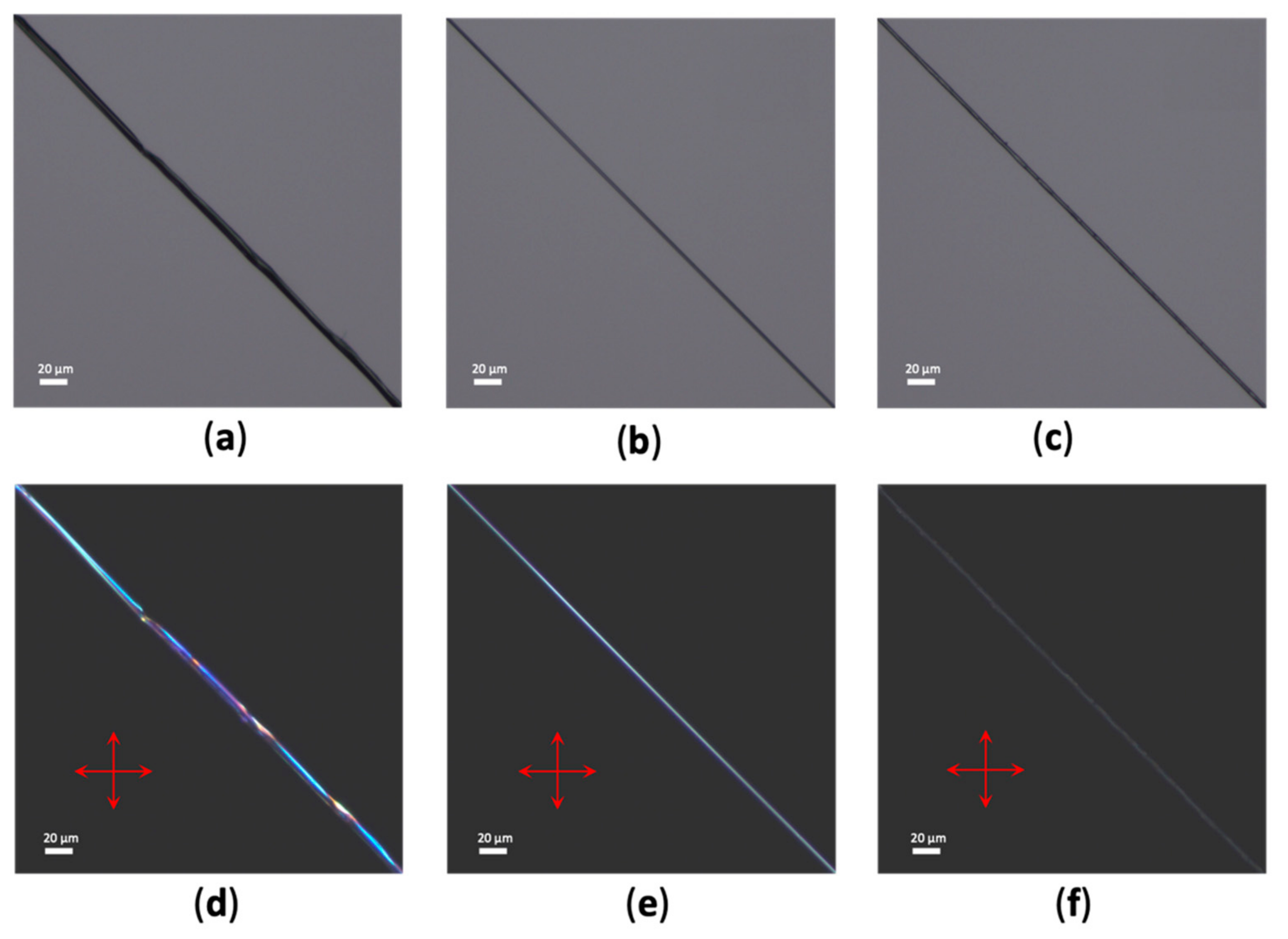
2.3. Light Microscopy
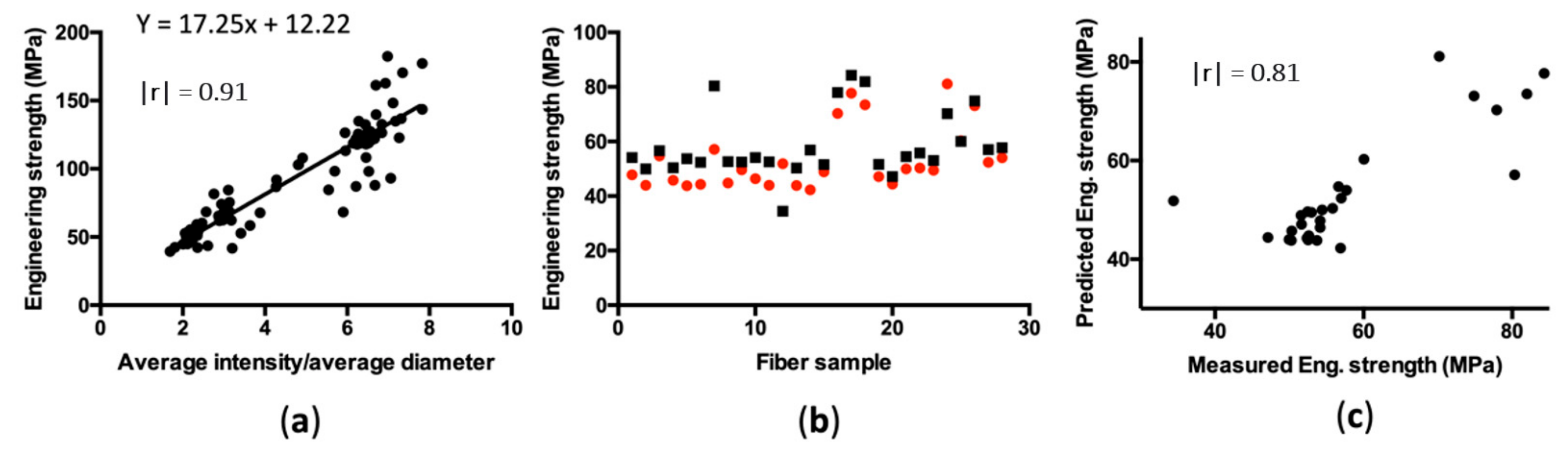
2.4. Image Analysis
2.5. Statistical Analysis
2.6. Tensile Testing
2.7. Scanning Electron Microscopy (SEM)
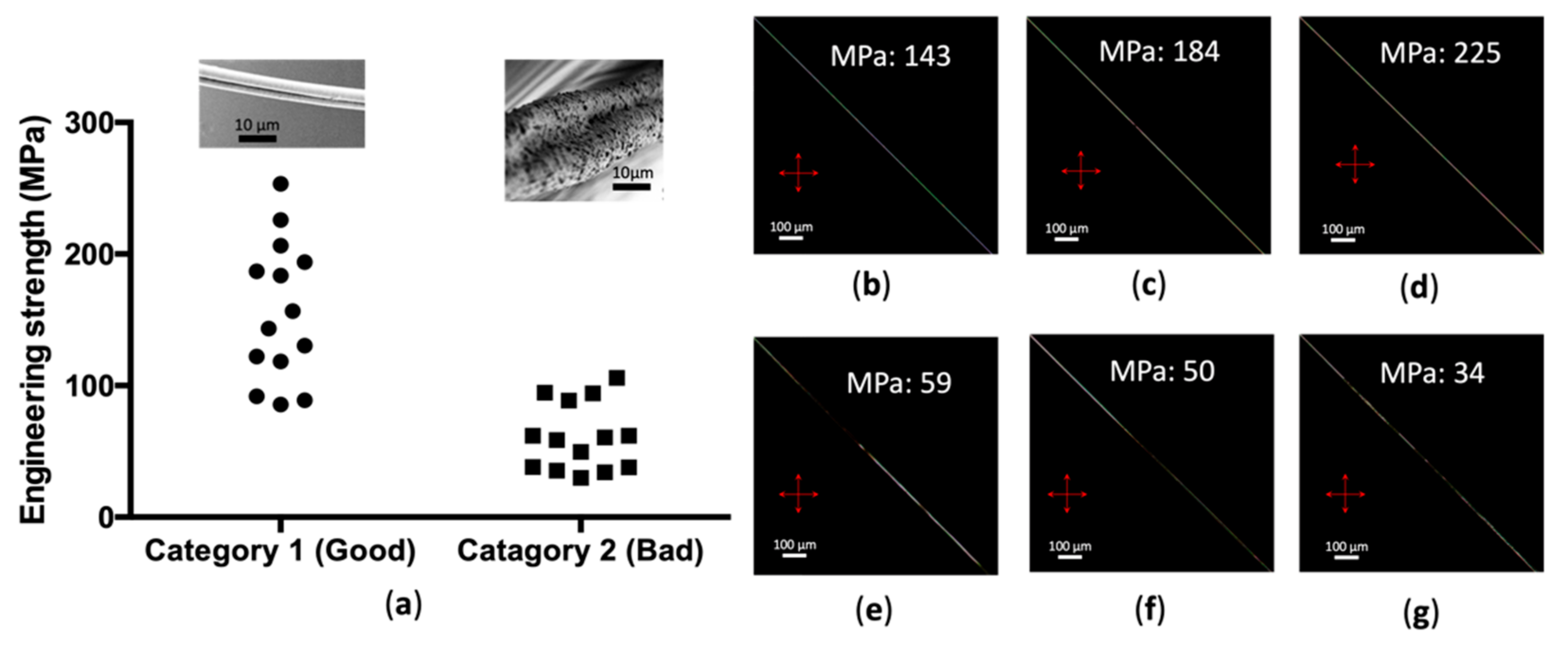
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, G.; Sapru, S.; Behera, S.; Yao, J.; Shao, Z.; Kundu, S.C.; Chen, X. Exploration of the Tight Structural-Mechanical Relationship in Mulberry and Non-Mulberry Silkworm Silks. J. Mater. Chem. B 2016, 4, 4337–4347. [Google Scholar] [CrossRef]
- Swanson, B.O.; Blackledge, T.A.; Beltrán, J.; Hayashi, C.Y. Variation in the Material Properties of Spider Dragline Silk across Species. Appl. Phys. A Mater. Sci. Process 2006, 82, 213–218. [Google Scholar] [CrossRef]
- Fu, C.; Porter, D.; Chen, X.; Vollrath, F.; Shao, Z. Understanding the Mechanical Properties of Antheraea Pernyi Silka-From Primary Structure to Condensed Structure of the Protein. Adv. Funct. Mater. 2011, 21, 729–737. [Google Scholar] [CrossRef]
- Gosline, J.M.; Guerette, P.A.; Ortlepp, C.S.; Savage, K.N. The Mechanical Design of Spider Silks: From Fibroin Sequence to Mechanical Function. J. Exp. Biol. 1999, 202, 3295–3303. [Google Scholar] [CrossRef] [PubMed]
- Fraternali, F.; Stehling, N.; Amendola, A.; Anrango, B.A.T.; Holland, C.; Rodenburg, C. Tensegrity Modelling and the High Toughness of Spider Dragline Silk. Nanomaterials 2020, 10, 1510. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhang, J.; Jordan, J.S.; Wang, X.; Henning, R.W.; Yarger, J.L. Structural Comparison of Various Silkworm Silks: An Insight into the Structure-Property Relationship. Biomacromolecules 2018, 19, 906–917. [Google Scholar] [CrossRef]
- Cranford, S.W.; Tarakanova, A.; Pugno, N.M.; Buehler, M.J. Nonlinear Material Behaviour of Spider Silk Yields Robust Webs. Nature 2012, 482, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Keten, S.; Xu, Z.; Ihle, B.; Buehler, M.J. Nanoconfinement Controls Stiffness, Strength and Mechanical Toughness of Β-Sheet Crystals in Silk. Nat. Mater. 2010, 9, 359–367. [Google Scholar] [CrossRef]
- Nova, A.; Keten, S.; Pugno, N.M.; Redaelli, A.; Buehler, M.J. Molecular and Nanostructural Mechanisms of Deformation, Strength and Toughness of Spider Silk Fibrils. Nano Lett. 2010, 10, 2626–2634. [Google Scholar] [CrossRef]
- Du, N.; Xiang, Y.L.; Narayanan, J.; Li, L.; Lim, M.L.M.; Li, D. Design of Superior Spider Silk: From Nanostructure to Mechanical Properties. Biophys. J. 2006, 91, 4528–4535. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Deng, Q.; Yang, Z.; Yang, D.; Han, M.Y.; Liu, X.Y. “Nano-Fishnet” Structure Making Silk Fibers Tougher. Adv. Funct. Mater. 2016, 26, 5534–5541. [Google Scholar] [CrossRef]
- Blackledge, T.A.; Pérez-Rigueiro, J.; Plaza, G.R.; Perea, B.; Navarro, A.; Guinea, G.V.; Elices, M. Sequential Origin in the High Performance Properties of Orb Spider Dragline Silk. Sci. Rep. 2012, 2, 650–662. [Google Scholar] [CrossRef]
- Swanson, B.O.; Anderson, S.P.; DiGiovine, C.; Ross, R.N.; Dorsey, J.P. The Evolution of Complex Biomaterial Performance: The Case of Spider Silk. Integr. Comp. Biol. 2009, 49, 21–31. [Google Scholar] [CrossRef][Green Version]
- Greco, G.; Wolff, J.O.; Pugno, N.M. Strong and Tough Silk for Resilient Attachment Discs: The Mechanical Properties of Piriform Silk in the Spider Cupiennius Salei (Keyserling, 1877). Front. Mater. 2020, 7, 138. [Google Scholar] [CrossRef]
- Greco, G.; Pugno, N.M. Mechanical Properties and Weibull Scaling Laws of Unknown Spider Silks. Molecules 2020, 25, 2938. [Google Scholar] [CrossRef] [PubMed]
- Omenetto, F.G.; Kaplan, D.L. New Opportunities for an Ancient Material. Science 2010, 329, 528–531. [Google Scholar] [CrossRef]
- Kluge, J.A.; Rabotyagova, O.; Leisk, G.G.; Kaplan, D.L. Spider Silks and Their Applications. Trends Biotechnol. 2008, 26, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Allmeling, C.; Jokuszies, A.; Reimers, K.; Kall, S.; Choi, C.Y.; Brandes, G.; Kasper, C.; Scheper, T.; Guggenheim, M.; Vogt, P.M. Spider Silk Fibres in Artificial Nerve Constructs Promote Peripheral Nerve Regeneration. Cell Prolif. 2008, 41, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Altman, G.H.; Diaz, F.; Jakuba, C.; Calabro, T.; Horan, R.L.; Chen, J.; Lu, H.; Richmond, J.; Kaplan, D.L. Silk-Based Biomaterials. Biomaterials 2003, 24, 401–416. [Google Scholar] [CrossRef]
- Chouhan, D.; Mandal, B.B. Silk Biomaterials in Wound Healing and Skin Regeneration Therapeutics: From Bench to Bedside. Acta Biomater. 2020, 103, 24–51. [Google Scholar] [CrossRef]
- Kim, S.; Mitropoulos, A.N.; Spitzberg, J.D.; Tao, H.; Kaplan, D.L.; Omenetto, F.G. Silk Inverse Opals. Nat. Photonics 2012, 6, 818–823. [Google Scholar] [CrossRef]
- Omenetto, F.G.; Kaplan, D.L. A New Route for Silk. Nat. Photonics 2008, 2, 641–643. [Google Scholar] [CrossRef]
- Foelix, R.F. (Ed.) Biology of Spider; Oxford University Press: Oxford, UK.
- Salehi, S.; Scheibel, T. Biomimetic Spider Silk Fibres: From Vision to Reality. Biochemist 2018, 40, 4–7. [Google Scholar] [CrossRef]
- Saric, M.; Scheibel, T. Engineering of Silk Proteins for Materials Applications. Curr. Opin. Biotechnol. 2019, 60, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, A.; Holland, C. Progress and Trends in Artificial Silk Spinning: A Systematic Review. ACS Biomater. Sci. Eng. 2017, 3, 226–237. [Google Scholar] [CrossRef]
- Menezes, G.M.; Teulé, F.; Lewis, R.V.; Silva, L.P.; Rech, E.L. Nanoscale Investigations of Synthetic Spider Silk Fibers Modified by Physical and Chemical Processes. Polym. J. 2013, 45, 997–1006. [Google Scholar] [CrossRef][Green Version]
- Ward, I.M. (Ed.) Structure and Properties of Oriented Polymers; Chapman and Hall: London, UK, 1997. [Google Scholar]
- Houck, M.M.; Siegel, J.A. Fundamentals of Forensic Science, 3rd ed.; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Jin, H.J.; Kaplan, D.L. Mechanism of Silk Processing in Insects and Spiders. Nature 2003, 424, 1057–1061. [Google Scholar] [CrossRef]
- Lazaris, A.; Arcidiacono, S.; Huang, Y.; Zhou, J.F.; Duguay, F.; Chretien, N.; Welsh, E.A.; Soares, J.W.; Karatzas, C.N. Spider Silk Fibers Spun from Soluble Recombinant Silk Produced in Mammalian Cells. Science 2002, 295, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Cannon, D.; Donald, A.M. Control of Liquid Crystallinity of Amyloid-Forming Systems. Soft Matter 2013, 9, 2852–2857. [Google Scholar] [CrossRef]
- Wang, L.; Bäcklund, F.G.; Yuan, Y.; Nagamani, S.; Hanczyc, P.; Sznitko, L.; Solin, N. Air-Water Interface Assembly of Protein Nanofibrils Promoted by Hydrophobic Additives. ACS Sustain. Chem. Eng. 2021, 9, 9289–9299. [Google Scholar] [CrossRef]
- Shu, T.; Zheng, K.; Zhang, Z.; Ren, J.; Wang, Z.; Pei, Y.; Yeo, J.; Gu, G.X.; Ling, S. Birefringent Silk Fibroin Hydrogel Constructed via Binary Solvent-Exchange-Induced Self-Assembly. Biomacromolecules 2021, 22, 1955–1965. [Google Scholar] [CrossRef] [PubMed]
- Bäcklund, F.G.; Elfwing, A.; Musumeci, C.; Ajjan, F.; Babenko, V.; Dzwolak, W.; Solin, N.; Inganäs, O. Conducting Microhelices from Self-Assembly of Protein Fibrils. Soft Matter 2017, 13, 4412–4417. [Google Scholar] [CrossRef]
- Shtukenberg, A.G.; Punin, Y.O.; Gunn, E.; Kahr, B. Spherulites. Chem. Rev. 2012, 112, 1805–1838. [Google Scholar] [CrossRef] [PubMed]
- Bäcklund, F.G.; Solin, N. Tuning the Aqueous Self-Assembly Process of Insulin by a Hydrophobic Additive. RSC Adv. 2015, 5, 92254–92262. [Google Scholar] [CrossRef]
- Bäcklund, F.G.; Pallbo, J.; Solin, N. Controlling Amyloid Fibril Formation by Partial Stirring. Biopolymers 2016, 105, 249–259. [Google Scholar] [CrossRef]
- Sipe, J.D.; Cohen, A.S. Review: History of the Amyloid Fibril. J. Struct. Biol. 2000, 130, 88–98. [Google Scholar] [CrossRef]
- Sokkar, T.Z.N.; El-Bakary, M.A. The Refractive Index Profile of Highly-Oriented Fibres. J. Phys. D Appl. Phys. 2001, 34, 373–378. [Google Scholar] [CrossRef]
- Carmichael, S.; Barghout, J.Y.J.; Viney, C. The Effect of Post-Spin Drawing on Spider Silk Microstructure: A Birefringence Model. Int. J. Biol. Macromol. 1999, 24, 219–226. [Google Scholar] [CrossRef]
- Carmichael, S.; Viney, C. Molecular Order in Spider Major Ampullate Silk (Dragline): Effects of Spinning Rate and Post-Spin Drawing. J. Appl. Polym. Sci. 1999, 72, 895–903. [Google Scholar] [CrossRef]
- Munro, T.; Putzeys, T.; Copeland, C.G.; Xing, C.; Lewis, R.V.; Ban, H.; Glorieux, C.; Wubbenhorst, M. Investigation of Synthetic Spider Silk Crystallinity and Alignment via Electrothermal, Pyroelectric, Literature XRD, and Tensile Techniques. Macromol. Mater. Eng. 2017, 302, 1600480. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Shao, H.; Hu, X.; Zhang, Y. Role of Humidity on the Structures and Properties of Regenerated Silk Fibers. Prog. Nat. Sci. Mater. Int. 2015, 25, 430–436. [Google Scholar] [CrossRef]
- Work, R.W. Dimensions, Birefringences, and Force-Elongation Behavior of Major and Minor Ampullate Silk Fibers from Orb-Web-Spinning Spiders—The Effects of Wetting on These Properties. Text. Res. J. 1977, 47, 650–662. [Google Scholar] [CrossRef]
- Augsten, K.; Mühlig, P.; Herrmann, C. Glycoproteins and Skin-Core Structure in Nephila Clavipes Spider Silk Observed by Light and Electron Microscopy. Scanning 2000, 22, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Poza, P.; Pérez-Rigueiro, J.; Elices, M.; LLorca, J. Fractographic Analysis of Silkworm and Spider Silk. Eng. Fract. Mech. 2002, 69, 1035–1048. [Google Scholar] [CrossRef]
- Kujala, S.; Mannila, A.; Karvonen, L.; Kieu, K.; Sun, Z. Natural Silk as a Photonics Component: A Study on Its Light Guiding and Nonlinear Optical Properties. Sci. Rep. 2016, 6, 22358. [Google Scholar] [CrossRef]
- Bowen, C.H.; Dai, B.; Sargent, C.J.; Bai, W.; Ladiwala, P.; Feng, H.; Huang, W.; Kaplan, D.L.; Galazka, J.M.; Zhang, F. Recombinant Spidroins Fully Replicate Primary Mechanical Properties of Natural Spider Silk. Biomacromolecules 2018, 19, 3853–3860. [Google Scholar] [CrossRef]
- Liu, W.; Huang, C.; Jin, X. Electrospinning of Grooved Polystyrene Fibers: Effect of Solvent Systems. Nanoscale Res. Lett. 2015, 10, 237. [Google Scholar] [CrossRef]
- Dou, Y.; Wang, Z.P.; He, W.; Jia, T.; Liu, Z.; Sun, P.; Wen, K.; Gao, E.; Zhou, X.; Hu, X.; et al. Artificial Spider Silk from Ion-Doped and Twisted Core-Sheath Hydrogel Fibres. Nat. Commun. 2019, 10, 5293. [Google Scholar] [CrossRef]
- Schmuck, B.; Greco, G.; Barth, A.; Pugno, N.M.; Johansson, J.; Rising, A. High-Yield Production of a Super-Soluble Miniature Spidroin for Biomimetic High-Performance Materials. Mater. Today 2021, 50, 16–23. [Google Scholar] [CrossRef]
- Greco, G.; Francis, J.; Arndt, T.; Schmuck, B.; Bäcklund, F.G.; Barth, A.; Johansson, J.; Pugno, N.M.; Rising, A. Properties of Biomimetic Artificial Spider Silk Fibers Tuned by PostSpin Bath Incubation. Molecules 2020, 25, 3248. [Google Scholar] [CrossRef]
- Bucciarelli, A.; Greco, G.; Corridori, I.; Pugno, N.M.; Motta, A. A Design of Experiment Rational Optimization of the Degumming Process and Its Impact on the Silk Fibroin Properties. ACS Biomater. Sci. Eng. 2021, 7, 1374–1393. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.-C.; Chang, F.-J.; Chang, S. A New Criterion for Automatic Multilevel Thresholding. IEEE Trans. Image Process. 1995, 4, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Leymarie, F.; Levine, M.D. Fast Raster Scan Distance Propagation on the Discrete Rectangular Lattice. CVGIP Image Underst. 1992, 55, 84–94. [Google Scholar] [CrossRef]
- Dong, Z.; Lewis, R.V.; Middaugh, C.R. Molecular Mechanism of Spider Silk Elasticity. Arch. Biochem. Biophys. 1991, 284, 53–57. [Google Scholar] [CrossRef]
- Greco, G.; Arndt, T.; Schmuck, B.; Francis, J.; Bäcklund, F.G.; Shilkova, O.; Barth, A.; Gonska, N.; Seisenbaeva, G.; Kessler, V.; et al. Tyrosine Residues Mediate Supercontraction in Biomimetic Spider Silk. Commun. Mater. 2021, 2, 43. [Google Scholar] [CrossRef]
- Ha, N.S.; Jin, T.L.; Goo, N.S.; Park, H.C. Anisotropy and Non-Homogeneity of an Allomyrina Dichotoma Beetle Hind Wing Membrane. Bioinspiration Biomim. 2011, 6, 046003. [Google Scholar] [CrossRef]
- Knight, D.P.; Vollrath, F. Liquid Crystalline Spinning of Spider Silk. Nature 2001, 410, 541–548. [Google Scholar]
- Cheng, Y.; Koh, L.D.; Li, D.; Ji, B.; Han, M.Y.; Zhang, Y.W. On the Strength of β-Sheet Crystallites of Bombyx Mori Silk Fibroin. J. R. Soc. Interface 2014, 11, 20140305. [Google Scholar] [CrossRef]
- Andersson, M.; Jia, Q.; Abella, A.; Lee, X.-Y.; Landreh, M.; Purhonen, P.; Hebert, H.; Tenje, M.; Robinson, C.V.; Meng, Q.; et al. Biomimetic Spinning of Artificial Spider Silk from a Chimeric Minispidroin. Nat. Chem. Biol. 2017, 13, 262–264. [Google Scholar] [CrossRef]
- Rim, N.G.; Roberts, E.G.; Ebrahimi, D.; Dinjaski, N.; Jacobsen, M.M.; Martín-Moldes, Z.; Buehler, M.J.; Kaplan, D.L.; Wong, J.Y. Predicting Silk Fiber Mechanical Properties through Multiscale Simulation and Protein Design. ACS Biomater. Sci. Eng. 2017, 3, 1542–1556. [Google Scholar] [CrossRef] [PubMed]
- Weibull, W. A Statistical Theory of the Strength of Materials. Handlingar 151: Ingeniörs Vetenskaps Akademien, 151st ed.; Generalstabens Litografiska Anstalts Förlag: Stockholm, Sweden, 1939; Volume 6. [Google Scholar]
- Phoenix, S.L. Stochastic Strength and Fatigue of Fiber Bundles. Int. J. Fract. 1978, 14, 327–344. [Google Scholar] [CrossRef]
- Yang, W.; Yu, Y.; Ritchie, R.O.; Meyers, M.A. On the Strength of Hair across Species. Matter 2020, 2, 136–149. [Google Scholar] [CrossRef]
- Hayashi, C.Y.; Shipley, N.H.; Lewis, R.V. Hypotheses That Correlate the Sequence, Structure, and Mechanical Properties of Spider Silk Proteins. Int. J. Biol. Macromol. 1999, 24, 271–275. [Google Scholar] [CrossRef]
- Rathore, O.; Sogah, D.Y. Self-Assembly of β-Sheets into Nanostructures by Poly(Alanine) Segments Incorporated in Multiblock Copolymers Inspired by Spider Silk. J. Am. Chem. Soc. 2001, 123, 5231–5239. [Google Scholar] [CrossRef]
- Gonska, N.; López, P.A.; Lozano-Picazo, P.; Thorpe, M.; Guinea, G.V.; Johansson, J.; Barth, A.; Pérez-Rigueiro, J.; Rising, A. Structure-Function Relationship of Artificial Spider Silk Fibers Produced by Straining Flow Spinning. Biomacromolecules 2020, 21, 2116–2124. [Google Scholar] [CrossRef] [PubMed]
- Htut, K.Z.; Alicea-Serrano, A.M.; Singla, S.; Agnarsson, I.; Garb, J.E.; Kuntner, M.; Gregorič, M.; Haney, R.A.; Marhabaie, M.; Blackledge, T.A.; et al. Correlation between Protein Secondary Structure and Mechanical Performance for the Ultra-Tough Dragline Silk of Darwin’s Bark Spider. J. R. Soc. Interface 2021, 18, 20210320. [Google Scholar] [CrossRef]
- Dicko, C.; Knight, D.; Kenney, J.M.; Vollrath, F. Secondary Structures and Conformational Changes in Flagelliform, Cylindrical, Major, and Minor Ampullate Silk Proteins. Temperature and Concentration Effects. Biomacromolecules 2004, 5, 2105–2115. [Google Scholar] [CrossRef]
- Sampath, S.; Isdebski, T.; Jenkins, J.E.; Ayon, J.V.; Henning, R.W.; Orgel, J.P.R.O.; Antipoa, O.; Yarger, J.L. X-ray Diffraction Study of Nanocrystalline and Amorphous Structure within Major and Minor Ampullate Dragline Spider Silks. Soft Matter 2012, 8, 6713–6722. [Google Scholar] [CrossRef]
- Lefèvre, T.; Rousseau, M.E.; Pézolet, M. Protein Secondary Structure and Orientation in Silk as Revealed by Raman Spectromicroscopy. Biophys. J. 2007, 92, 2885–2895. [Google Scholar] [CrossRef] [PubMed]
- Carpinteri, A.; Pugnø, N. Are Scaling Laws on Strength of Solids Related to Mechanics or to Geometry? Nat. Mater. 2005, 4, 421–423. [Google Scholar] [CrossRef] [PubMed]
- The GIMP Development Team. GIMP, Version 2.10.12. 2019. Available online: https://www.gimp.org (accessed on 29 October 2021).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bäcklund, F.G.; Schmuck, B.; Miranda, G.H.B.; Greco, G.; Pugno, N.M.; Rydén, J.; Rising, A. An Image-Analysis-Based Method for the Prediction of Recombinant Protein Fiber Tensile Strength. Materials 2022, 15, 708. https://doi.org/10.3390/ma15030708
Bäcklund FG, Schmuck B, Miranda GHB, Greco G, Pugno NM, Rydén J, Rising A. An Image-Analysis-Based Method for the Prediction of Recombinant Protein Fiber Tensile Strength. Materials. 2022; 15(3):708. https://doi.org/10.3390/ma15030708
Chicago/Turabian StyleBäcklund, Fredrik G., Benjamin Schmuck, Gisele H. B. Miranda, Gabriele Greco, Nicola M. Pugno, Jesper Rydén, and Anna Rising. 2022. "An Image-Analysis-Based Method for the Prediction of Recombinant Protein Fiber Tensile Strength" Materials 15, no. 3: 708. https://doi.org/10.3390/ma15030708
APA StyleBäcklund, F. G., Schmuck, B., Miranda, G. H. B., Greco, G., Pugno, N. M., Rydén, J., & Rising, A. (2022). An Image-Analysis-Based Method for the Prediction of Recombinant Protein Fiber Tensile Strength. Materials, 15(3), 708. https://doi.org/10.3390/ma15030708







