First-Principles Simulation of Dielectric Function in Biomolecules
Abstract
:1. Introduction
2. Methods
2.1. Model Construction
2.2. Structure Optimization Using VASP
2.3. DFT Calculations Using OLCAO
2.4. Optical Transition and Random Phase Approximation
2.5. Past Record of Using above Methods
3. Results on RGD (1FUV) Peptide
3.1. RGD (1FUV) Peptide
3.2. Local Environmental Effect on RGD (1FUV) Peptide
3.3. Role of Partial Charge
4. SD1 of SARS-CoV-2 Spike-Protein
5. Amino Acid Bond Pairing—AABP
- The AAs in 1FUV models have far more off-diagonal contributions to AABP than the AAs in SD1, as expected from the very different 3D structure of these two proteins.
- The solvated models exhibit a reduced total AABP, mainly from the reduced NN interactions.
- The five off-diagonal pairs in dry 1FUV are reduced to four pairs when solvated by 80 H2O molecules. The missing pair is between Ala1-Phe9. Surprisingly, when solvated by 100 H2O molecules, the number of pairs increases again to eight. The new pairs are Ala1-Asp3, Cys2-Cys8, Asp3-Cys8, Gly6-Phe9.
- For SD1, the two off-diagonal pairs remain the same when solvated by 300 H2O molecules, but the total AABP values for all AAs are decreased by about 13% on average.
- The decrease in total AABP in solvated models in comparison to the dry model is due to the interactions between AAs modified by the presence of H2O molecules.
- It can then be concluded that the total AABP values can be changed in a rather complicated fashion depending on the nature of the protein and the amount of water molecules surrounding them.
6. Discussion
7. Conclusions
- We introduced the QMRPA method for dielectric spectra for small proteins such as RGD (1FUV) peptide and the SD1 subdomain of the spike protein of SARS-CoV-2 virus.
- We pointed out the possible connections between atomic scale partial charges of AAs in proteins and their specific role in the electrostatic interaction.
- We described the role of non-local AA-AA interactions via AABP values in the 3D structure of the protein comparing dry and solvated models.
- We laid out the roadmap to use QMRPA method for applications to electrostatic interactions in spike protein and other biomolecular systems in general.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schutz, C.N.; Warshel, A. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins Struct. Funct. Bioinform. 2001, 44, 400–417. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-X.; Pang, X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef]
- Woods, L.; Dalvit, D.A.R.; Tkatchenko, A.; Rodriguez-Lopez, P.; Rodriguez, A.W.; Podgornik, R. Materials perspective on Casimir and van der Waals interactions. Rev. Mod. Phys. 2016, 88, 045003. [Google Scholar] [CrossRef]
- Dryden, D.M.; Hopkins, J.C.; Denoyer, L.K.; Poudel, L.; Steinmetz, N.F.; Ching, W.-Y.; Podgornik, R.; Parsegian, A.; French, R.H. van der Waals interactions on the mesoscale: Open-science implementation, anisotropy, retardation, and solvent effects. Langmuir 2015, 31, 10145–10153. [Google Scholar] [CrossRef]
- Bai, C.; Warshel, A. Critical Differences Between the Binding Features of the Spike Proteins of SARS-CoV-2 and SARS-CoV. J. Phys. Chem. B 2020, 124, 5907–5912. [Google Scholar] [CrossRef]
- Javidpour, L.; Bozic, A.; Naji, A.; Podgornik, R. Electrostatic interaction between SARS-CoV-2 virus and charged electret fibre. arXiv 2020, arXiv:2012.07160. [Google Scholar]
- McLaughlin, S. The electrostatic properties of membranes. Annu. Rev. Biophys. Biophys. Chem. 1989, 18, 113–136. [Google Scholar] [CrossRef] [PubMed]
- Warshel, A.; Sharma, P.K.; Kato, M.; Parson, W.W. Modeling electrostatic effects in proteins. Biochim. Biophys. Acta BBA-Proteins Proteom. 2006, 1764, 1647–1676. [Google Scholar] [CrossRef]
- Li, L.; Li, C.; Zhang, Z.; Alexov, E. On the dielectric “constant” of proteins: Smooth dielectric function for macromolecular modeling and its implementation in DelPhi. J. Chem. Theory Comput. 2013, 9, 2126–2136. [Google Scholar] [CrossRef]
- García-Moreno, B.E.; Dwyer, J.J.; Gittis, A.G.; Lattman, E.E.; Spencer, D.S.; Stites, W.E. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys. Chem. 1997, 64, 211–224. [Google Scholar] [CrossRef]
- Li, C.; Jia, Z.; Chakravorty, A.; Pahari, S.; Peng, Y.; Basu, S.; Koirala, M.; Panday, S.K.; Petukh, M.; Li, L. DelPhi suite: New developments and review of functionalities. J. Comput. Chem. 2019, 40, 2502–2508. [Google Scholar] [CrossRef]
- Koehl, P. Electrostatics calculations: Latest methodological advances. Curr. Opin. Struct. Biol. 2006, 16, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Simonson, T. Electrostatics and dynamics of proteins. Rep. Prog. Phys. 2003, 66, 737. [Google Scholar] [CrossRef]
- Gudarzi, M.M.; Aboutalebi, S.H. Self-consistent dielectric functions of materials: Toward accurate computation of Casimir–van der Waals forces. Sci. Adv. 2021, 7, eabg2272. [Google Scholar] [CrossRef]
- Parsegian, V.; Ninham, B. Application of the Lifshitz theory to the calculation of van der Waals forces across thin lipid films. Nature 1969, 224, 1197–1198. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. [Google Scholar] [CrossRef] [PubMed]
- Stöhr, M.; Tkatchenko, A. Quantum mechanics of proteins in explicit water: The role of plasmon-like solute-solvent interactions. Sci. Adv. 2019, 5, eaax0024. [Google Scholar] [CrossRef] [Green Version]
- Velichko, E.; Baranov, M.; Mostepanenko, V. Change of sign in the Casimir interaction of peptide films deposited on a dielectric substrate. Mod. Phys. Lett. A 2020, 35, 2040020. [Google Scholar] [CrossRef]
- Baranov, M.; Klimchitskaya, G.; Mostepanenko, V.; Velichko, E. Fluctuation-induced free energy of thin peptide films. Phys. Rev. E 2019, 99, 022410. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, P.; Wen, A.M.; French, R.H.; Parsegian, V.A.; Steinmetz, N.F.; Podgornik, R.; Ching, W.-Y. Electronic structure, dielectric response, and surface charge distribution of RGD (1FUV) peptide. Sci. Rep. 2014, 4, 5605. [Google Scholar] [CrossRef]
- Bloch, F. Über die quantenmechanik der elektronen in kristallgittern. Z. Phys. 1929, 52, 555–600. [Google Scholar] [CrossRef]
- Ehrenreich, H.; Cohen, M.H. Self-consistent field approach to the many-electron problem. Phys. Rev. 1959, 115, 786–790. [Google Scholar] [CrossRef]
- Ching, W.-Y.; Adhikari, P.; Jawad, B.; Podgornik, R. Ultra-Large-Scale Ab Initio Quantum Chemical Computation of Bio-Molecular Systems: The Case of Spike Protein of SARS-CoV-2 Virus. Comput. Struct. Biotechnol. J. 2021, 19, 1288–1301. [Google Scholar] [CrossRef]
- Millefiori, S.; Alparone, A.; Millefiori, A.; Vanella, A. Electronic and vibrational polarizabilities of the twenty naturally occurring amino acids. Biophys. Chem. 2008, 132, 139–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swart, M.; Snijders, J.G.; van Duijnen, P.T. Polarizabilities of amino acid residues. J. Comput. Methods Sci. Eng. 2004, 4, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Abeyrathne, C.D.; Halgamuge, M.N.; Farrell, P.M.; Skafidas, E. An ab-initio computational method to determine dielectric properties of biological materials. Sci. Rep. 2013, 3, 1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stöhr, M.; Van Voorhis, T.; Tkatchenko, A. Theory and practice of modeling van der Waals interactions in electronic-structure calculations. Chem. Soc. Rev. 2019, 48, 4118–4154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- 1FUV Solution Structure of an RGD Peptide ISOMER-A. Available online: https://www.rcsb.org/structure/1FUV (accessed on 20 March 2021).
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Pearlman, D.A.; Case, D.A.; Caldwell, J.W.; Ross, W.S.; Cheatham III, T.E.; DeBolt, S.; Ferguson, D.; Seibel, G.; Kollman, P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VASP—Vienna Ab initio Simulation Package. Available online: https://www.vasp.at/ (accessed on 20 March 2021).
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865. [Google Scholar] [CrossRef] [Green Version]
- Ching, W.-Y.; Rulis, P. Electronic Structure Methods for Complex Materials: The Orthogonalized Linear Combination of Atomic Orbitals; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Poudel, L.; Steinmetz, N.F.; French, R.H.; Parsegian, V.A.; Podgornik, R.; Ching, W.-Y. Implication of the solvent effect, metal ions and topology in the electronic structure and hydrogen bonding of human telomeric G-quadruplex DNA. Phys. Chem. Chem. Phys. 2016, 18, 21573–21585. [Google Scholar] [CrossRef]
- Poudel, L.; Twarock, R.; Steinmetz, N.F.; Podgornik, R.; Ching, W.-Y. Impact of hydrogen bonding in the binding site between capsid protein and MS2 bacteriophage ssRNA. J. Phys. Chem. B 2017, 121, 6321–6330. [Google Scholar] [CrossRef]
- Eifler, J.; Podgornik, R.; Steinmetz, N.F.; French, R.H.; Parsegian, V.A.; Ching, W.Y. Charge distribution and hydrogen bonding of a collagen α2-chain in vacuum, hydrated, neutral, and charged structural models. Int. J. Quantum Chem. 2016, 116, 681–691. [Google Scholar] [CrossRef]
- Adhikari, P.; Li, N.; Shin, M.; Steinmetz, N.F.; Twarock, R.; Podgornik, R.; Ching, W.-Y. Intra- and intermolecular atomic-scale interactions in the receptor binding domain of SARS-CoV-2 spike protein: Implication for ACE2 receptor binding. Phys. Chem. Chem. Phys. 2020, 22, 18272–18283. [Google Scholar] [CrossRef]
- Adhikari, P.; Ching, W.-Y. Amino acid interacting network in the receptor-binding domain of SARS-CoV-2 spike protein. RSC Adv. 2020, 10, 39831–39841. [Google Scholar] [CrossRef]
- Mulliken, R.S. Electronic population analysis on LCAO–MO molecular wave functions. I. J. Chem. Phys. 1955, 23, 1833–1840. [Google Scholar] [CrossRef] [Green Version]
- Mulliken, R. Electronic population analysis on LCAO–MO molecular wave functions. II. Overlap populations, bond orders, and covalent bond energies. J. Chem. Phys. 1955, 23, 1841–1846. [Google Scholar] [CrossRef]
- Dereka, B.; Yu, Q.; Lewis, N.H.; Carpenter, W.B.; Bowman, J.M.; Tokmakoff, A. Crossover from hydrogen to chemical bonding. Science 2021, 371, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.C. Sum rules, Kramers-Kronig relations, and transport coefficients in charged systems. Phys. Rev. 1967, 161, 143. [Google Scholar] [CrossRef]
- Ching, W.; Xu, Y.; Zhao, G.-L.; Wong, K.; Zandiehnadem, F. Electronic structure and excitonic-enhanced superconducting mechanism in YBa2Cu3O7−δ. Phys. Rev. Lett. 1987, 59, 1333. [Google Scholar] [CrossRef]
- Zhao, G.-L.; Xu, Y.; Ching, W.; Wong, K. Theoretical calculation of optical properties of Y-Ba-Cu-O superconductors. Phys. Rev. B 1987, 36, 7203. [Google Scholar] [CrossRef]
- Ching, W.; Huang, M.-Z.; Xu, Y.-N.; Harter, W.; Chan, F. First-principles calculation of optical properties of C 60 in the fcc lattice. Phys. Rev. Lett. 1991, 67, 2045. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-N.; Huang, M.-Z.; Ching, W. Optical properties of superconducting K 3 C 60 and insulating K 6 C 60. Phys. Rev. B 1991, 44, 13171. [Google Scholar] [CrossRef]
- Baral, K.; Adhikari, P.; Ching, W.Y. Ab initio Modeling of the Electronic Structures and Physical Properties of a-Si1−xGexO2 Glass (x = 0 to 1). J. Am. Ceram. Soc. 2016, 99, 3677–3684. [Google Scholar] [CrossRef]
- Adhikari, P.; Xiong, M.; Li, N.; Zhao, X.; Rulis, P.; Ching, W.-Y. Structure and electronic properties of a continuous random network model of an amorphous zeolitic imidazolate framework (a-ZIF). J. Phys. Chem. C 2016, 120, 15362–15368. [Google Scholar] [CrossRef]
- Adhikari, P.; Li, N.; Rulis, P.; Ching, W.-Y. Deformation behavior of an amorphous zeolitic imidazolate framework–from a supersoft material to a complex organometallic alloy. Phys. Chem. Chem. Phys. 2018, 20, 29001–29011. [Google Scholar] [CrossRef]
- Ouyang, L.; Randaccio, L.; Rulis, P.; Kurmaev, E.; Moewes, A.; Ching, W. Electronic structure and bonding in vitamin B12, cyanocobalamin. J. Mol. Struct. THEOCHEM 2003, 622, 221–227. [Google Scholar] [CrossRef]
- Ouyang, L.; Rulis, P.; Ching, W.; Nardin, G.; Randaccio, L. Accurate redetermination of the X-ray structure and electronic bonding in adenosylcobalamin. Inorg. Chem. 2004, 43, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Rulis, P.; Ouyang, L.; Ching, W. Electronic structure and bonding in calcium apatite crystals: Hydroxyapatite, fluorapatite, chlorapatite, and bromapatite. Phys. Rev. B 2004, 70, 155104. [Google Scholar] [CrossRef]
- Rulis, P.; Yao, H.; Ouyang, L.; Ching, W. Electronic structure, bonding, charge distribution, and x-ray absorption spectra of the (001) surfaces of fluorapatite and hydroxyapatite from first principles. Phys. Rev. B 2007, 76, 245410. [Google Scholar] [CrossRef]
- Kahr, B.; Freudenthal, J.; Phillips, S.; Kaminsky, W. Herapathite. Science 2009, 324, 1407. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Rulis, P.; Kahr, B.; Ching, W. Theoretical study of the large linear dichroism of herapathite. Phys. Rev. B 2009, 80, 235132. [Google Scholar] [CrossRef]
- French, R.H.; Parsegian, V.A.; Podgornik, R.; Rajter, R.F.; Jagota, A.; Luo, J.; Asthagiri, D.; Chaudhury, M.K.; Chiang, Y.-M.; Granick, S.; et al. Long range interactions in nanoscale science. Rev. Mod. Phys. 2010, 82, 1887. [Google Scholar] [CrossRef]
- Hopkins, J.C.; Dryden, D.M.; Ching, W.-Y.; French, R.H.; Parsegian, V.A.; Podgornik, R. Dielectric response variation and the strength of van der Waals interactions. J. Colloid Interface Sci. 2014, 417, 278–284. [Google Scholar] [CrossRef]
- Schimelman, J.B.; Dryden, D.M.; Poudel, L.; Krawiec, K.E.; Ma, Y.; Podgornik, R.; Parsegian, V.A.; Denoyer, L.K.; Ching, W.-Y.; Steinmetz, N.F.; et al. Optical properties and electronic transitions of DNA oligonucleotides as a function of composition and stacking sequence. Phys. Chem. Chem. Phys. 2015, 17, 4589–4599. [Google Scholar] [CrossRef]
- Gong, Y.; Adhikari, P.; Liu, Q.; Wang, T.; Gong, M.; Chan, W.-L.; Ching, W.-Y.; Wu, J. Designing the interface of carbon nanotube/biomaterials for high-performance ultra-broadband photodetection. ACS Appl. Mater. Interfaces 2017, 9, 11016–11024. [Google Scholar] [CrossRef]
- Gong, M.; Adhikari, P.; Gong, Y.; Wang, T.; Liu, Q.; Kattel, B.; Ching, W.Y.; Chan, W.L.; Wu, J.Z. Polarity-Controlled Attachment of Cytochrome C for High-Performance Cytochrome C/Graphene van der Waals Heterojunction Photodetectors. Adv. Funct. Mater. 2018, 28, 1704797. [Google Scholar] [CrossRef]
- Donev, R. Protein and Peptide Nanoparticles for Drug Delivery; Academic Press: Waltham, MA, USA, 2015. [Google Scholar]
- Wang, F.; Li, Y.; Shen, Y.; Wang, A.; Wang, S.; Xie, T. The functions and applications of RGD in tumor therapy and tissue engineering. Int. J. Mol. Sci. 2013, 14, 13447–13462. [Google Scholar] [CrossRef] [Green Version]
- Avraamides, C.J.; Garmy-Susini, B.; Varner, J.A. Integrins in angiogenesis and lymphangiogenesis. Nat. Rev. Cancer 2008, 8, 604–617. [Google Scholar] [CrossRef] [Green Version]
- Garanger, E.; Boturyn, D.; Dumy, P. Tumor targeting with RGD peptide ligands-design of new molecular conjugates for imaging and therapy of cancers. Anti-Cancer Agents Med. Chem. 2007, 7, 552–558. [Google Scholar] [CrossRef]
- Metcalfe, A.D.; Ferguson, M.W. Tissue engineering of replacement skin: The crossroads of biomaterials, wound healing, embryonic development, stem cells and regeneration. J. R. Soc. Interface 2007, 4, 413–437. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Chakraborty, S.; Liu, S. Radiolabeled cyclic RGD peptides as radiotracers for imaging tumors and thrombosis by SPECT. Theranostics 2011, 1, 58. [Google Scholar] [CrossRef] [Green Version]
- Meyers, S.R.; Grinstaff, M.W. Biocompatible and bioactive surface modifications for prolonged in vivo efficacy. Chem. Rev. 2012, 112, 1615–1632. [Google Scholar] [CrossRef] [Green Version]
- Bellis, S.L. Advantages of RGD peptides for directing cell association with biomaterials. Biomaterials 2011, 32, 4205–4210. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Yang, S.-J.; Zheng, Y.-C.; Cui, Y.; Zhang, Z.; Zhong, J.-Y.; Zhou, J. Electrostatic Effect of Functional Surfaces on the Activity of Adsorbed Enzymes: Simulations and Experiments. ACS Appl. Mater. Interfaces 2020, 12, 35676–35687. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, C.-M.; Sehnal, D.; Falginella, F.L.; Pant, P.; Pravda, L.; Bouchal, T.; Vařeková, R.S.; Geidl, S.; Koča, J. AtomicChargeCalculator: Interactive web-based calculation of atomic charges in large biomolecular complexes and drug-like molecules. J. Cheminformatics 2015, 7, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; Edwards, R.J.; Mansouri, K.; Janowska, K.; Stalls, V.; Gobeil, S.M.; Kopp, M.; Li, D.; Parks, R.; Hsu, A.L.; et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020, 27, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.; Duan, J.-Z.; Wang, Z.-H.; Chen, Y.-Y.; Zhang, P.-H.; Zhan, L.; Yan, X.-Y.; Cao, W.-C.; Jin, G. Investigation of interaction between two neutralizing monoclonal antibodies and SARS virus using biosensor based on imaging ellipsometry. Biomed. Microdevices 2006, 8, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Orooji, Y.; Sohrabi, H.; Hemmat, N.; Oroojalian, F.; Baradaran, B.; Mokhtarzadeh, A.; Mohaghegh, M.; Karimi-Maleh, H. An overview on SARS-CoV-2 (COVID-19) and other human coronaviruses and their detection capability via amplification assay, chemical sensing, biosensing, immunosensing, and clinical assays. Nano-Micro Lett. 2021, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Spinello, A.; Saltalamacchia, A.; Magistrato, A. Is the rigidity of SARS-CoV-2 spike receptor-binding motif the hallmark for its enhanced infectivity? Insights from all-atom simulations. J. Phys. Chem. Lett. 2020, 11, 4785–4790. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Karki, C.B.; Du, D.; Li, H.; Wang, J.; Sobitan, A.; Teng, S.; Tang, Q.; Li, L. Spike proteins of SARS-CoV and SARS-CoV-2 utilize different mechanisms to bind with human ACE2. Front. Mol. Biosci. 2020, 7, 392. [Google Scholar] [CrossRef]
- Peng, C.; Zhu, Z.; Shi, Y.; Wang, X.; Mu, K.; Yang, Y.; Zhang, X.; Xu, Z.; Zhu, W. Computational Insights into the Conformational Accessibility and Binding Strength of SARS-CoV-2 Spike Protein to Human Angiotensin-Converting Enzyme 2. J. Phys. Chem. Lett. 2020, 11, 10482–10488. [Google Scholar] [CrossRef]
- Amin, M.; Sorour, M.K.; Kasry, A. Comparing the binding interactions in the receptor binding domains of SARS-CoV-2 and SARS-CoV. J. Phys. Chem. Lett. 2020, 11, 4897–4900. [Google Scholar] [CrossRef]


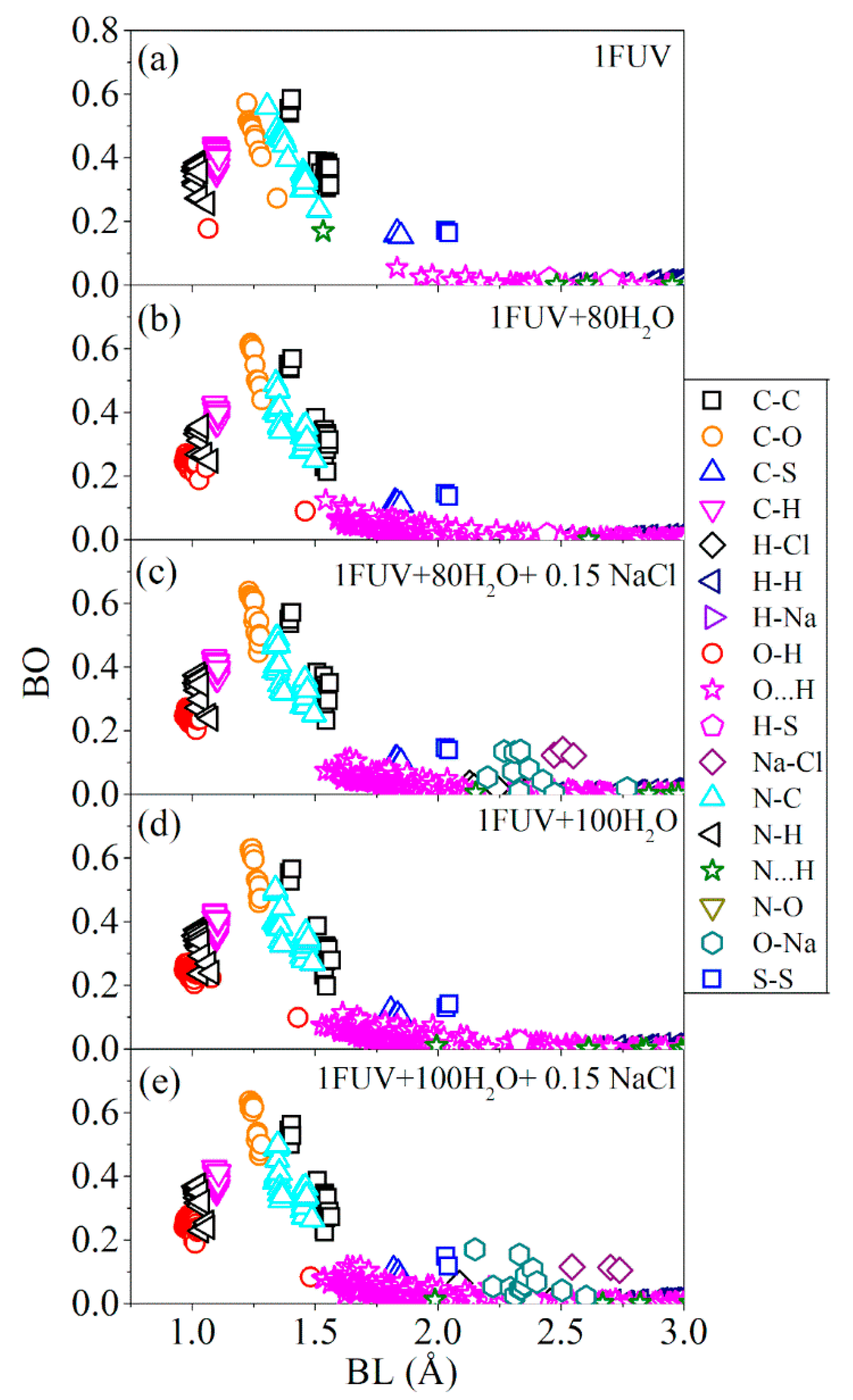
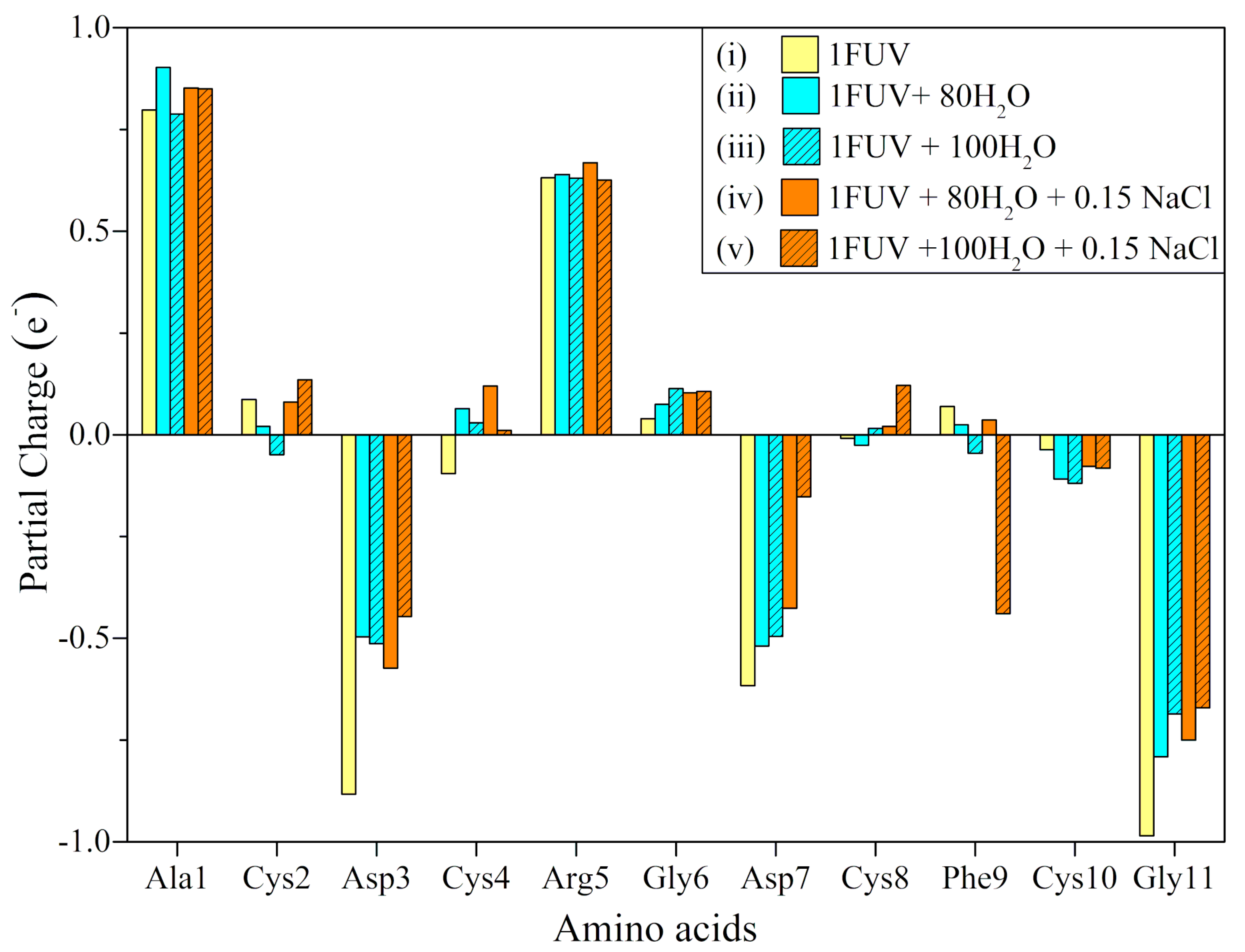


| Models | TBO (e) | Eg(eV) |
|---|---|---|
| 1FUV | 54.27 | 3.61 |
| 1FUV + 80 H2O | 98.49 | 0.68 |
| 1FUV + 80 H2O + NaCl | 99.95 | 0.47 |
| 1FUV + 100 H2O | 109.64 | 0.57 |
| 1FUV + 100 H2O + NaCl | 110.30 | 0.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adhikari, P.; Podgornik, R.; Jawad, B.; Ching, W.-Y. First-Principles Simulation of Dielectric Function in Biomolecules. Materials 2021, 14, 5774. https://doi.org/10.3390/ma14195774
Adhikari P, Podgornik R, Jawad B, Ching W-Y. First-Principles Simulation of Dielectric Function in Biomolecules. Materials. 2021; 14(19):5774. https://doi.org/10.3390/ma14195774
Chicago/Turabian StyleAdhikari, Puja, Rudolf Podgornik, Bahaa Jawad, and Wai-Yim Ching. 2021. "First-Principles Simulation of Dielectric Function in Biomolecules" Materials 14, no. 19: 5774. https://doi.org/10.3390/ma14195774
APA StyleAdhikari, P., Podgornik, R., Jawad, B., & Ching, W.-Y. (2021). First-Principles Simulation of Dielectric Function in Biomolecules. Materials, 14(19), 5774. https://doi.org/10.3390/ma14195774






