Electrospun Polylactic Acid (PLLA) Microtube Array Membrane (MTAM)—An Advanced Substrate for Anticancer Drug Screening
Abstract
:1. Introduction
2. Materials and Methods
2.1. MTAM Preparation
2.2. Cell Culture
2.3. Cell Proliferation Assay
2.4. Cytotoxicity Study
2.5. Preparation and Implantation of Cell-Loaded MTAM
2.6. Patients
2.7. Response Evaluation Criteria In Solid Tumors (RECIST)
2.8. Statistics
3. Results
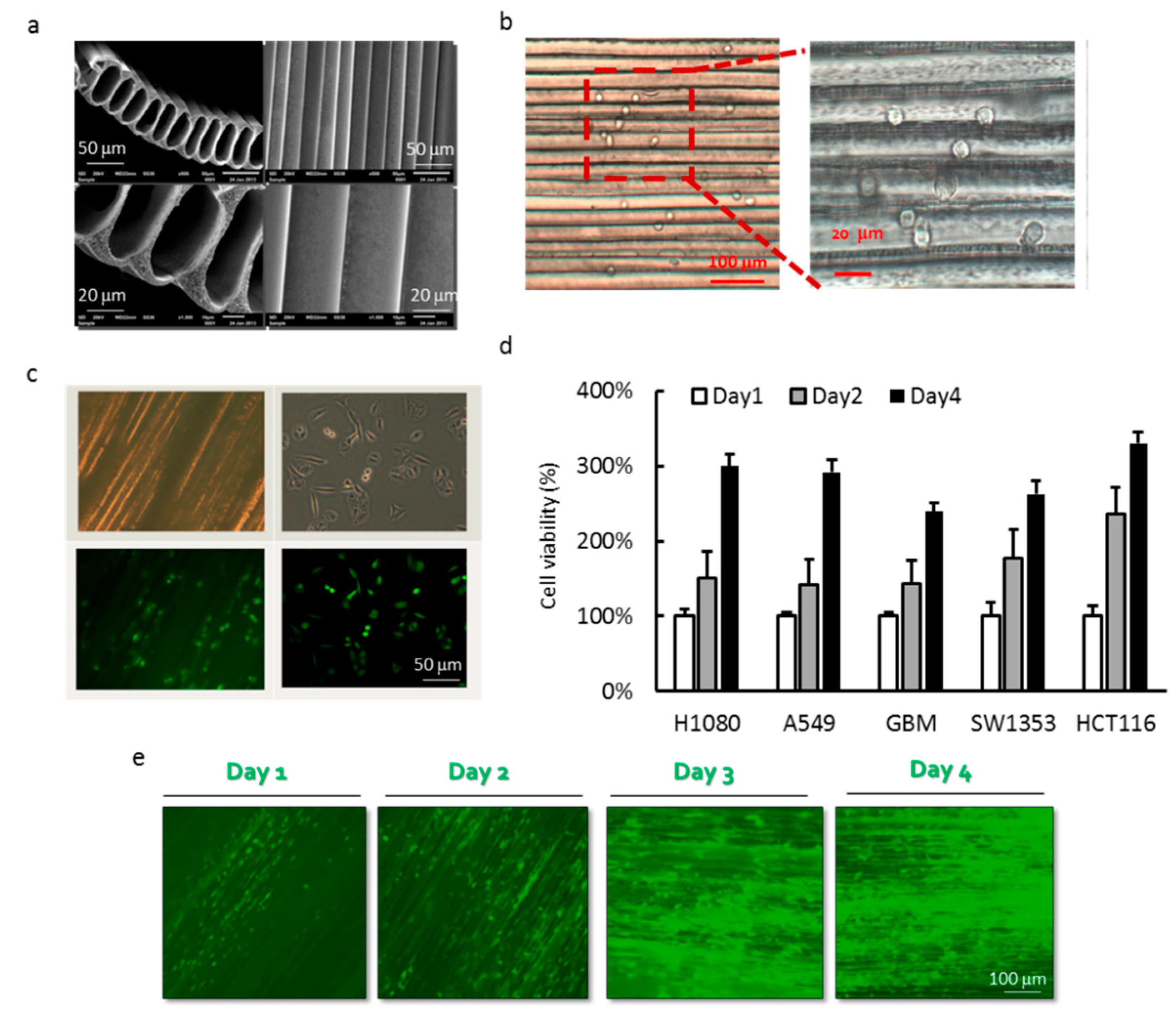
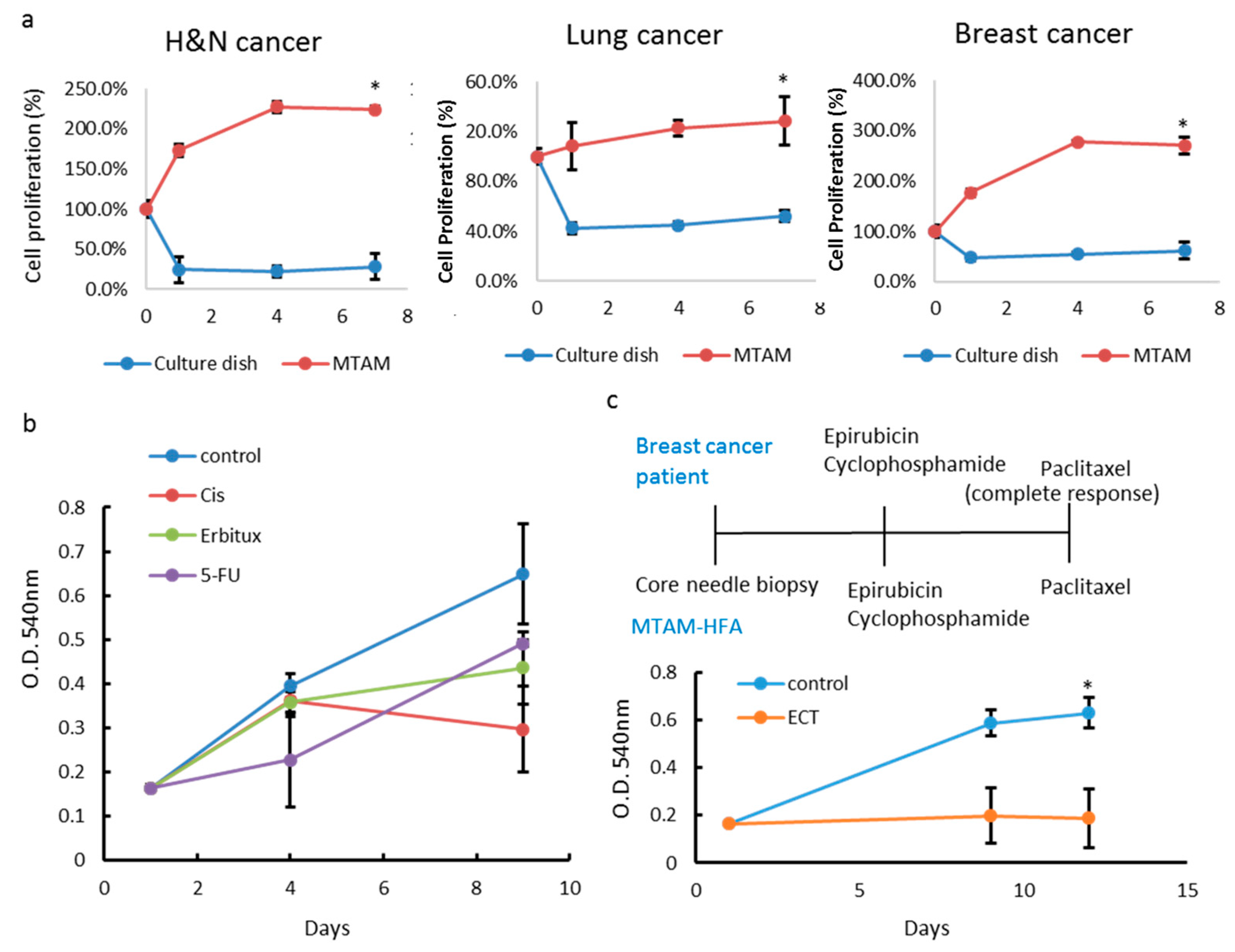
3.1. Cell Culture of Cancer Cell Lines in PLLA MTAM
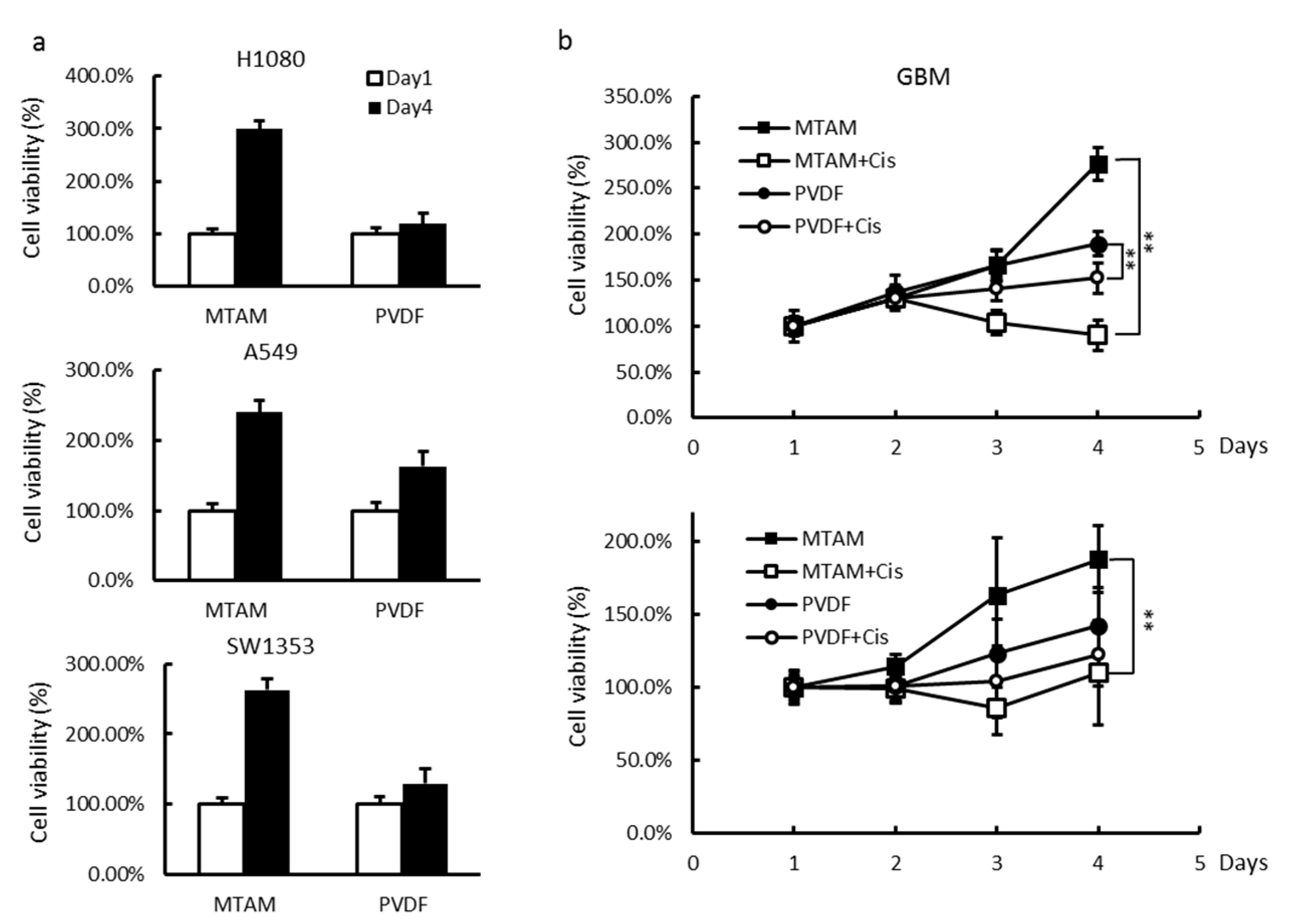
3.2. Effects of MTAM/HFA System on the Cytotoxicity of Cancer Cells
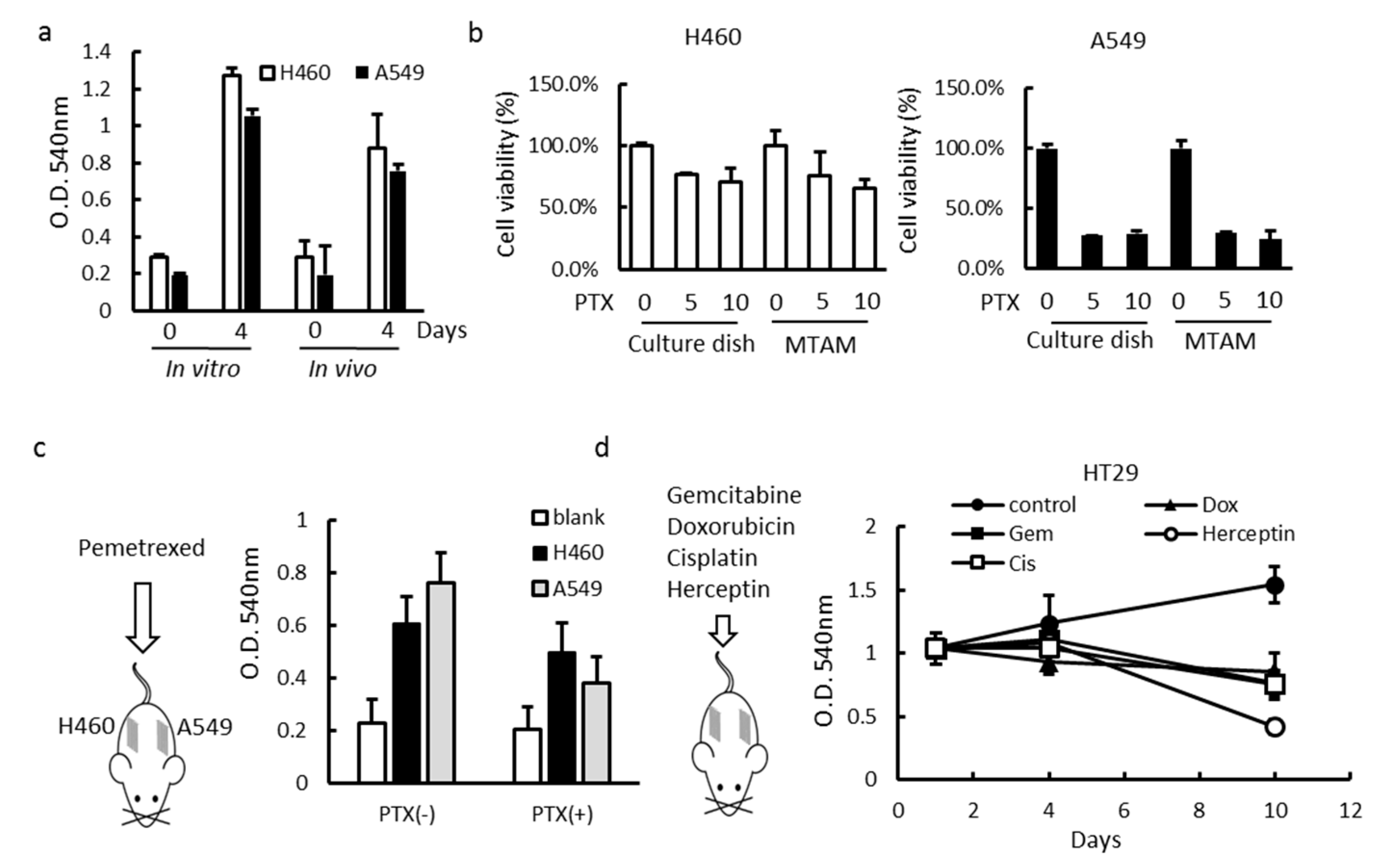
3.3. In Vitro and In Vivo Studies of Drug Sensitivity of Cancer Cell Lines Cultured within MTAM/HFA and PVDF(Polyvinylidene Fluoride) HFA
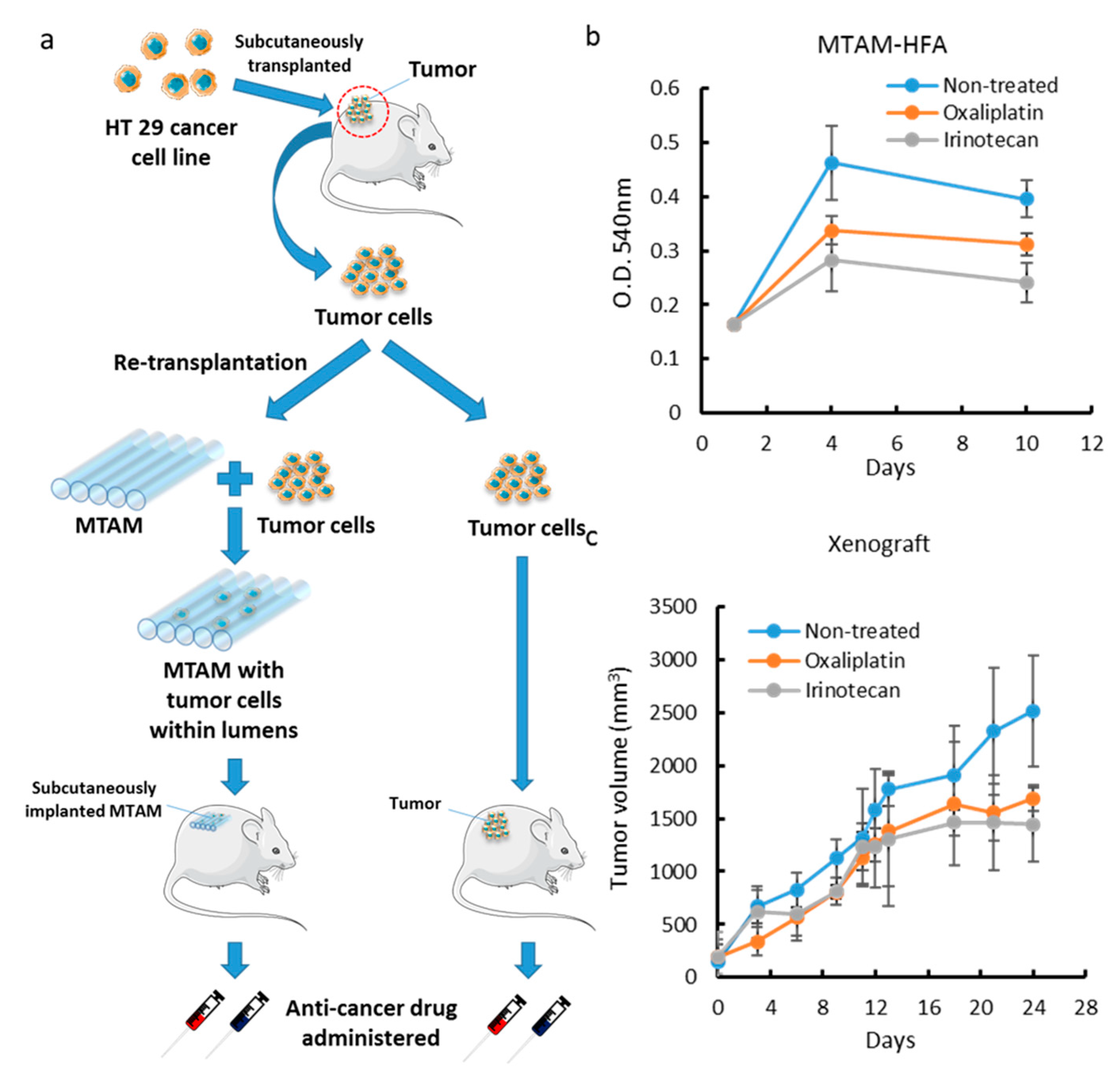
3.4. Comparison of MTAM vs. PDX as a Primary Culture Substrate
3.5. MTAM/HFA as a Potential Platform for Anticancer Drug Screening in Personalized Medicine
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Denison, T.A.; Bae, Y.H. Tumor heterogeneity and its implication for drug delivery. J. Controll. Release 2012, 164, 187–191. [Google Scholar] [CrossRef]
- Jonkers, J.; Berns, A. Conditional mouse models of sporadic cancer. Nat. Rev. Cancer 2002, 2, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Huse, J.T.; Holland, E.C. Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol. 2009, 19, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M.; Bruckheimer, E.; Rajeshkumar, N.V.; Garrido-Laguna, I.; De Oliveira, E.; Rubio-Viqueira, B.; Strawn, S.; Wick, M.J.; Martell, J.; Sidransky, D. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol. Cancer Ther. 2011, 10, 1311. [Google Scholar] [CrossRef] [PubMed]
- Garralda, E.; Paz, K.; Lopez-Casas, P.P.; Jones, S.; Katz, A.; Kann, L.M.; Lopez-Rios, F.; Sarno, F.; Al-Shahrour, F.; Vasquez, D.; et al. Integrated next-generation sequencing and avatar mouse models for personalized cancer treatment. Clin. Cancer Res. 2014, 20, 2476–2484. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, M.C.; Rajeshkumar, N.V.; Garrido-Laguna, I.; De Jesus-Acosta, A.; Jones, S.; Maitra, A.; Hruban, R.H.; Eshleman, J.R.; Klein, A.; Laheru, D.; et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol. Cancer Ther. 2011, 10, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, J.; Paz, K.; Schwartz, G.K.; Wexler, L.H.; Maki, R.; Pollock, R.E.; Morris, R.; Cohen, R.; Shankar, A.; Blackman, G.; et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer 2014, 120, 2006–2015. [Google Scholar] [CrossRef]
- Bruna, A.; Rueda, O.M.; Greenwood, W.; Batra, A.S.; Callari, M.; Batra, R.N.; Pogrebniak, K.; Sandoval, J.; Cassidy, J.W.; Tufegdzic-Vidakovic, A.; et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell 2016, 167, 260–274. [Google Scholar] [CrossRef]
- Bian, B.; Bigonnet, M.; Gayet, O.; Loncle, C.; Maignan, A.; Gilabert, M.; Moutardier, V.; Garcia, S.; Turrini, O.; Delpero, J.R.; et al. Gene expression profiling of patient-derived pancreatic cancer xenografts predicts sensitivity to the BET bromodomain inhibitor JQ1: implications for individualized medicine efforts. EMBO Mol. Med. 2017, 9, 482–497. [Google Scholar] [CrossRef]
- Bertotti, A.; Migliardi, G.; Galimi, F.; Sassi, F.; Torti, D.; Isella, C.; Cora, D.; Di Nicolantonio, F.; Buscarino, M.; Petti, C.; et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011, 1, 508–523. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinska, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Maelandsmo, G.M.; et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Julien, S.; Merino-Trigo, A.; Lacroix, L.; Pocard, M.; Goere, D.; Mariani, P.; Landron, S.; Bigot, L.; Nemati, F.; Dartigues, P.; et al. Characterization of a Large Panel of Patient-Derived Tumor Xenografts Representing the Clinical Heterogeneity of Human Colorectal Cancer. Clin. Cancer Res. 2012, 18, 5314–5328. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.P.; Calvo, E.; Ordonez, E.; Wick, M.J.; Viqueira, B.R.; Lopez-Casas, P.P.; Bruckheimer, E.; Calles-Blanco, A.; Sidransky, D.; Hidalgo, M. Prioritizing Phase I Treatment Options Through Preclinical Testing on Personalized Tumorgraft. J. Clin. Oncol. 2012, 30, E45–E48. [Google Scholar] [CrossRef] [PubMed]
- Jimeno, A.; Tan, A.C.; Coffa, J.; Rajeshkumar, N.V.; Kulesza, P.; Rubio-Viqueira, B.; Wheelhouse, J.; Diosdado, B.; Messersmith, W.A.; Lacobuzio-Donahue, C.; et al. Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer Res. 2008, 68, 2841–2849. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.Z.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.Z.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Decker, S.; Hollingshead, M.; Bonomi, C.A.; Carter, J.P.; Sausville, E.A. The hollow fibre model in cancer drug screening: the NCI experience. Eur. J. Cancer 2004, 40, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Haber, D.A.; Settleman, J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer 2010, 10, 241–253. [Google Scholar] [CrossRef]
- Suggitt, M.; Cooper, P.A.; Shnyder, S.D.; Bibby, M.C. The hollow fibre model—facilitating anticancer pre-clinical pharmacodynamics and improving animal welfare. Int. J. Oncol. 2006, 29, 1493–1499. [Google Scholar]
- Shnyder, S.D.; Cooper, P.A.; Scally, A.J.; Bibby, M.C. Reducing the cost of screening novel agents using the hollow fibre assay. Anticancer Res. 2006, 26, 2049–2052. [Google Scholar]
- Ou, K.L.; Chen, C.S.; Lin, L.H.; Lu, J.C.; Shu, Y.C.; Tseng, W.C.; Yang, J.C.; Lee, S.Y.; Chen, C.C. Membranes of epitaxial-like packed, super aligned electrospun micron hollow poly(L-lactic acid) (PLLA) fibers. Eur. Polym. J. 2011, 47, 882–892. [Google Scholar] [CrossRef]
- Yang, J.C.; Lee, S.Y.; Tseng, W.C.; Shu, Y.C.; Lu, J.C.; Shie, H.S.; Chen, C.C. Formation of Highly Aligned, Single-Layered, Hollow Fibrous Assemblies and the Fabrication of Large Pieces of PLLA Membranes. Macromol. Mater. Eng. 2012, 297, 115–122. [Google Scholar] [CrossRef]
- Lin, L.C.; Shu, Y.C.; Yang, J.C.; Shie, H.S.; Lee, S.Y.; Chen, C.C. Nano-porous Poly-L-lactic Acid Microtube Array Membranes. Curr. Nanosci. 2014, 10, 227–234. [Google Scholar] [CrossRef]
- Hung, W.C.; Lin, L.H.; Tsen, W.C.; Shie, H.S.; Chiu, H.L.; Yang, T.C.K.; Chen, C.C. Permeation of biological compounds through porous poly((L)-lactic acid) (PLLA) microtube array membranes (MTAM). Eur. Polym. J. 2015, 67, 166–173. [Google Scholar] [CrossRef]
- Yang, S.H.; Lin, H.Y.; Chang, V.H.S.; Chen, C.C.; Liu, Y.R.; Wang, J.H.; Zhang, K.Q.; Jiang, X.Q.; Yen, Y. Lovastatin overcomes gefitinib resistance through TNF-alpha signaling in human cholangiocarcinomas with different LKB1 statuses in vitro and in vivo. Oncotarget 2015, 6, 23857–23873. [Google Scholar] [PubMed]
- Tseng, V.C.H.; Chew, C.H.; Huang, W.T.; Wang, Y.K.; Chen, K.S.; Chou, S.Y.; Chen, C.C. An Effective Cell Coculture Platform Based on the Electrospun Microtube Array Membrane for Nerve Regeneration. Cells Tissues Organs 2017, 204, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Wu, C.H.; Wu, J.J.; Chiu, C.C.; Wong, C.H.; Tsai, M.L.; Lin, H.T.V. Accelerated bioethanol fermentation by using a novel yeast immobilization technique: Microtube array membrane. Process. Biochem. 2015, 50, 1509–1515. [Google Scholar] [CrossRef]
- Chew, C.; Wu, C.; Chen, C. A novel electrospun Microtube Array Membrane (MTAM) based low cost conceptual tubular Microbial Fuel Cell (MFC). Eur. Polym. J. 2016, 83, 138–147. [Google Scholar] [CrossRef]
- Yang, A.-J.; Marito, S.; Yang, J.-J.; Keshari, S.; Chew, C.-H.; Chen, C.-C.; Huang, C.-M. A Microtube Array Membrane (MTAM) Encapsulated Live Fermenting Staphylococcus epidermidis as a Skin Probiotic Patch against Cutibacterium acnes. Int. J. Mol. Sci. 2019, 20, 14. [Google Scholar] [CrossRef]
- Izumchenko, E.; Paz, K.; Ciznadija, D.; Sloma, I.; Katz, A.; Vasquez-Dunddel, D.; Ben-Zvi, I.; Stebbing, J.; McGuire, W.; Harris, W.; et al. Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Ann. Oncol. 2017, 28, 2595–2605. [Google Scholar] [CrossRef]
- Barutello, G.; Rolih, V.; Arigoni, M.; Tarone, L.; Conti, L.; Quaglino, E.; Buracco, P.; Cavallo, F.; Riccardo, F. Strengths and weaknesses of pre-clinical models for human melanoma treatment: dawn of dogs’ revolution for immunotherapy. Int. J. Mol. Sci. 2018, 19, 799. [Google Scholar] [CrossRef]
- Sadar, M.D.; Akopian, V.A.; Beraldi, E. Characterization of a new in vivo hollow fiber model for the study of progression of prostate cancer to androgen independence. Mol. Cancer Ther. 2002, 1, 629–637. [Google Scholar] [PubMed]
- Davidson, A.J.; Ellis, M.J.; Chaudhuri, J.B. A theoretical approach to zonation in a bioartificial liver. Biotechnol. Bioeng. 2012, 109, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Chapman, L.A.C.; Shipley, R.J.; Whiteley, J.P.; Ellis, M.J.; Byrne, H.M.; Waters, S.L. Optimising Cell Aggregate Expansion in a Perfused Hollow Fibre Bioreactor via Mathematical Modelling. PLoS ONE 2014, 9, e105813. [Google Scholar] [CrossRef] [PubMed]
- Khajavi, R.; Abbasipour, M. Electrospinning as a versatile method for fabricating coreshell, hollow and porous nanofibers. Sci. Iran. 2012, 19, 2029–2034. [Google Scholar] [CrossRef]
- Gloeckner, H.; Lemke, H.D. New miniaturized hollow-fiber bioreactor for in vivo like cell culture, cell expansion, and production of cell-derived products. Biotechnol. Progr. 2001, 17, 828–831. [Google Scholar] [CrossRef]
- Izumchenko, E.; Meir, J.; Bedi, A.; Wysocki, P.T.; Hoque, M.O.; Sidransky, D. Patient-Derived Xenografts as Tools in Pharmaceutical Development. Clin. Pharmacol. Ther. 2016, 99, 612–621. [Google Scholar] [CrossRef]
- Kreso, A.; O’Brien, C.A.; van Galen, P.; Gan, O.I.; Notta, F.; Brown, A.M.K.; Ng, K.; Ma, J.; Wienholds, E.; Dunant, C.; et al. Variable Clonal Repopulation Dynamics Influence Chemotherapy Response in Colorectal Cancer. Science 2013, 339, 543–548. [Google Scholar] [CrossRef]
- Peterson, J.K.; Houghton, P.J. Integrating pharmacology and in vivo cancer models in preclinical and clinical drug development. Eur. J. Cancer 2004, 40, 837–844. [Google Scholar] [CrossRef]
- Aparicio, S.; Hidalgo, M.; Kung, A.L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer 2015, 15, 311–316. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, C.-H.; Huang, W.-T.; Chew, C.H.; Lai, J.-K.; Tu, S.-H.; Wei, P.-L.; Lee, K.-Y.; Lai, G.-M.; Chen, C.-C. Electrospun Polylactic Acid (PLLA) Microtube Array Membrane (MTAM)—An Advanced Substrate for Anticancer Drug Screening. Materials 2019, 12, 569. https://doi.org/10.3390/ma12040569
Tseng C-H, Huang W-T, Chew CH, Lai J-K, Tu S-H, Wei P-L, Lee K-Y, Lai G-M, Chen C-C. Electrospun Polylactic Acid (PLLA) Microtube Array Membrane (MTAM)—An Advanced Substrate for Anticancer Drug Screening. Materials. 2019; 12(4):569. https://doi.org/10.3390/ma12040569
Chicago/Turabian StyleTseng, Chia-Hsuan, Wan-Ting Huang, Chee Ho Chew, Jun-Kai Lai, Shih-Hsin Tu, Po-Li Wei, Kang-Yun Lee, Gi-Ming Lai, and Chien-Chung Chen. 2019. "Electrospun Polylactic Acid (PLLA) Microtube Array Membrane (MTAM)—An Advanced Substrate for Anticancer Drug Screening" Materials 12, no. 4: 569. https://doi.org/10.3390/ma12040569
APA StyleTseng, C.-H., Huang, W.-T., Chew, C. H., Lai, J.-K., Tu, S.-H., Wei, P.-L., Lee, K.-Y., Lai, G.-M., & Chen, C.-C. (2019). Electrospun Polylactic Acid (PLLA) Microtube Array Membrane (MTAM)—An Advanced Substrate for Anticancer Drug Screening. Materials, 12(4), 569. https://doi.org/10.3390/ma12040569



