Revealing the Nanoparticle-Protein Corona with a Solid-State Nanopore
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of the Gold Nanoparticle-Protein Mixtures
2.3. Solid-State Nanopore Drilling and Functionalization
2.4. Dynamic Light Scattering Measurements
2.5. Gold Nanoparticle-Protein Complex Detection through SiNx Nanopore
3. Results and Discussion
3.1. Influence of Different Proteins on Nanoparticle Aggregation
3.2. Characterization of the Species by Single Nanopore Technique
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bhakta, S.A.; Evans, E.; Benavidez, T.E.; Garcia, C.D. Protein adsorption onto nanomaterials for the development of biosensors and analytical devices: A review. Anal. Chim. Acta 2015, 872, 7–25. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Bombelli, F.B.; Dawson, K.A. Physical-chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef]
- Lynch, I.; Dawson, K.A. Protein-nanoparticle interactions. Nano Today 2008, 3, 40–47. [Google Scholar] [CrossRef]
- Coglitore, D.; Janot, J.-M.; Balme, S. Protein at liquid solid interfaces: Toward a new paradigm to change the approach to design hybrid protein/solid-state materials. Adv. Colloid Interface Sci. 2019, 270, 278–292. [Google Scholar] [CrossRef]
- Lepoitevin, M.; Lemouel, M.; Bechelany, M.; Janot, J.-M.; Balme, S. Gold nanoparticles for the bare-eye based and spectrophotometric detection of proteins, polynucleotides and DNA. Microchim. Acta 2015, 182, 1223–1229. [Google Scholar] [CrossRef]
- Sabela, M.; Balme, S.; Bechelany, M.; Janot, J.-M.; Bisetty, K. A review of gold and silver nanoparticle-based colorimetric sensing assays. Adv. Eng. Mater. 2017, 19, 1700270. [Google Scholar] [CrossRef]
- Piella, J.; Bastús, N.G.; Puntes, V. Size-dependent protein-nanoparticle interactions in citrate-stabilized gold nanoparticles: The emergence of the protein corona. Bioconjug. Chem. 2017, 28, 88–97. [Google Scholar] [CrossRef]
- Casals, E.; Pfaller, T.; Duschl, A.; Oostingh, G.J.; Puntes, V. Time evolution of the nanoparticle protein corona. ACS Nano 2010, 4, 3623–3632. [Google Scholar] [CrossRef]
- Del Pino, P.; Pelaz, B.; Zhang, Q.; Maffre, P.; Nienhaus, G.U.; Parak, W.J. Protein corona formation around nanoparticles—From the past to the future. Mater. Horiz. 2014, 1, 301–313. [Google Scholar] [CrossRef]
- Gebauer, J.S.; Malissek, M.; Simon, S.; Knauer, S.K.; Maskos, M.; Stauber, R.H.; Peukert, W.; Treuel, L. Impact of the nanoparticle-protein corona on colloidal stability and protein structure. Langmuir 2012, 28, 9673–9679. [Google Scholar] [CrossRef]
- Milani, S.; Bombelli, F.B.; Pitek, A.S.; Dawson, K.A.; Rädler, J. Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: Soft and hard corona. ACS Nano 2012, 6, 2532–2541. [Google Scholar] [CrossRef]
- Ke, P.C.; Lin, S.; Parak, W.J.; Davis, T.P.; Caruso, F. A Decade of the Protein Corona. ACS Nano 2017, 11, 11773–11776. [Google Scholar] [CrossRef]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef]
- Howorka, S.; Cheley, S.; Bayley, H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat. Biotechnol. 2001, 19, 636–639. [Google Scholar] [CrossRef]
- Clarke, J.; Wu, H.-C.; Jayasinghe, L.; Patel, A.; Reid, S.; Bayley, H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009, 4, 265–270. [Google Scholar] [CrossRef]
- Bayley, H. Nanopore sequencing: From imagination to reality. Clin. Chem. 2015, 61, 25–31. [Google Scholar] [CrossRef]
- Piguet, F.; Ouldali, H.; Pastoriza-Gallego, M.; Manivet, P.; Pelta, J.; Oukhaled, A. Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat. Commun. 2018, 9, 966. [Google Scholar] [CrossRef]
- Cressiot, B.; Ouldali, H.; Pastoriza-Gallego, M.; Bacri, L.; van der Goot, F.G.; Pelta, J. Aerolysin, a powerful protein sensor for fundamental studies and development of upcoming applications. ACS Sens. 2019, 4, 530–548. [Google Scholar] [CrossRef]
- Larkin, J.; Henley, R.Y.; Muthukumar, M.; Rosenstein, J.K.; Wanunu, M. High-bandwidth protein analysis using solid-state nanopores. Biophys. J. 2014, 106, 696–704. [Google Scholar] [CrossRef]
- Oukhaled, G.; Mathé, J.; Biance, A.-L.; Bacri, L.; Betton, J.-M.; Lairez, D.; Pelta, J.; Auvray, L. Unfolding of proteins and long transient conformations detected by single nanopore recording. Phys. Rev. Lett. 2007, 98, 158101. [Google Scholar] [CrossRef] [PubMed]
- Oukhaled, A.; Cressiot, B.; Bacri, L.; Pastoriza-Gallego, M.; Betton, J.-M.; Bourhis, E.; Jede, R.; Gierak, J.; Auvray, L.; Pelta, J. Dynamics of completely unfolded and native proteins through solid-state nanopores as a function of electric driving force. ACS Nano 2011, 5, 3628–3638. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Ying, Y.-L.; Li, Y.; Kraatz, H.-B.; Long, Y.-T. Nanopore analysis of β-amyloid peptide aggregation transition induced by small molecules. Anal. Chem. 2011, 83, 1746–1752. [Google Scholar] [CrossRef]
- Giamblanco, N.; Coglitore, D.; Janot, J.-M.; Coulon, P.E.; Charlot, B.; Balme, S. Detection of protein aggregate morphology through single antifouling nanopore. Sens. Actuators B Chem. 2018, 260, 736–745. [Google Scholar] [CrossRef]
- Balme, S.; Coulon, P.E.; Lepoitevin, M.; Charlot, B.; Yandrapalli, N.; Favard, C.; Muriaux, D.; Bechelany, M.; Janot, J.-M. Influence of adsorption on proteins and amyloid detection by silicon nitride nanopore. Langmuir 2016, 32, 8916–8925. [Google Scholar] [CrossRef]
- Houghtaling, J.; List, J.; Mayer, M. Nanopore-based, rapid characterization of individual amyloid particles in solution: Concepts, challenges, and prospects. Small 2018, 14, e1802412. [Google Scholar] [CrossRef]
- Yusko, E.C.; Prangkio, P.; Sept, D.; Rollings, R.C.; Li, J.; Mayer, M. Single-particle characterization of Aβ oligomers in solution. ACS Nano 2012, 6, 5909–5919. [Google Scholar] [CrossRef]
- Coglitore, D.; Merenda, A.; Giamblanco, N.; Dumée, L.F.; Janot, J.-M.; Balme, S. Metal alloy solid-state nanopores for single nanoparticle detection. Phys. Chem. Chem. Phys. 2018, 20, 12799–12807. [Google Scholar] [CrossRef]
- Balme, S.; Lepoitevin, M.; Dumée, L.F.; Bechelany, M.; Janot, J.-M. Diffusion dynamics of latex nanoparticles coated with ssDNA across a single nanopore. Soft Matter 2017, 13, 496–502. [Google Scholar] [CrossRef]
- Bacri, L.; Oukhaled, A.G.; Schiedt, B.; Patriarche, G.; Bourhis, E.; Gierak, J.; Pelta, J.; Auvray, L. Dynamics of colloids in single solid-state nanopores. J. Phys. Chem. B 2011, 115, 2890–2898. [Google Scholar] [CrossRef]
- Yusko, E.C.; Johnson, J.M.; Majd, S.; Prangkio, P.; Rollings, R.C.; Li, J.; Yang, J.; Mayer, M. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 2011, 6, 253–260. [Google Scholar] [CrossRef]
- Yusko, E.C.; Bruhn, B.R.; Eggenberger, O.M.; Houghtaling, J.; Rollings, R.C.; Walsh, N.C.; Nandivada, S.; Pindrus, M.; Hall, A.R.; Sept, D.; et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2017, 12, 360–367. [Google Scholar] [CrossRef]
- Coglitore, D.; Giamblanco, N.; Kizalaité, A.; Coulon, P.E.; Charlot, B.; Janot, J.-M.; Balme, S. Unexpected hard protein behavior of BSA on gold nanoparticle caused by resveratrol. Langmuir 2018, 34, 8866–8874. [Google Scholar] [CrossRef]
- Orengo, C.A.; Pearl, F.M.; Bray, J.E.; Todd, A.E.; Martin, A.C.; Lo Conte, L.; Thornton, J.M. The CATH database provides insights into protein structure/function relationships. Nucleic Acids Res. 1999, 27, 275–279. [Google Scholar] [CrossRef]
- Orengo, C.A.; Michie, A.D.; Jones, S.; Jones, D.T.; Swindells, M.B.; Thornton, J.M. CATH—A hierarchic classification of protein domain structures. Structure 1997, 5, 1093–1109. [Google Scholar] [CrossRef]
- Plesa, C.; Dekker, C. Data analysis methods for solid-state nanopores. Nanotechnology 2015, 26, 84003. [Google Scholar] [CrossRef]
- Lundqvist, M.; Augustsson, C.; Lilja, M.; Lundkvist, K.; Dahlbäck, B.; Linse, S.; Cedervall, T. The nanoparticle protein corona formed in human blood or human blood fractions. PLoS ONE 2017, 12, e0175871. [Google Scholar] [CrossRef]
- Brewer, S.H.; Glomm, W.R.; Johnson, M.C.; Knag, M.K.; Franzen, S. Probing BSA binding to citrate-coated gold nanoparticles and surfaces. Langmuir 2005, 21, 9303–9307. [Google Scholar] [CrossRef]
- Roman, J.; Français, O.; Jarroux, N.; Patriarche, G.; Pelta, J.; Bacri, L.; Le Pioufle, B. Solid-state nanopore easy chip integration in a cheap and reusable microfluidic device for ion transport and polymer conformation sensing. ACS Sens. 2018, 3, 2129–2137. [Google Scholar] [CrossRef]
- Roman, J.; Jarroux, N.; Patriarche, G.; Français, O.; Pelta, J.; Le Pioufle, B.; Bacri, L. Functionalized solid-state nanopore integrated in a reusable microfluidic device for a better stability and nanoparticle detection. ACS Appl. Mater. Interfaces 2017, 9, 41634–41640. [Google Scholar] [CrossRef]
- Plesa, C.; Kowalczyk, S.W.; Zinsmeester, R.; Grosberg, A.Y.; Rabin, Y.; Dekker, C. Fast translocation of proteins through solid state nanopores. Nano Lett. 2013, 13, 658–663. [Google Scholar] [CrossRef]
- Servagent-Noinville, S.; Revault, M.; Quiquampoix, H.; Baron, M.-H. Conformational changes of bovine serum albumin induced by adsorption on different clay surfaces: FTIR analysis. J. Colloid Interface Sci. 2000, 221, 273–283. [Google Scholar] [CrossRef]
- Staunton, S.; Quiquampoix, H. Adsorption and conformation of bovine serum albumin on montmorillonite: Modification of the balance between hydrophobic and electrostatic interactions by protein methylation and pH variation. J. Colloid Interface Sci. 1994, 166, 89–94. [Google Scholar] [CrossRef]
- Arai, T.; Norde, W. The behavior of some model proteins at solid-liquid interfaces 1. Adsorption from single protein solutions. Colloids Surf. 1990, 51, 1–15. [Google Scholar] [CrossRef]
- Norde, W. Energy and entropy of protein adsorption. J. Dispers. Sci. Technol. 1992, 13, 363–377. [Google Scholar] [CrossRef]
- Norde, W. Driving forces for protein adsorption at solid surfaces. Macromol. Symp. 1996, 103, 5–18. [Google Scholar] [CrossRef]
- Norde, W.; Lyklema, J. Interfacial behaviour of proteins, with special reference to immunoglobulins. A physicochemical study. Adv. Colloid Interface Sci. 2012, 179–182, 5–13. [Google Scholar] [CrossRef]

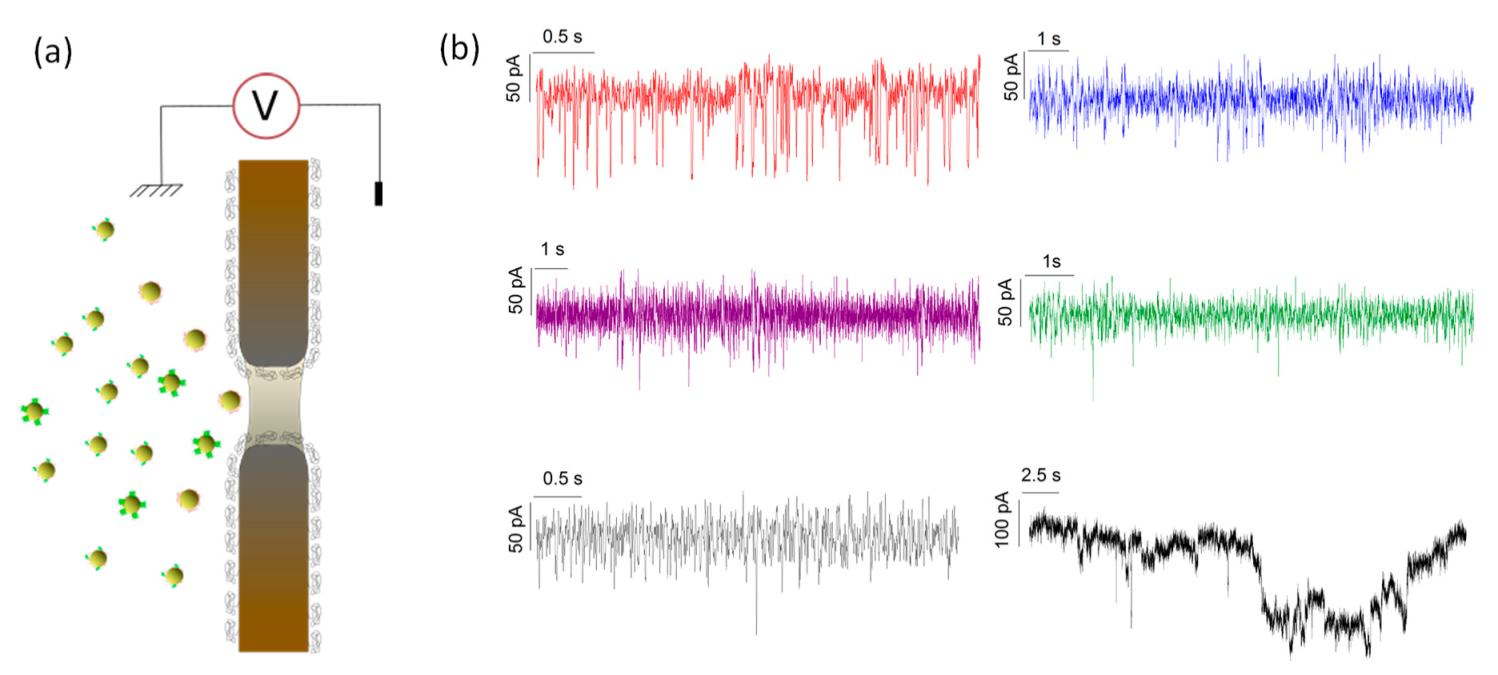

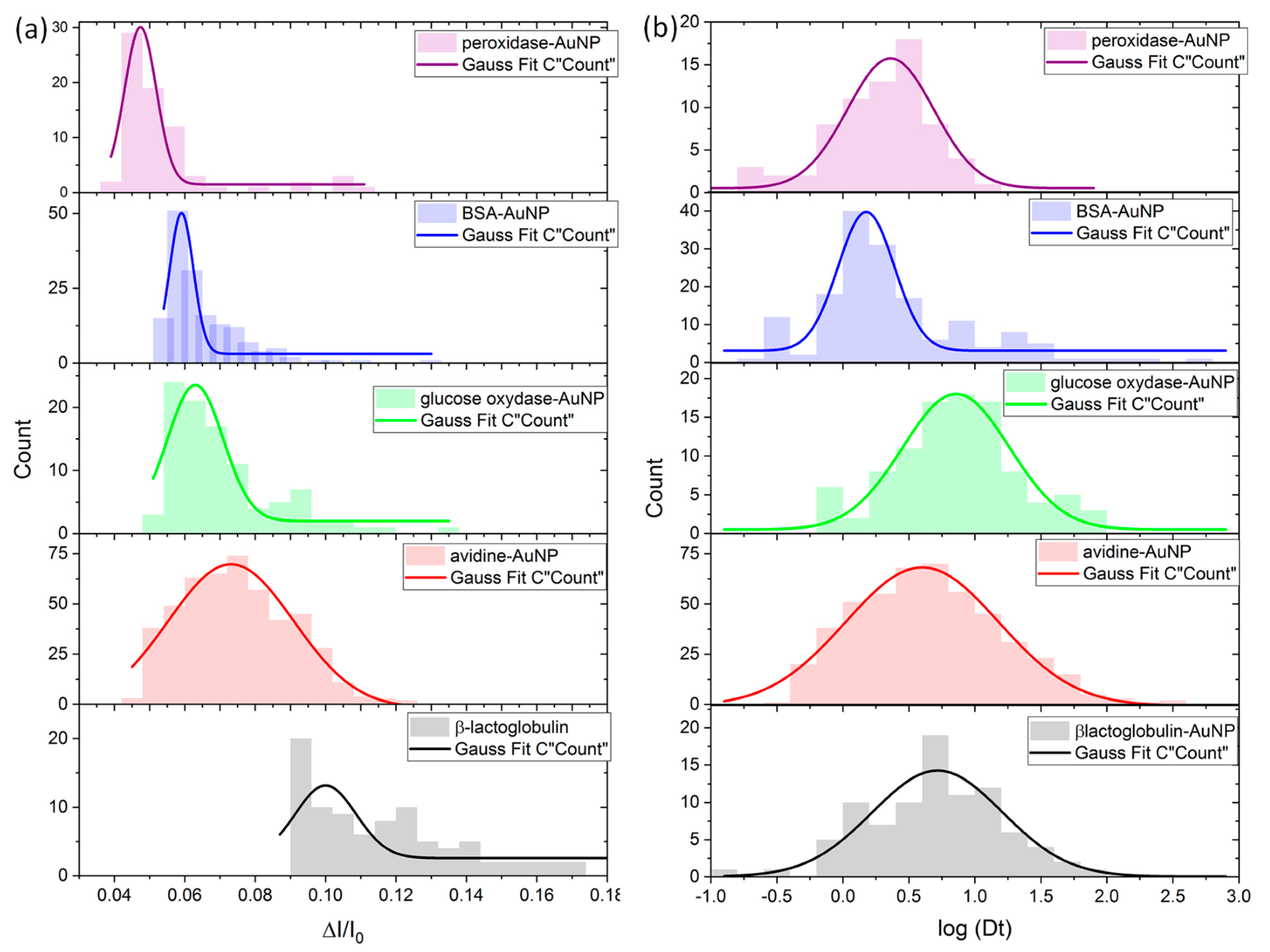
| ΔI/I0 | Colloids Volume (nm3) | Corona Volume (nm3) | Hydrodynamic Radius of Protein (nm) | |
|---|---|---|---|---|
| peroxidase-AuNP | 0.047 ± 0.011 | 1433 ± 73 | 909 | 3 |
| BSA-AuNP | 0.059 ± 0.008 | 1512 ± 53 | 989 | 3.5 |
| glucose oxidase-AuNP | 0.062 ± 0.0062 | 1532 ± 409 | 1008 | 4.3 |
| avidin-AuNP | 0.074 ± 0.041 | 1611 ± 277 | 1087 | 3.9 |
| β-lactoglobulin-AuNP | 0.100 ± 0.020 | 1783 ± 132 | 1259 | 2.9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coglitore, D.; Coulon, P.E.; Janot, J.-M.; Balme, S. Revealing the Nanoparticle-Protein Corona with a Solid-State Nanopore. Materials 2019, 12, 3524. https://doi.org/10.3390/ma12213524
Coglitore D, Coulon PE, Janot J-M, Balme S. Revealing the Nanoparticle-Protein Corona with a Solid-State Nanopore. Materials. 2019; 12(21):3524. https://doi.org/10.3390/ma12213524
Chicago/Turabian StyleCoglitore, Diego, Pierre Eugene Coulon, Jean-Marc Janot, and Sébastien Balme. 2019. "Revealing the Nanoparticle-Protein Corona with a Solid-State Nanopore" Materials 12, no. 21: 3524. https://doi.org/10.3390/ma12213524
APA StyleCoglitore, D., Coulon, P. E., Janot, J.-M., & Balme, S. (2019). Revealing the Nanoparticle-Protein Corona with a Solid-State Nanopore. Materials, 12(21), 3524. https://doi.org/10.3390/ma12213524





