Adaptive Laboratory Evolution for Multistress Tolerance, including Fermentability at High Glucose Concentrations in Thermotolerant Candida tropicalis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Media and Growth Conditions
2.3. Evolutionary Adaptation by RLCGT
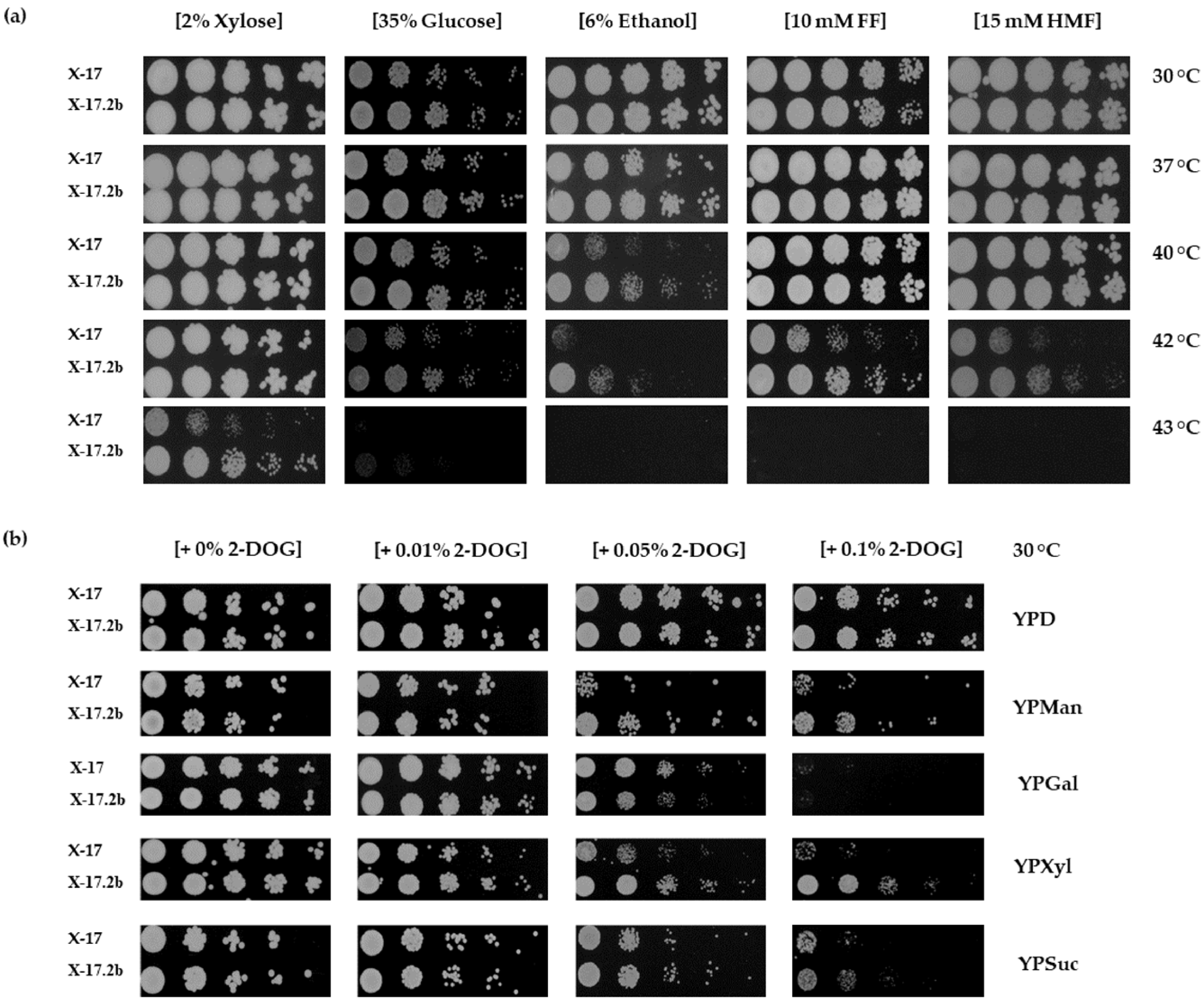
2.4. Analysis of Stress Resistance and Effects of 2-DOG on Utilization of Various Sugars
2.5. Analysis of Ethanol Fermentation
2.6. Preparation of Genomic DNA, Genomic Sequencing and Determination of Mutations
2.7. RNA-Seq Analysis
2.8. Hydropathy Analysis
3. Results
3.1. Evolutionary Adaptation of C. tropicalis X-17 by RLCGT
3.2. Effects of Various Stresses on Growth of X-17.2b
3.3. Ethanol Fermentation Ability of X-17.2b
3.4. Transcriptome Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moreno, A.D.; Carbone, A.; Pavone, R.; Olsson, L.; Geijer, C. Evolutionary engineered Candida tropicalis exhhibits improved xylose utilization and robustness to lignocelluloses-derived inhibitors and ethanol. Appl. Microbiol. Biotechnol. 2019, 103, 1405–1406. [Google Scholar] [CrossRef] [Green Version]
- Hoekman, S.K. Biofuels in the U.S.-challenges and opportunities. Renew. Energy 2009, 34, 14–22. [Google Scholar] [CrossRef]
- Li, G.; Lu, Z.; Zhang, J.; Li, H.; Zhou, Y.; Zayan, A.M.I.; Huang, Z. Life cycle assessment of biofuel production from microalgae cultivated in anaerobic digested wastewater. Int. J. Agric. Biol. 2020, 13, 241–246. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Bai, X.; Huo, S.; Huang, Z. Fast pyrolysis of LERDADEs for renewable biofuels. IET Renew. Power Gener. 2020, 14, 959–967. [Google Scholar] [CrossRef]
- Manyuchi, M.M.; Chiutsi, P.; Mbohwa, C.; Muzenda, E.; Mutusva, T. Bio ethanol from sewage sludge: A bio fuel alternative. S. Afr. J. Chem. Eng. 2018, 25, 123–127. [Google Scholar] [CrossRef]
- Patel, A.; Shah, A.R. Integrated lignocellulosic biorefinery: Gateway for production of second generation ethanol and value assed products. J. Bioresour. Bioprod. 2021, 6, 108–128. [Google Scholar] [CrossRef]
- Hamouda, H.I.; Nassar, H.N.; Madian, H.R.; Amr, S.A.; EI-Gendy, N.S. Statistical optimization of batch ethanol fermentation of sugarcane molasses by Candida tropicalis strain HSC-24. Int. J. Chemtech. 2015, 8, 878–889. [Google Scholar]
- Nweze, J.E.; Ndubuisi, I.; Murata, Y.; Omae, H.; Ogbonna, J.C. Isolation and evaluation of xylose-fermening thermotolerant yeasts for bioethanol production. Biofuels 2019, 12, 961–970. [Google Scholar] [CrossRef]
- Rodrussamee, N.; Sattayawat, P.; Yamada, M. Highly efficient conversion of xylose to ethanol without glucose repression by newly isolated thermotolerant Spathaspora passalidarum CMUWF1–2. BMC Microbiol. 2018, 18, 73. [Google Scholar] [CrossRef] [PubMed]
- Panwar, N.L.; Kaushik, S.C.; Kothari, S. Role of renewable energy sources in environmental protection: A review. Renew. Sust. Energ. Rev. 2011, 15, 1513–1524. [Google Scholar] [CrossRef]
- Chen, H.Z.; Fu, X.G. Indrustrial technologies for bioethanol production from lignocellulosic biomass. Renew. Sustain. Energy Rev. 2016, 57, 468–478. [Google Scholar] [CrossRef]
- Mano, M.C.R.; Neri-Numa, I.A.; Silva, J.B.; Paulino, B.N.; Pessoa, M.G.; Pastore, G.M. Oligosaccharide biotechnology: An approach of prebiotic revolution on the industry. Appl. Microbiol. Biotechol. 2018, 102, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Triwahyuni, E.; Muryanto; Sudiyani, Y.; Abimanyu, H. The effect of substrate loading on simultaneous saccharification and fermentation process for bioethanol production from oil palm empty fruit bunches. Energy Procedia. 2015, 68, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Nitiyon, S.; Keo-oudone, C.; Murata, M.; Lertwattanasakul, N.; Limtong, S.; Kosaka, T.; Yamada, M. Efficient conversion of xylose to ethanol by stress-tolerant Kluyveromyces marxianus BUNL-21. SprigerPlus 2016, 5, 185. [Google Scholar] [CrossRef] [Green Version]
- Nguyen Tran, P.H.; Ko, J.K.; Gong, G.; Um, Y.; Lee, S.M. Improved simultaneous co-fermentation of glucose and xylose by Saccharomyces cerevisiae for efficient lignocellulosic biorefinery. Biotechnol. Biofuels 2020, 13, 12. [Google Scholar] [CrossRef]
- Kim, D. Physico-chemical conversion of lignocellulose: Inhibitor effects and detoxification strategies: A mini review. Molecules 2018, 23, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, Ó.J.; Cardona, C.A. Trends in biotechnological production of fuel ethanol from different feedstocks. Bioresour. Technol. 2008, 99, 5270–5295. [Google Scholar] [CrossRef]
- Gibson, B.R.; Lawrence, S.J.; Leclaire, J.P.R.; Powell, C.D.; Smart, K.A. Yeast responses to stresses associated with industrial brewery Handling. FEMS Microbiol. Rev. 2007, 31, 535–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.Q.; Bai, F.W. Mechanisms of yeast stress tolerance and its manipulation for efficient fuel ethanol production. J. Biotechnol. 2009, 144, 23–30. [Google Scholar] [CrossRef]
- Puligundla, P.; Smogrovicova, D.; Obulam, V.S.R.; Ko, S. Very high gravity (VHG) ethanolic brewing and fermentation: A research update. J. Ind. Microbiol. Biotechnol. 2011, 38, 1133–1144. [Google Scholar] [CrossRef]
- Auesukaree, C. Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J. Biosci. Bioeng. 2017, 124, 133–142. [Google Scholar] [CrossRef]
- Azhar, S.H.M.; Abdulla, R.; Jambo, S.A.; Marbawi, H.; Gansau, J.A.; Faik, A.A.M.; Rodrigues, K.F. Yeast in sustainable bioethanol production: A review. Biochem. Biophys. Rep. 2017, 10, 52–61. [Google Scholar]
- Kamoldeen, A.A.; Lee, C.K.; Abdullah, W.N.W.; Leh, C.P. Enhanced ethanol production from mild alkali-treated oil-palm empty fruit bunches via co-fermentation of glucose and xylose. Renew. Energy 2017, 107, 113–123. [Google Scholar] [CrossRef]
- Greetham, D.; Hart, A.J.; Tucker, G.A. Presence of low concentrations of acetic acid improves yeast tolerance to hydroxymethylfurfural (HMF) and furfural. Biomass Bioenergy 2016, 85, 53–60. [Google Scholar] [CrossRef]
- Cheng, N.G.; Hasan, M.; Kumoro, A.C.; Ling, C.F.; Tham, M. Production of ethanol by fed-batch fermentation. Pertanika J. Sci. Technol. 2009, 17, 399–408. [Google Scholar]
- Alba, M.G.; Parra, M.A.M.; Olmo, M.D. Response of yeast cells to high glucose involves molecular and physiological difference when compared to other osmostress conditions. FEMS Yeast Res. 2015, 15, fov039. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, B.; Wang, S.P.; Xia, Z.Y.; Gou, M.; Tang, Y.Q. Improving multiple stress tolerance of a flocculating industrial Saccharomyces cerivisiae strain by random mutagenesis and hybridization. Process Biochem. 2021, 102, 275–285. [Google Scholar] [CrossRef]
- Limtong, S.; Sringiew, C.; Yongmanitchai, W. Production of fuel ethanol at high temperature from sugar cane juice by a newly isolated Kluyveromyces marxianus. Bioresour. Technol. 2007, 17, 3367–3374. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Preparation and analysis of eukaryotic genomic DNA. In Molecular Cloning a Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 3, pp. 1–62. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrow-wheeler transform. J. Bioinform. 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Nurcholis, M.; Nitiyon, S.; Suprayogi; Rodrussamee, N.; Lertwattanasakul, N.; Limtong, S.; Kosaka, T.; Yamada, M. Functional analysis of Mig1 and Rag5 as expressional regulators in thermotolerant yeast Kluyveromyces marxianus. Appl. Microbiol. Biotechnol. 2019, 103, 395–410. [Google Scholar] [CrossRef]
- Lertwattanasakul, N.; Kosaka, T.; Hosoyama, A.; Suzuki, Y.; Rodrussamee, N.; Matsutani, M.; Murata, M.; Fujimoto, N.; Suprayogi; Tsuchikane, K.; et al. Genetic basis of the highly efficient yeast Kluyveromyces marxianus: Complete genome sequence and transcriptome analyses. Biotechnol. Biofuels 2015, 8, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, I.S.; Kim, Y.S.; Kim, H.; Jin, I.; Yoon, H.S. Saccharomyces cerevisiae KNU5377 stress response during high-temperature ethanol fermentation. Mol. Cells. 2013, 35, 210–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andars, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010, 11, R106. [Google Scholar]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, D.; Schwarz, E.; Komaromy, M.; Wall, R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984, 179, 125–142. [Google Scholar] [CrossRef]
- Rodrussamee, N.; Lertwattanasakul, N.; Hirata, K.; Suprayogi; Limtong, S.; Kosaka, T.; Yamada, M. Growth and ethanol fermentation ability on hexose and pentose sugars and glucose effect under various conditions in thermotolerant yeast Kluyveromyces marxianus. Biomass Bioenergy 2011, 90, 1573–1586. [Google Scholar] [CrossRef]
- Guragain, Y.N.; Chitta, D.; Karanjikar, M.; Vadlani, P.V. Appropriate lignocellulosic biomass processing strategies for efficient 2,3-butanediol production from biomass derived sugars using Bacillus licheniformis DSM 8785. Food Bioprod. Process. 2017, 104, 147–158. [Google Scholar] [CrossRef]
- Ji, X.J.; Huang, H.; Du, J.; Zhu, J.G.; Ren, L.J.; Li, S.; Nie, Z.K. Development of an industrial medium for economical 2,3-butanediol production through co-fermentation of glucose and xylose by Klebsiella oxytoca. Bioresour. Technol. 2009, 100, 5214–5218. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, H.; Chen, J.; Lin, Z.; Jin, Q.; Jia, H.; Huang, H. Dilute sulfuric acid cycle spray flow-through pretreatment of corn stover for enhancement of sugar recovery. Bioresour. Technol. 2009, 100, 1803–1808. [Google Scholar] [CrossRef]
- Stephanopoulos, G. Challenges in engineering microbes for biofuels production. Science 2007, 315, 801–804. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.K.; Wu, J.; Lin, Z.N.; Zhang, J.A. Aerobic and sequential anaerobic fermentation to produce xylitol and ethanol using non-detoxified acid pretreated corncob. Biotechnol. Biofuels 2014, 7, 166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattam, A.J.; Kuila, A.; Suralikerimath, N.; Choudary, N.; Rao, P.V.C.; Velankar, H.R. Cellulolytic enzyme expression and simultaneous conversion of lignocellulosic sugars into ethanol and xylitol by a new Candida tropicalis strain. Biotechnol. Biofuels 2016, 9, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antunes, F.A.F.; Thomé, L.C.; Santos, J.C.; Ingle, A.P.; Costa, C.B.; Anjos, V.D.; Bell, M.J.V.; Rosa, C.A.; Silva, S.S. Multi-scale study of the integrated use of the carbohydrate fraction of sugarcane bagasse for ethanol and xylitol production. Renew. Energy 2021, 163, 1343–1355. [Google Scholar] [CrossRef]
- Li, X.; Deng, Y.; Yang, Y.; Wei, Z.; Cheng, J.; Cao, L.; Mu, D.; Luo, S.; Zheng, Z.; Jiang, S.; et al. Fermentation process and metabolic flux of ethanol production from the detoxified hydrolyzate of cassava residue. Microbiology 2017, 8, 1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Name | Log2 Fold Change | Product |
|---|---|---|
| CTRG_06057 | 4.95 | Hypothetical protein |
| CTRG_06055 | 4.07 | Hypothetical protein |
| CTRG_06056 | 3.84 | Hypothetical protein |
| CTRG_06100 | 3.63 | Maltose permease |
| CTRG_06311 | 3.31 | tRNA |
| CTRG_00691 | 3.31 | Hypothetical protein |
| CTRG_04447 | 3.23 | Hypothetical protein |
| CTRG_02794 | 3.12 | Hypothetical protein |
| CTRG_03584 | 3.08 | Opaque-phase-specific protein OP4 precursor |
| CTRG_06401 | 3.06 | tRNA |
| CTRG_02358 | 3.05 | Resistance to glucose repression protein 1 |
| CTRG_00749 | 3.02 | Hypothetical protein |
| CTRG_03295 | 2.86 | Hypothetical protein |
| CTRG_06346 | 2.82 | tRNA |
| CTRG_05401 | 2.77 | Ornitine carbamoyltransferase |
| CTRG_05272 | 2.73 | Aga1p |
| CTRG_00349 | 2.71 | Cell wall protein RHD3 |
| CTRG_05402 | 2.61 | Methylglyoxal reductase (NADPH-dependent) |
| CTRG_00711 | 2.60 | White-opaque regulator 3 |
| CTRG_01483 | 2.57 | Hypothetical protein |
| CTRG_05080 | 2.56 | Hypothetical protein |
| CTRG_04732 | 2.55 | Histone H3 |
| CTRG_01407 | 2.55 | Hypothetical protein |
| CTRG_00808 | 2.54 | Hypothetical protein |
| CTRG_06299 | 2.54 | tRNA |
| CTRG_05490 | 2.52 | Hypothetical protein |
| CTRG_06416 | 2.51 | tRNA |
| CTRG_03294 | 2.51 | Hypothetical protein |
| CTRG_04755 | 2.51 | Hypothetical protein |
| CTRG_06250 | 2.50 | Glucose transporter of major facilitator superfamily |
| CTRG_02210 | 2.42 | Acetylornithine aminotransferase, mitochondrial precursor |
| CTRG_03885 | 2.29 | Lipase 8 |
| CTRG_00519 | 2.28 | Hypothetical protein |
| CTRG_00291 | 2.28 | Hypothetical protein |
| CTRG_03791 | 2.27 | Hypothetical protein |
| CTRG_00298 | 2.27 | Hypothetical protein |
| CTRG_05031 | 2.25 | Hypothetical protein |
| CTRG_00623 | 2.24 | Hypothetical protein |
| CTRG_02946 | 2.24 | Peroxiredoxin HYR1 |
| CTRG_06103 | 2.23 | Hypothetical protein |
| CTRG_00350 | 2.22 | Cell wall protein PGA31 |
| CTRG_06301 | 2.20 | tRNA |
| CTRG_05078 | 2.17 | Hypothetical protein |
| CTRG_03785 | 2.16 | Cell wall protein PGA31 |
| CTRG_00604 | 2.15 | Hypothetical protein |
| CTRG_01139 | 2.14 | Hypothetical protein |
| CTRG_02833 | 2.12 | Vacuolar basic amino acid transporter 5 |
| CTRG_00842 | 2.11 | Peroxisomal membrane protein LPX1 |
| CTRG_00266 | 2.10 | Hypothetical protein |
| CTRG_00233 | 2.10 | Hypothetical protein |
| CTRG_02278 | 2.10 | Thiol-specific monooxygenase |
| CTRG_01965 | 2.10 | Hypothetical protein |
| CTRG_01779 | 2.09 | 4-hydroxyphenylpyruvate dioxygenase |
| CTRG_04145 | 2.07 | Hypothetical protein |
| CTRG_00102 | 2.07 | Hypothetical protein |
| CTRG_03730 | 2.07 | NAG4 |
| CTRG_06102 | 2.07 | Hypothetical protein |
| CTRG_02773 | 2.06 | Hypothetical protein |
| CTRG_03597 | 2.05 | Hypothetical protein |
| CTRG_06383 | 2.05 | tRNA |
| CTRG_05709 | 2.04 | Carboxylic acid transporter |
| CTRG_00500 | 2.03 | Hypothetical protein |
| CTRG_06026 | 2.02 | Hypothetical protein |
| CTRG_00590 | 2.02 | Stress response regulator protein 1 |
| CTRG_04524 | 2.01 | Hypothetical protein |
| CTRG_06404 | 2.01 | tRNA |
| Strains | Temp. (°C) | Sugars Conc. (g·L−1) | Time (h) | Sugars Consumption (g·L−1) | Ethanol Production (g·L−1) | Xylitol Production (g·L−1) | Glycerol Production (g·L−1) | Acetic Acid Production (g·L−1) | Ethanol Yield (g·g−1) | Ethanol Productivity (g·L−1·h−1) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C. tropicalis X-17 | 37 | Glc 20 | 6 | Glc 15.6 ± 2.9 | 8.2 ± 0.6 | - | - | 0.0 ± 0.0 | 0.4 ± 0.0 | 1.37 ± 0.1 | This study |
| 37 | Glc 160 | 24 | Glc 107.2 ± 1.3 | 46.5 ± 0.7 | - | 11.4 ± 0.2 | 0.0 ± 0.0 | 0.3 ± 0.0 | 1.94 ± 0.0 | This study | |
| 37 | Xyl 20 | 36 | Xyl 17.1 ± 3.1 | 2.0 ± 0.3 | 8.0 ± 0.2 | - | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.06 ± 0.0 | This study | |
| 37 | Xyl 50 | 60 | Xyl 37.9 ± 1.6 | 3.1 ± 0.1 | 25.2 ± 0.1 | 0.5 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.05 ± 0.0 | This study | |
| 37 | Glc 2 + Xyl 20 | 36 | Glc 2.1 ± 0.0 Xly 15.6 ± 0.1 | 2.8 ± 0.2 | 8.3 ± 0.2 | - | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.08 ± 0.0 | This study | |
| 37 | Glc 20 + Xyl 20 | 60 | Glc 19.2 ± 2.0 Xyl 16.7 ± 1.1 | 8.0 ± 0.6 | 7.2 ± 0.6 | - | 0.3 ± 0.2 | 0.2 ± 0.0 | 0.13 ± 0.0 | This study | |
| 37 | Glc 20 + Xyl 50 | 72 | Glc 21.6 ± 1.7 Xyl 45.1 ± 1.0 | 10.6 ± 0.7 | 30.3 ± 5.9 | 0.4 ± 0.2 | 0.1 ± 0.1 | 0.1 ± 0.0 | 0.15 ± 0.0 | This study | |
| 35 | Glc 50 + Xyl 50 | 72 | Glc 50.0 ± 0.0 Xyl 39.1 ± 0.0 | 25.2 ± 0.0 | 20.0 ± 0.0 | - | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.42 ± 0.0 | This study | |
| C. tropicalis X-17.2b | 37 | Glc 20 | 6 | Glc 16.2 ± 4.2 | 8.1 ± 0.9 | - | - | 0.0 ± 0.0 | 0.4 ± 0.0 | 1.35 ± 0.1 | This study |
| 37 | Glc 160 | 24 | Glc 132.2 ± 1.4 | 60.7 ± 2.1 | - | 4.5 ± 0.1 | 0.1 ± 0.0 | 0.4 ± 0.0 | 2.53 ± 0.1 | This study | |
| 37 | Xyl 20 | 36 | Xyl 17.8 ± 2.7 | 2.9 ± 0.1 | 7.1 ± 0.4 | - | 0.2 ± 0.1 | 0.1 ± 0.0 | 0.08 ± 0.0 | This study | |
| 37 | Xyl 50 | 60 | Xyl 41.1 ± 1.4 | 4.4 ± 0.1 | 26.9 ± 1.5 | 0.8 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.07 ± 0.0 | This study | |
| 37 | Glc 2 + Xyl 20 | 36 | Glc 2.1 ± 0.0 Xyl 16.0 ± 0.1 | 3.8 ± 0.3 | 6.6 ± 0.4 | - | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.10 ± 0.0 | This study | |
| 37 | Glc 20 + Xyl 20 | 60 | Glc 18.1 ± 1.9 Xyl 17.8 ± 1.2 | 10.1 ± 0.9 | 6.9 ± 1.0 | - | 0.6 ± 0.1 | 0.3 ± 0.0 | 0.17 ± 0.0 | This study | |
| 37 | Glc 20 + Xyl 50 | 72 | Glc 20.8 ± 0.7 Xyl 45.7 ± 1.0 | 13.6 ± 0.5 | 25.4 ± 2.3 | 0.4 ± 0.3 | 0.7 ± 0.7 | 0.2 ± 0.0 | 0.19 ± 0.0 | This study | |
| 35 | Glc 50 + Xyl 50 | 72 | Glc 50.00 ± 0.0 Xyl 40.6 ± 0.0 | 24.6 ± 0.9 | 22.7 ± 0.0 | - | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.40 ± 0.0 | This study | |
| K. marxianus DMKU 3-1042 | 37 | Glc 20 | 6 | Glc 6.6 ± 3.2 | 3.9 ± 0.7 | - | - | 0.4 ± 0.0 | 0.2 ± 0.0 | 0.65 ± 0.1 | This study |
| 37 | Glc 160 | 24 | Glc 132.4 ± 1.2 | 56.8 ± 0.8 | - | 6.5 ± 0.0 | 1.0 ± 0.1 | 0.3 ± 0.0 | 2.37 ± 0.0 | This study | |
| 37 | Xyl 20 | 36 | Xyl 12.8 ± 2.3 | 1.0 ± 0.1 | 6.6 ± 1.4 | - | 0.9 ± 0.2 | 0.1 ± 0.0 | 0.03 ± 0.0 | This study | |
| 37 | Xyl 50 | 60 | Xly 36.3 ± 0.4 | 3.1 ± 0.1 | 27.6 ± 0.7 | 0.1 ± 0.0 | 1.4 ± 0.0 | 0.1 ± 0.0 | 0.05 ± 0.0 | This study | |
| 37 | Glc 2 + Xyl 20 | 36 | Glc 2.1 ± 0.0 Xyl 10.3 ± 0.2 | 0.5 ± 0.0 | 10.2 ± 1.7 | - | 1.5 ± 0.0 | 0.0 ± 0.00 | 0.01 ± 0.0 | This study | |
| 37 | Glc 20 + Xyl 20 | 60 | Glc 18.3 ± 1.9 Xyl 5.7 ± 2.3 | 3.8 ± 0.9 | 3.3 ± 0.5 | - | 4.5 ± 0.3 | 0.1 ± 0.0 | 0.06 ± 0.0 | This study | |
| 37 | Glc 20 + Xyl 50 | 72 | Glc 22.0 ± 1.7 Xyl 15.5 ± 5.5 | 3.3 ± 1.5 | 3.5 ± 1.1 | 0.9 ± 0.1 | 4.6 ± 0.7 | 0.1 ± 0.02 | 0.05 ± 0.0 | This study | |
| 30 | Xyl 20 | 48 | Xyl 19.2 ± 1.09 | 1.7 ± 0.4 | 2.2 ± 0.5 | NR | 7.2 ± 0.4 | 0.1 ± 0.0 | ~0.03 | [14] | |
| 30 | Glc 20: Xyl 20 | 60 | Glc~0.0 Xyl~7 | ~8.0 | ~2.00 | NR | NR | ~0.20 | ~0.13 | [14] | |
| C. tropicalis W103 | 35 | Glc 50 + Xyl 50 | 75 | Glc~50 Xyl~25 | ~20 | NR | NR | NR | ~0.2 | ~0.26 | [43] |
| C. tropicalis MTCC 25057 | 32 | Glc 100 | 48 | Glc~100 | 36 | NR | NR | NR | ~0.36 | ~0.75 | [44] |
| 32 | Glc 50 + Xyl 50 | 24 | Glc~50 Xyl~00 | 18.8 ± 0.8 | NR | NR | NR | ~0.2 | ~0.78 | [44] | |
| C. tropicalis UFMGBX12-a | 30 | Glc 18 + Xyl 2 | 30 | Glc~00 Xyl~0.7 | 1.5 | 12 | NR | NR | 0.1 | 0.05 | [45] |
| C. tropicalis CICC1779 | 34 | Glc 30 + Xyl 30 | 72 | Glc 27.5 Xyl 16.7 | 17.6 | NR | NR | NR | ~0.3 | ~0.24 | [46] |
| C. tropicalis M9 | 42 | Xyl 50 | 168 | NR | ~12 | NR | NR | NR | ~0.2 | ~0.07 | [8] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phommachan, K.; Keo-oudone, C.; Nurcholis, M.; Vongvilaisak, N.; Chanhming, M.; Savanhnaly, V.; Bounphanmy, S.; Matsutani, M.; Kosaka, T.; Limtong, S.; et al. Adaptive Laboratory Evolution for Multistress Tolerance, including Fermentability at High Glucose Concentrations in Thermotolerant Candida tropicalis. Energies 2022, 15, 561. https://doi.org/10.3390/en15020561
Phommachan K, Keo-oudone C, Nurcholis M, Vongvilaisak N, Chanhming M, Savanhnaly V, Bounphanmy S, Matsutani M, Kosaka T, Limtong S, et al. Adaptive Laboratory Evolution for Multistress Tolerance, including Fermentability at High Glucose Concentrations in Thermotolerant Candida tropicalis. Energies. 2022; 15(2):561. https://doi.org/10.3390/en15020561
Chicago/Turabian StylePhommachan, Koudkeo, Chansom Keo-oudone, Mochamad Nurcholis, Nookhao Vongvilaisak, Mingkhuan Chanhming, Vanhnavong Savanhnaly, Somchanh Bounphanmy, Minenosuke Matsutani, Tomoyuki Kosaka, Savitree Limtong, and et al. 2022. "Adaptive Laboratory Evolution for Multistress Tolerance, including Fermentability at High Glucose Concentrations in Thermotolerant Candida tropicalis" Energies 15, no. 2: 561. https://doi.org/10.3390/en15020561
APA StylePhommachan, K., Keo-oudone, C., Nurcholis, M., Vongvilaisak, N., Chanhming, M., Savanhnaly, V., Bounphanmy, S., Matsutani, M., Kosaka, T., Limtong, S., & Yamada, M. (2022). Adaptive Laboratory Evolution for Multistress Tolerance, including Fermentability at High Glucose Concentrations in Thermotolerant Candida tropicalis. Energies, 15(2), 561. https://doi.org/10.3390/en15020561








