Effect of Temporal Variation in Chemical Composition on Methane Yields of Rendering Plant Wastewater
Abstract
1. Introduction
2. Materials and Methods
2.1. Substrate and Inoculum
2.2. Experimental Set-Up
2.3. Kinetic Modelling
2.4. Analitical Methods
2.5. Calculations
2.6. Statistical Analysis
3. Results and Discussion
3.1. Temporal Variation in Chemical Composition of Rendering Plant Wastewater
3.2. Batch Experiments
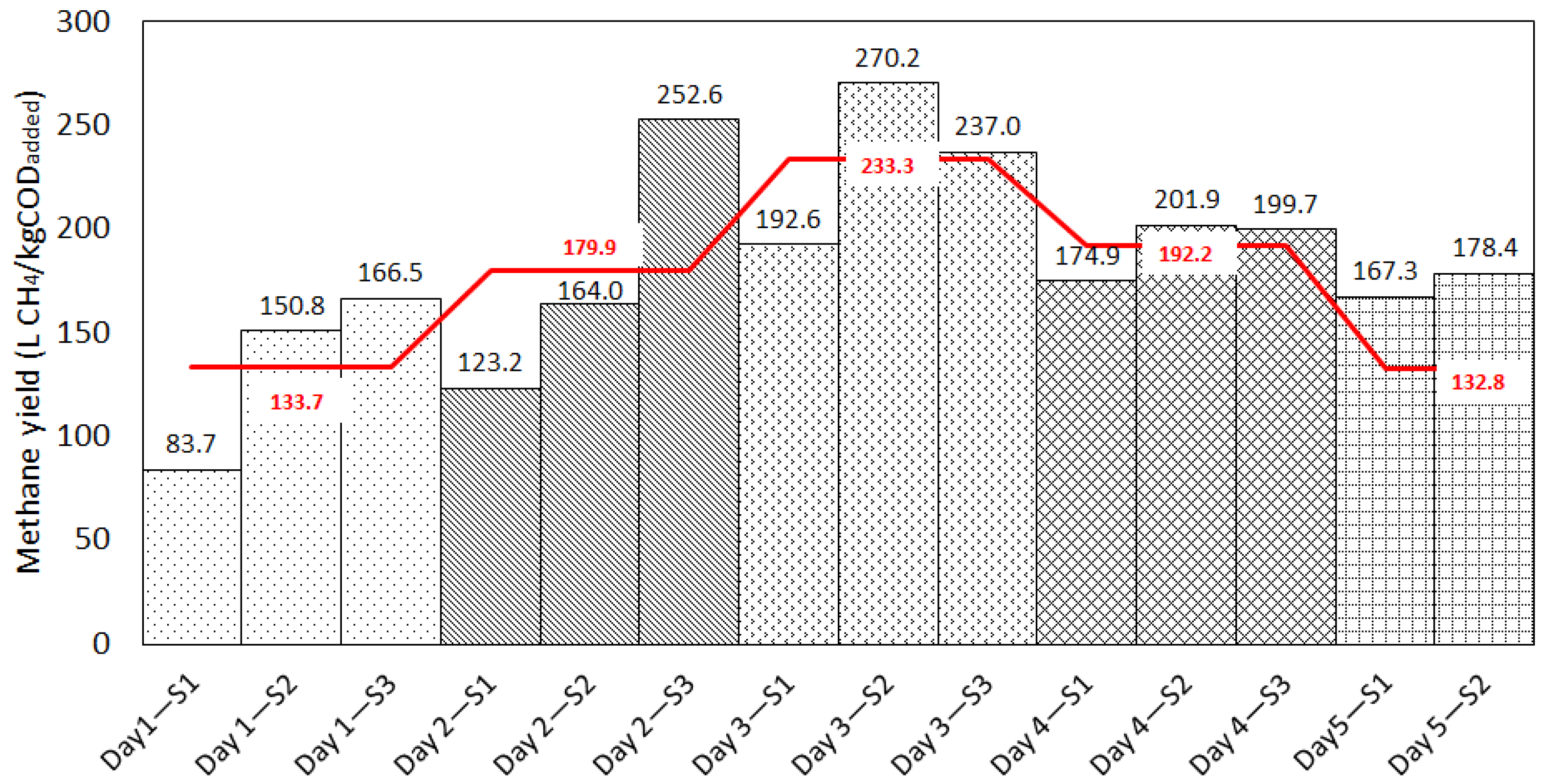
3.2.1. Effect of Temporal Variation in Chemical Composition on Methane Yields from Rendering Wastewater
3.2.2. Chemical Composition of the Digestates
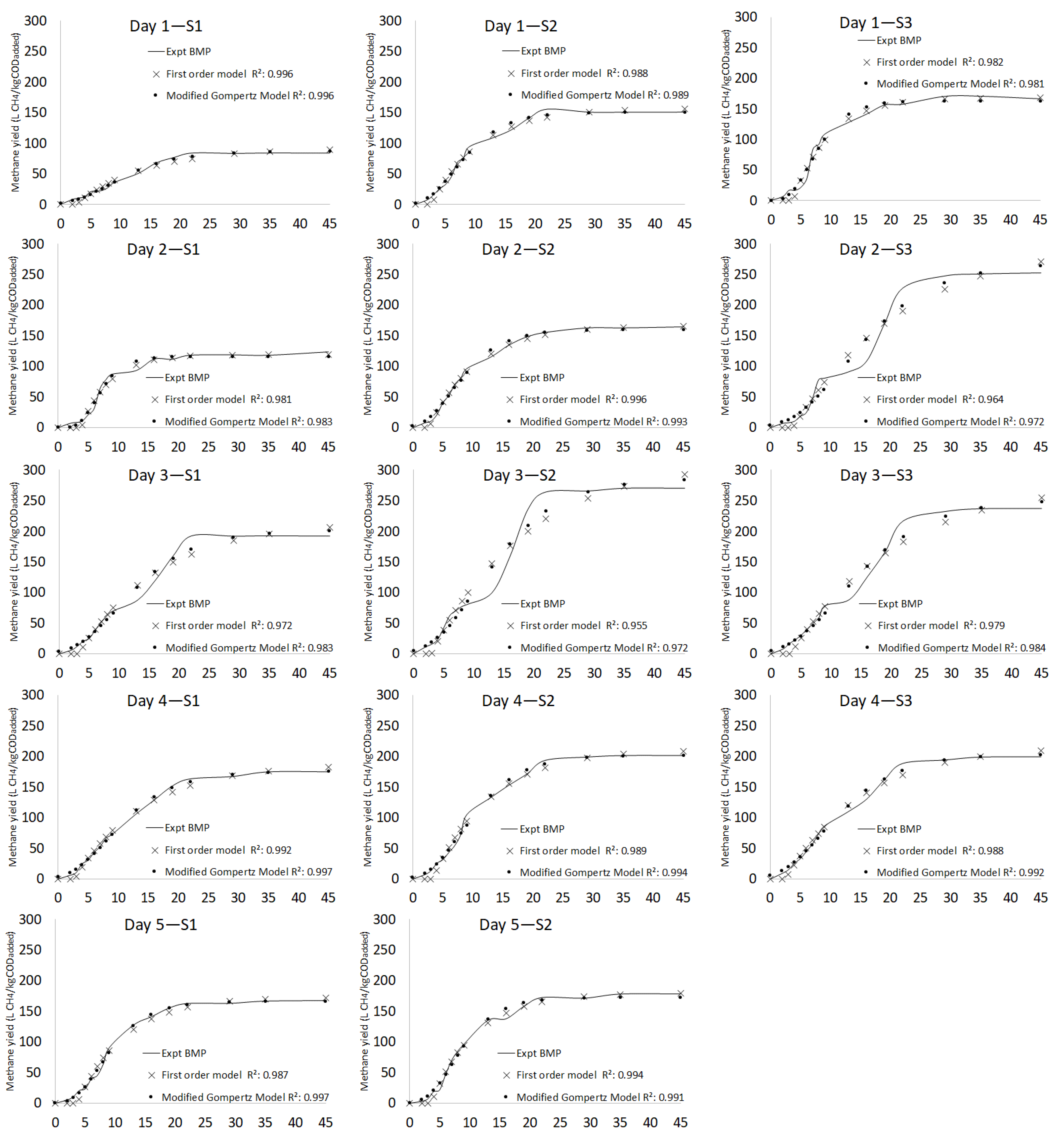
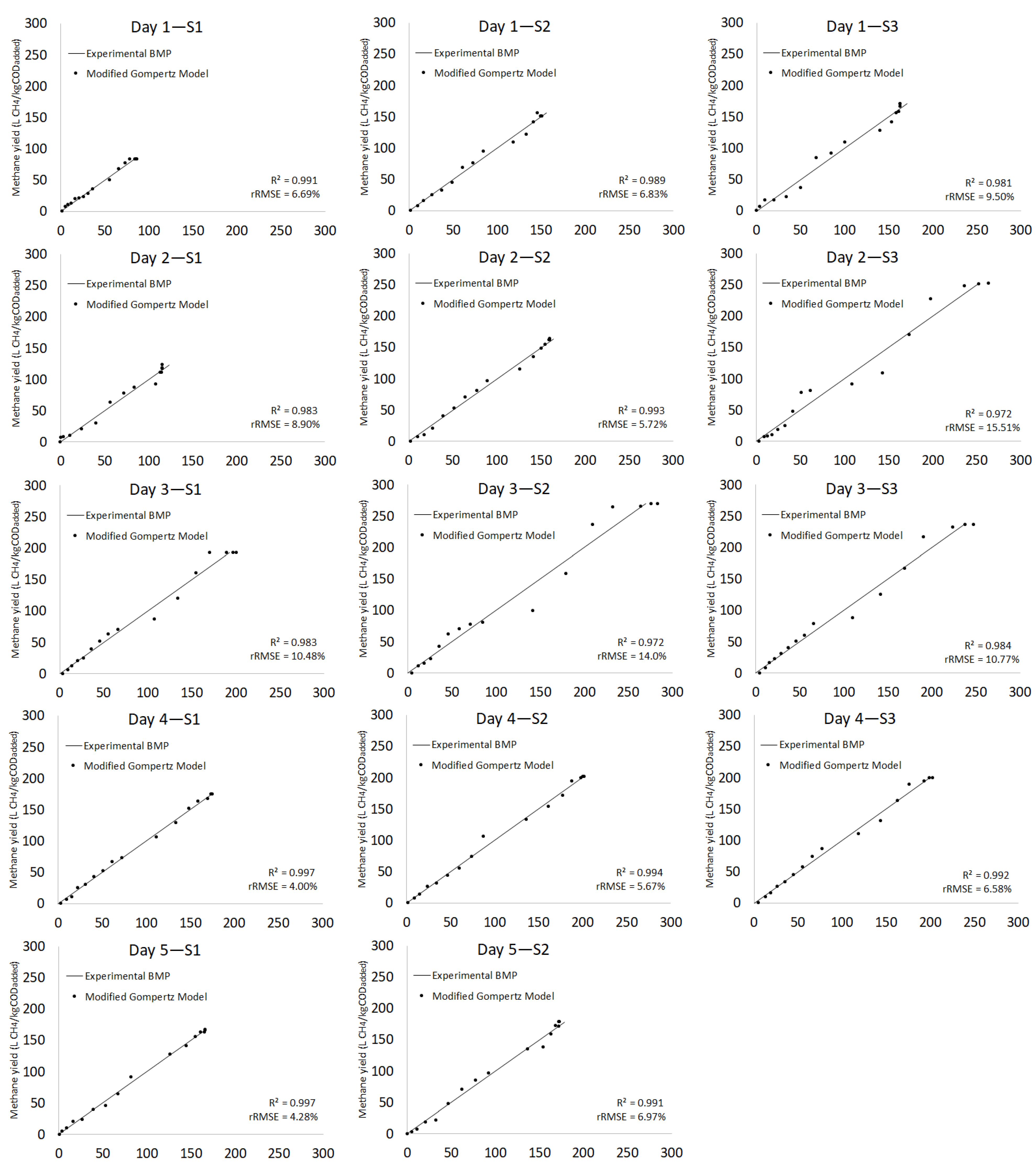
3.2.3. Kinetic Model
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- OECD-FAO. OECD-FAO Agricultural Outlook 2022–2030; OECD Publishing: Paris, France, 2022. [Google Scholar]
- MLA. Industry Projections 2021—Australian Cattle; MLA: New York, NY, USA, 2021. [Google Scholar]
- NSW Environment Protection Authority. Future Use of Household Waste and Mixed Waste Organic Outputs; NSW Environment Protection Authority: Parramatta, Australia, 2019.
- AMPC; MLA. 2020 Environmental Performance Review (EPR) for the Red Meat Processing (RMP) Industry; MLA: New York, NY, USA, 2021. [Google Scholar]
- Xie, A.; Deaver, J.A.; Miller, E.; Popat, S.C. Effect of feed-to-inoculum ratio on anaerobic digestibility of high-fat content animal rendering wastewater. Biochem. Eng. J. 2021, 176, 108215. [Google Scholar] [CrossRef]
- Sindt, G. Environmental issues in the rendering industry. In Essential Rendering: All About The Animal By-Products Industry; Meeker, D.L., Ed.; The Animal Protein Producers Industry: Alexandria, VA, USA, 2006; pp. 245–272. [Google Scholar]
- Aziz, A.; Basheer, F.; Sengar, A.; Khan, S.U.; Farooqi, I.H. Biological wastewater treatment (anaerobic-aerobic) technologies for safe discharge of treated slaughterhouse and meat processing wastewater. Sci. Total Environ. 2019, 686, 681–708. [Google Scholar] [CrossRef] [PubMed]
- Laginestra, M.; van-Oorschot, R. Wastewater Treatment Pond Systems—An Australian Experience. In Proceedings of the Australian Water Association Conference, Melboure, Australia, 16–18 March 2009; 2009. [Google Scholar]
- Masse, L.; Masse, D. Effect of soluble organic, particulate organic, and hydraulic shock loads on anaerobic sequencing batch reactors treating slaughterhouse wastewater at 20 C. Process Biochem. 2005, 40, 1225–1232. [Google Scholar] [CrossRef]
- Shilton, A.; Harrison, J. Development of guidelines for improved hydraulic design of waste stabilisation ponds. Water Sci. Technol. 2003, 48, 173–180. [Google Scholar] [CrossRef]
- Harris, P.W.; McCabe, B.K. Review of pre-treatments used in anaerobic digestion and their potential application in high-fat cattle slaughterhouse wastewater. Appl. Energy 2015, 155, 560–575. [Google Scholar] [CrossRef]
- Harris, P.W.; McCabe, B.K. Process optimisation of anaerobic digestion treating high-strength wastewater in the Australian red meat processing industry. Appl. Sci. 2020, 10, 7947. [Google Scholar] [CrossRef]
- APMC. Wastewater Management in the Australian Red Meat Processing Industry; The Australian Meat Processor: Sydney, NSW, Australia, 2017. [Google Scholar]
- McCabe, B.K.; Harris, P.; Baillie, C.; Pittaway, P.; Yusaf, T. Assessing a new approach to covered anaerobic pond design in the treatment of abattoir wastewater. Aust. J. Multi-Discip. Eng. 2013, 10, 81–93. [Google Scholar] [CrossRef]
- McCabe, B.K.; Harris, P.; Antille, D.L.; Schmidt, T.; Lee, S.; Hill, A.; Baillie, C. Toward profitable and sustainable bioresource management in the Australian red meat processing industry: A critical review and illustrative case study. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2415–2439. [Google Scholar] [CrossRef]
- Shende, A.D.; Pophali, G.R. Anaerobic treatment of slaughterhouse wastewater: A review. Environ. Sci. Pollut. Res. 2021, 28, 35–55. [Google Scholar] [CrossRef]
- Johns, M. Developments in Waste Treatment in the Meat Processing Industry: A Review of Literature 1979–1993; Meat Research Corporation: Sydney, NSW, Australia, 1993. [Google Scholar]
- White, T.; Johns, M.; Butler, B. Methane Recovery and Use at a Meat Processing Facility—King Island; Rural Industries Research and Development Corporation Australian Government: Barton, Australia, 2013.
- UNSW, C. Treatment of Abattoir Wastewater Using a Covered Anaerobic Lagoon; UNSW CRC for Waste Management & Pollution Control: Sydney, Australia, 1998. [Google Scholar]
- Filer, J.; Ding, H.H.; Chang, S. Biochemical Methane Potential (BMP) Assay Method for Anaerobic Digestion Research. Water 2019, 11, 921. [Google Scholar] [CrossRef]
- Holliger, C.; Alves, M.; Andrade, D.; Angelidaki, I.; Astals, S.; Baier, U.; Bougrier, C.; Buffière, P.; Carballa, M.; De Wilde, V.; et al. Towards a standardization of biomethane potential tests. Water Sci. Technol. 2016, 74, 2515–2522. [Google Scholar] [CrossRef] [PubMed]
- Angelidaki, I.; Alves, M.; Bolzonella, D.; Borzacconi, L.; Campos, J.L.; Guwy, A.J.; Kalyuzhnyi, S.; Jenicek, P.; Lier, J.B.V. Defining the biomethane potential (BMP) of solid organic wastes and energy crops: A proposed protocol for batch assays. Water Sci. Technol. 2009, 59, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Pererva, Y.; Miller, C.D.; Sims, R.C. Existing empirical kinetic models in biochemical methane potential (BMP) testing, their selection and numerical solution. Water 2020, 12, 1831. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- Latif, M.; Mehta, C.; Batstone, D. Influence of low pH on continuous anaerobic digestion of waste activated sludge. Water Res. 2017, 113, 42–49. [Google Scholar] [CrossRef]
- Lachat, A.G. QuikChem® Method 10-107-06-2-A. Determination of Ammonia by Flow Injection Analysis. 2015. Available online: https://support.hach.com/ci/okcsFattach/get/1028334_4 (accessed on 1 June 2022).
- Sarojam, P. Analysis of Wastewater for Metals using ICP-OES. Perkin Elmer Instrum. 2010, 11. Available online: https://www.perkinelmer.com.cn/lab-solutions/resources/docs/APP_MetalsinWastewater.pdf (accessed on 1 June 2019).
- Beckett, N.M.; Cresswell, S.L.; Grice, D.I.; Carter, J.F. Isotopic profiling of seized benzylpiperazine and trifluoromethylphenylpiperazine tablets using δ13C and δ15N stable isotopes. Sci. Justice 2015, 55, 51–56. [Google Scholar] [CrossRef]
- Paulose, P.; Kaparaju, P. Anaerobic mono-digestion of sugarcane trash and bagasse with and without pretreatment. Ind. Crops Prod. 2021, 170, 113712. [Google Scholar] [CrossRef]
- Valero, D.; Montes, J.A.; Rico, J.L.; Rico, C. Influence of headspace pressure on methane production in Biochemical Methane Potential (BMP) tests. Waste Manag. 2016, 48, 193–198. [Google Scholar] [CrossRef]
- Astals, S.; Esteban-Gutiérrez, M.; Fernández-Arévalo, T.; Aymerich, E.; García-Heras, J.; Mata-Alvarez, J. Anaerobic digestion of seven different sewage sludges: A biodegradability and modelling study. Water Res. 2013, 47, 6033–6043. [Google Scholar] [CrossRef]
- Odirile, P.T.; Marumoloa, P.M.; Manali, A.; Gikas, P. Anaerobic Digestion for Biogas Production from Municipal Sewage Sludge: A Comparative Study between Fine Mesh Sieved Primary Sludge and Sedimented Primary Sludge. Water 2021, 13, 3532. [Google Scholar] [CrossRef]
- Auterská, P.; Novák, L. Successful solution for high nitrogen content wastewater treatment from rendering plants. Water Sci. Technol. 2006, 54, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Brennan, B.; Gunes, B.; Jacobs, M.R.; Lawler, J.; Regan, F. Potential viable products identified from characterisation of agricultural slaughterhouse rendering wastewater. Water 2021, 13, 352. [Google Scholar] [CrossRef]
- Bustillo-Lecompte, C.F.; Mehrvar, M. Slaughterhouse wastewater characteristics, treatment, and management in the meat processing industry: A review on trends and advances. J. Environ. Manag. 2015, 161, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.A.; Khodadoost, F. Effects of detergents on natural ecosystems and wastewater treatment processes: A review. Environ. Sci. Pollut. Res. 2019, 26, 26439–26448. [Google Scholar] [CrossRef] [PubMed]
- Mensah, K.A.; Forster, C.F. An examination of the effects of detergents on anaerobic digestion. Bioresour. Technol. 2003, 90, 133–138. [Google Scholar] [CrossRef]
- Bayr, S.; Ojanperä, M.; Kaparaju, P.; Rintala, J. Long-term thermophilic mono-digestion of rendering wastes and co-digestion with potato pulp. Waste Manag. 2014, 34, 1853–1859. [Google Scholar] [CrossRef]
- Pitk, P.; Palatsi, J.; Kaparaju, P.; Fernández, B.; Vilu, R. Mesophilic co-digestion of dairy manure and lipid rich solid slaughterhouse wastes: Process efficiency, limitations and floating granules formation. Bioresour. Technol. 2014, 166, 168–177. [Google Scholar] [CrossRef]
- Bayr, S.; Rantanen, M.; Kaparaju, P.; Rintala, J. Mesophilic and thermophilic anaerobic co-digestion of rendering plant and slaughterhouse wastes. Bioresour. Technol. 2012, 104, 28–36. [Google Scholar] [CrossRef]
- Gutu, L.; Basitere, M.; Harding, T.; Ikumi, D.; Njoya, M.; Gaszynski, C. Multi-Integrated Systems for Treatment of Abattoir Wastewater: A Review. Water 2021, 13, 2462. [Google Scholar] [CrossRef]
- Kafle, G.K.; Kim, S.H. Anaerobic treatment of apple waste with swine manure for biogas production: Batch and continuous operation. Appl. Energy 2013, 103, 61–72. [Google Scholar] [CrossRef]
- Kafle, G.K.; Kim, S.H.; Sung, K.I. Ensiling of fish industry waste for biogas production: A lab scale evaluation of biochemical methane potential (BMP) and kinetics. Bioresour. Technol. 2013, 127, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.H.; Mattioli, A.; Gil, A.; Frison, N.; Battista, F.; Bolzonella, D. Evaluation of the methane potential of different agricultural and food processing substrates for improved biogas production in rural areas. Renew. Sustain. Energy Rev. 2019, 112, 1–10. [Google Scholar] [CrossRef]

| Parameter | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | |
| TS (%) a | 0.13 ± 0.0 | 0.28 ± 0.0 | 0.61 ± 0.1 | 0.44 ± 0.1 | 0.60 ± 0.1 | 1.82 ± 0.1 | 0.76 ± 0.1 | 0.66 ± 0.1 | 0.81 ± 0.2 | 1.02 ± 0.1 | 0.67 ± 0.1 | 0.75 ± 0.2 | 0.57 ± 0.0 | 0.54 ± 0.1 |
| VS (%) a | 0.11 ± 0.0 | 0.21 ± 0.0 | 0.54 ± 0.1 | 0.34 ± 0.0 | 0.44 ± 0.1 | 1.44 ± 0.1 | 0.66 ± 0.1 | 0.56 ± 0.0 | 0.60 ± 0.2 | 0.72 ± 0.1 | 0.61 ± 0.1 | 0.67 ± 0.1 | 0.49 ± 0.1 | 0.45 ± 0.0 |
| VS/TS | 0.80 | 0.74 | 0.88 | 0.78 | 0.73 | 0.79 | 0.86 | 0.86 | 0.74 | 0.71 | 0.91 | 0.90 | 0.87 | 0.83 |
| COD (mg/L) a | 3747.0 ± 1.0 | 3966.5 ± 1.2 | 7586.6 ± 0.5 | 6873.3 ± 0.6 | 7605.3 ± 1.1 | 9047.9 ± 1.2 | 8153.3 ± 1.1 | 10,469.1 ± 1.0 | 9291.0 ± 0.9 | 7663.5 ± 0.9 | 8029.5 ± 1.1 | 8844.2 ± 0.9 | 7037.5 ± 1.0 | 6722.5 ± 0.8 |
| NH4-N (mg/L) a | 52.2 ± 0.8 | 42.9 ± 0.9 | 45.4 ± 0.7 | 71.3 ± 0.9 | 63.3 ± 0.7 | 66.2 ± 0.3 | 61.2 ± 0.9 | 58.8 ± 0.8 | 58.2 ± 0.8 | 51.2 ± 0.2 | 65.9 ± 0.5 | 53.9 ± 0.7 | 59.9 ± 0.6 | 56.9 ± 0.6 |
| PO4-P (mg/L) a | 52.2 ± 1.0 | 60.4 ± 1.2 | 33.1 ± 1.2 | 76.3 ± 2.1 | 60.2 ± 1.5 | 50.3 ± 1.7 | 63.2 ± 1.4 | 40.3 ± 1.5 | 33.5 ± 1.0 | 50.9 ± 0.9 | 60.2 ± 1.2 | 54.0 ± 1.0 | 51.4 ± 1.6 | 52.2 ± 2.3 |
| TKP (mg/kg) a | 20.4 ± 2.5 | 25.7 ± 1.9 | 25.3 ± 2.2 | 20.4 ± 2.3 | 21.9 ± 2.0 | 20.6 ± 2.6 | 25.4 ± 2.2 | 31.0 ± 2.4 | 25.3 ± 2.1 | 30.4 ± 2.3 | 33.5 ± 2.4 | 21.7 ± 2.3 | 33.2 ± 2.2 | 32.2 ± 2.2 |
| TKN (mg/kg) a | 112.7 ± 1.9 | 265.7 ± 2.2 | 151.0 ± 2.0 | 60.9 ± 2.1 | 70.6 ± 2.3 | 90.5 ± 2.5 | 151.0 ± 2.3 | 60.9 ± 2.2 | 70.6 ± 2.1 | 40.5 ± 2.2 | 43.6 ± 2.2 | 98.6 ± 2.1 | 80.7 ± 2.1 | 98.7 ± 2.2 |
| TVFA (mg/L) a | 149.1 ± 3.3 | 189.5 ± 2.6 | 288.8 ± 2.5 | 177.8 ± 2.1 | 198.1 ± 2.3 | 267.5 ± 2.4 | 217.6 ± 2.2 | 384.7 ± 2.0 | 455.4 ± 2.3 | 212.7 ± 2.3 | 227.0 ± 2.3 | 280.8 ± 2.4 | 241.4 ± 2.6 | 247.7 ± 1.9 |
| Acetic acid (mg/L) | 97.1 | 111.7 | 129.0 | 103.7 | 117.0 | 212.3 | 179.3 | 217.4 | 418.3 | 163.5 | 121.1 | 146.2 | 139.6 | 243.8 |
| Propionic acid (mg/L) | 30.5 | 77.6 | 94.9 | 50.1 | 60.1 | 28.6 | 30.4 | 141.6 | 11.1 | 12.7 | 91.2 | 95.6 | 93.4 | 0.0 |
| iso-Butyric acid (mg/L) | 12.8 | 0.0 | 13.2 | 6.9 | 0.8 | 16.2 | 0.0 | 3.4 | 8.8 | 12.2 | 0.2 | 4.6 | 1.1 | 3.9 |
| Butyric acid (mg/L) | 0.0 | 0.0 | 15.7 | 10.0 | 11.9 | 7.2 | 0.0 | 9.4 | 8.1 | 12.3 | 6.5 | 12.6 | 1.8 | 0.0 |
| iso-Valeric acid (mg/L) | 5.9 | 0.1 | 17.4 | 3.8 | 8.4 | 3.2 | 3.6 | 13.1 | 3.6 | 5.6 | 5.1 | 8.2 | 0.7 | 0.0 |
| Valeric acid (mg/L) | 2.0 | 0.0 | 7.7 | 0.9 | 0.0 | 0.0 | 4.1 | 0.0 | 4.1 | 1.9 | 1.4 | 5.6 | 0.3 | 0.0 |
| 4-Methyl valeric acid (mg/L) | 0.9 | 0.0 | 5.5 | 1.2 | 0.0 | 0.0 | 0.1 | 0.0 | 0.1 | 0.8 | 1.8 | 3.9 | 0.2 | 0.0 |
| Hexanoic acid (mg/L) | 0.0 | 0.0 | 5.0 | 1.1 | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 | 3.7 | 0.0 | 4.2 | 4.5 | 0.0 |
| Parameter | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | |
| COD (mg/L) | 1435.43 | 1203.75 | 2231.82 | 2560.52 | 2166.60 | 1966.83 | 2203.50 | 2011.57 | 2162.89 | 2317.65 | 2318.98 | 2544.03 | 2105.33 | 2011.04 |
| % COD degradation | 61.69 | 69.65 | 70.58 | 62.75 | 71.51 | 78.26 | 72.97 | 80.79 | 76.72 | 69.76 | 71.12 | 71.24 | 70.08 | 70.08 |
| TKP (mg/kg) | 0.23 | 0.24 | 0.22 | 0.23 | 0.22 | 0.21 | 0.21 | 0.22 | 0.22 | 0.25 | 0.24 | 0.25 | 0.26 | 0.23 |
| TKN (mg/kg) | 0.37 | 0.36 | 0.33 | 0.36 | 0.34 | 0.34 | 0.14 | 0.38 | 0.34 | 0.40 | 0.21 | 0.37 | 0.33 | 0.37 |
| TVFA (mg/L) | 15.66 | 16.18 | 16.52 | 17.13 | 17.31 | 18.31 | 16.98 | 18.74 | 17.43 | 18.14 | 17.61 | 17.56 | 16.99 | 17.98 |
| Acetic acid (mg/L) | 10.43 | 11.47 | 11.84 | 12.27 | 12.46 | 12.72 | 11.91 | 13.16 | 12.59 | 12.45 | 12.22 | 12.30 | 11.72 | 12.44 |
| Propionic acid (mg/L) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| iso-Butyric acid (mg/L) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Butyric acid (mg/L) | 1.78 | 1.87 | 1.72 | 1.84 | 1.81 | 1.91 | 1.87 | 1.83 | 1.69 | 1.93 | 1.96 | 1.68 | 1.72 | 1.88 |
| iso-Valeric acid (mg/L) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Valeric acid (mg/L) | 2.12 | 2.10 | 1.97 | 2.04 | 2.11 | 1.92 | 1.96 | 2.16 | 1.89 | 2.00 | 2.00 | 1.97 | 1.94 | 1.95 |
| 4-Methyl valeric acid (mg/L) | 0.33 | 0.73 | 0.99 | 0.99 | 0.94 | 1.38 | 1.24 | 0.97 | 1.27 | 1.76 | 1.43 | 1.61 | 1.30 | 1.70 |
| Hexanoic acid (mg/L) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.38 | 0.00 | 0.62 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Sample | Experimental Methane Yields (L/kgCODadded) | First-Order Kinetic Model | Gompertz Equation Model | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bo (L/kgCODadded) | Difference (%) | t (d−1) | khyd (d−1) | rRMSE (%) | R2 | Bo (L/kgCODadded) | Difference (%) | λ (d) | T90 (d) | Tef (d) | RMax (L/kgCODadded d−1) | rRMSE (%) | R2 | |||
| Day 1 | S1 | 83.7 ± 1.39 | 91.90 | 9.3 | 2.51 | 0.09 | 12.07 | 0.971 | 86.81 | 3.6 | 2.06 | 18.53 | 16.45 | 5.25 | 6.69 | 0.991 |
| S2 | 150.8 ± 1.41 | 156.80 | 3.9 | 2.59 | 0.12 | 7.38 | 0.988 | 150.72 | 0.1 | 2.00 | 18.15 | 16.15 | 12.24 | 6.83 | 0.989 | |
| S3 | 166.5 ± 1.39 | 168.50 | 1.2 | 3.74 | 0.17 | 9.37 | 0.982 | 163.00 | 2.1 | 3.15 | 17.67 | 14.52 | 17.69 | 9.50 | 0.981 | |
| Day 2 | S1 | 123.2 ± 1.71 | 119.19 | 3.3 | 3.82 | 0.21 | 9.22 | 0.981 | 115.68 | 6.3 | 3.64 | 15.89 | 12.25 | 16.99 | 8.90 | 0.983 |
| S2 | 164.0 ± 1.70 | 166.90 | 1.8 | 2.70 | 0.13 | 3.97 | 0.996 | 160.13 | 2.4 | 2.06 | 18.92 | 16.86 | 13.05 | 5.72 | 0.993 | |
| S3 | 252.6 ± 1.73 | 304.7 | 18.7 | 3.84 | 0.05 | 17.44 | 0.964 | 268.99 | 6.3 | 3.95 | 21.98 | 18.03 | 11.98 | 15.51 | 0.972 | |
| Day 3 | S1 | 192.6 ± 1.27 | 215.6 | 11.3 | 3.33 | 0.08 | 13.53 | 0.972 | 201.88 | 4.7 | 2.85 | 20.16 | 17.31 | 10.73 | 10.48 | 0.983 |
| S2 | 270.2 ± 1.25 | 310.5 | 13.9 | 3.30 | 0.07 | 17.55 | 0.956 | 285.85 | 5.6 | 3.09 | 19.75 | 16.66 | 14.35 | 14.00 | 0.972 | |
| S3 | 237.0 ± 1.00 | 283.7 | 17.9 | 3.26 | 0.06 | 14.73 | 0.970 | 252.04 | 6.2 | 3.14 | 21.78 | 16.64 | 11.23 | 10.77 | 0.984 | |
| Day 4 | S1 | 174.9 ± 1.17 | 186.7 | 6.5 | 2.75 | 0.09 | 6.42 | 0.992 | 175.45 | 0.3 | 2.31 | 20.49 | 18.18 | 10.84 | 4.00 | 0.997 |
| S2 | 201.9 ± 1.16 | 211.5 | 4.6 | 3.39 | 0.11 | 7.81 | 0.989 | 201.93 | 0.0 | 2.69 | 20.32 | 17.63 | 13.93 | 5.67 | 0.994 | |
| S3 | 199.7 ± 1.10 | 218.4 | 8.9 | 2.58 | 0.08 | 8.22 | 0.988 | 203.51 | 1.9 | 2.02 | 20.93 | 18.91 | 11.06 | 6.58 | 0.992 | |
| Day 5 | S1 | 167.3 ± 2.82 | 172.09 | 2.8 | 3.70 | 0.13 | 8.59 | 0.987 | 166.09 | 0.7 | 3.33 | 17.99 | 14.66 | 14.46 | 4.28 | 0.997 |
| S2 | 178.4 ± 280 | 179.90 | 0.8 | 3.54 | 0.14 | 5.66 | 0.994 | 172.75 | 3.2 | 2.99 | 19.47 | 16.48 | 15.53 | 6.97 | 0.991 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conde, E.; Kaparaju, P. Effect of Temporal Variation in Chemical Composition on Methane Yields of Rendering Plant Wastewater. Energies 2022, 15, 7252. https://doi.org/10.3390/en15197252
Conde E, Kaparaju P. Effect of Temporal Variation in Chemical Composition on Methane Yields of Rendering Plant Wastewater. Energies. 2022; 15(19):7252. https://doi.org/10.3390/en15197252
Chicago/Turabian StyleConde, Erika, and Prasad Kaparaju. 2022. "Effect of Temporal Variation in Chemical Composition on Methane Yields of Rendering Plant Wastewater" Energies 15, no. 19: 7252. https://doi.org/10.3390/en15197252
APA StyleConde, E., & Kaparaju, P. (2022). Effect of Temporal Variation in Chemical Composition on Methane Yields of Rendering Plant Wastewater. Energies, 15(19), 7252. https://doi.org/10.3390/en15197252







