Molecular Mechanisms Underlying Flax (Linum usitatissimum L.) Tolerance to Cadmium: A Case Study of Proteome and Metabolome of Four Different Flax Genotypes
Abstract
1. Introduction
2. Results
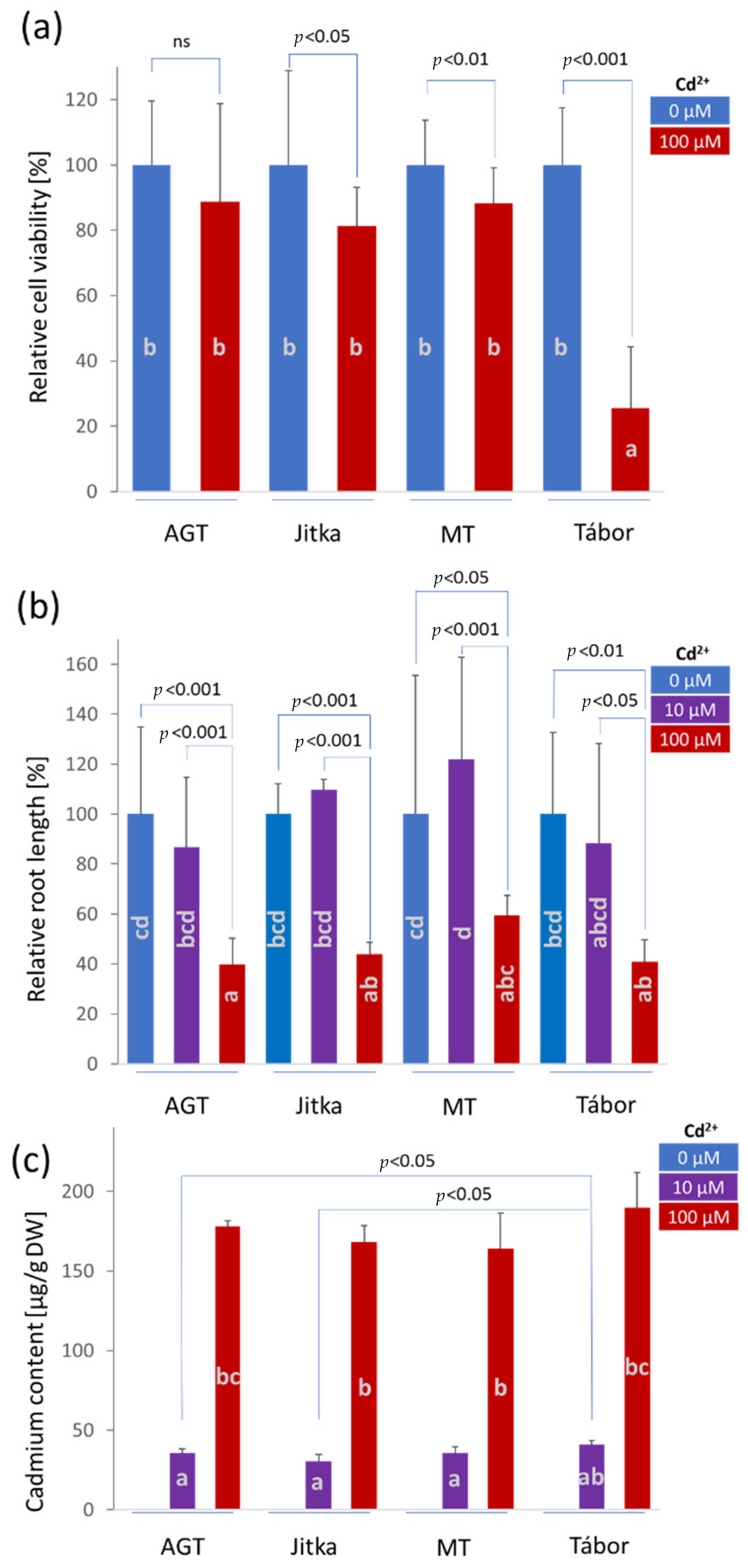
2.1. The Effects of Cadmium on Cell Viability and Root Growth
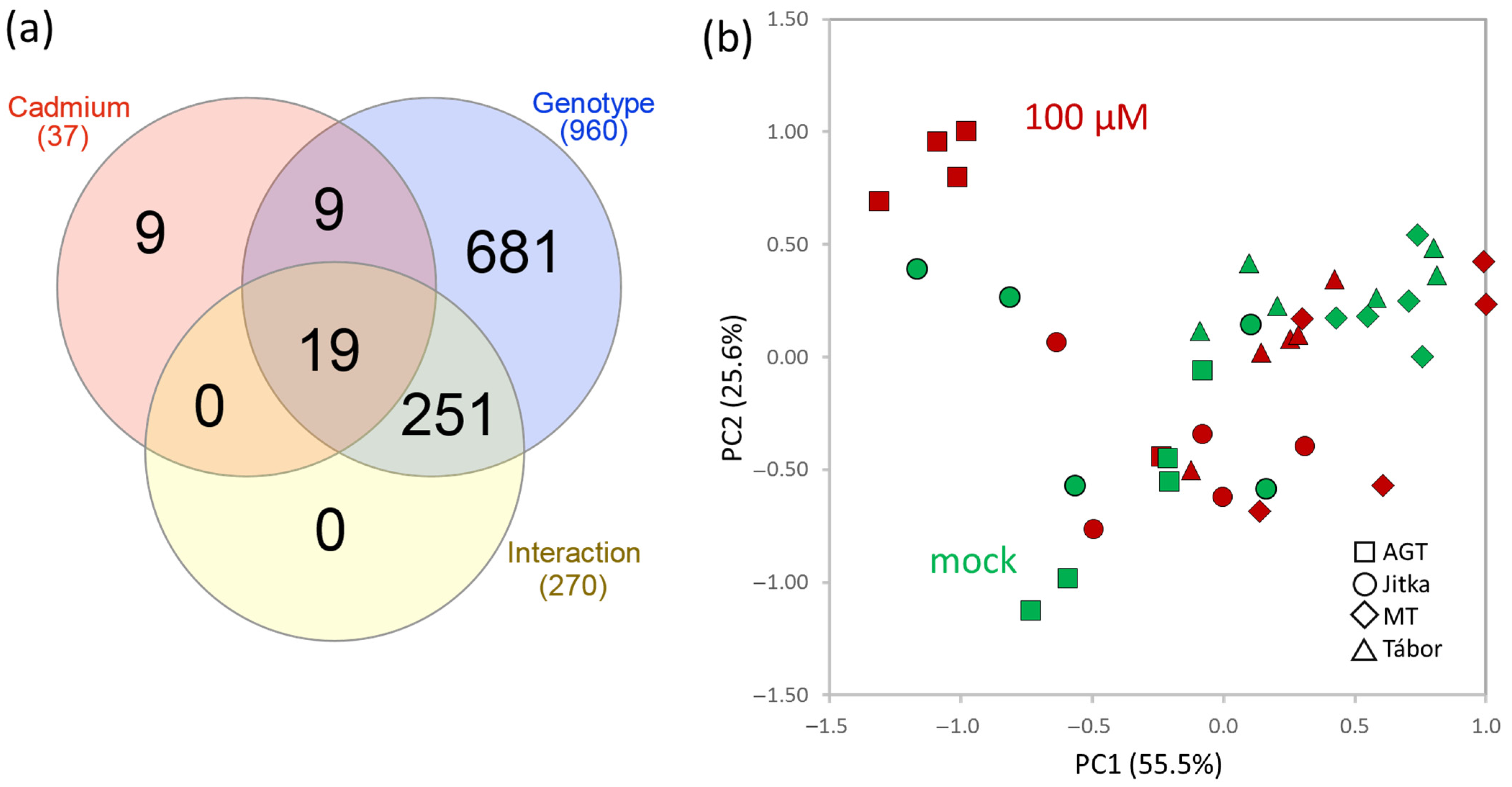
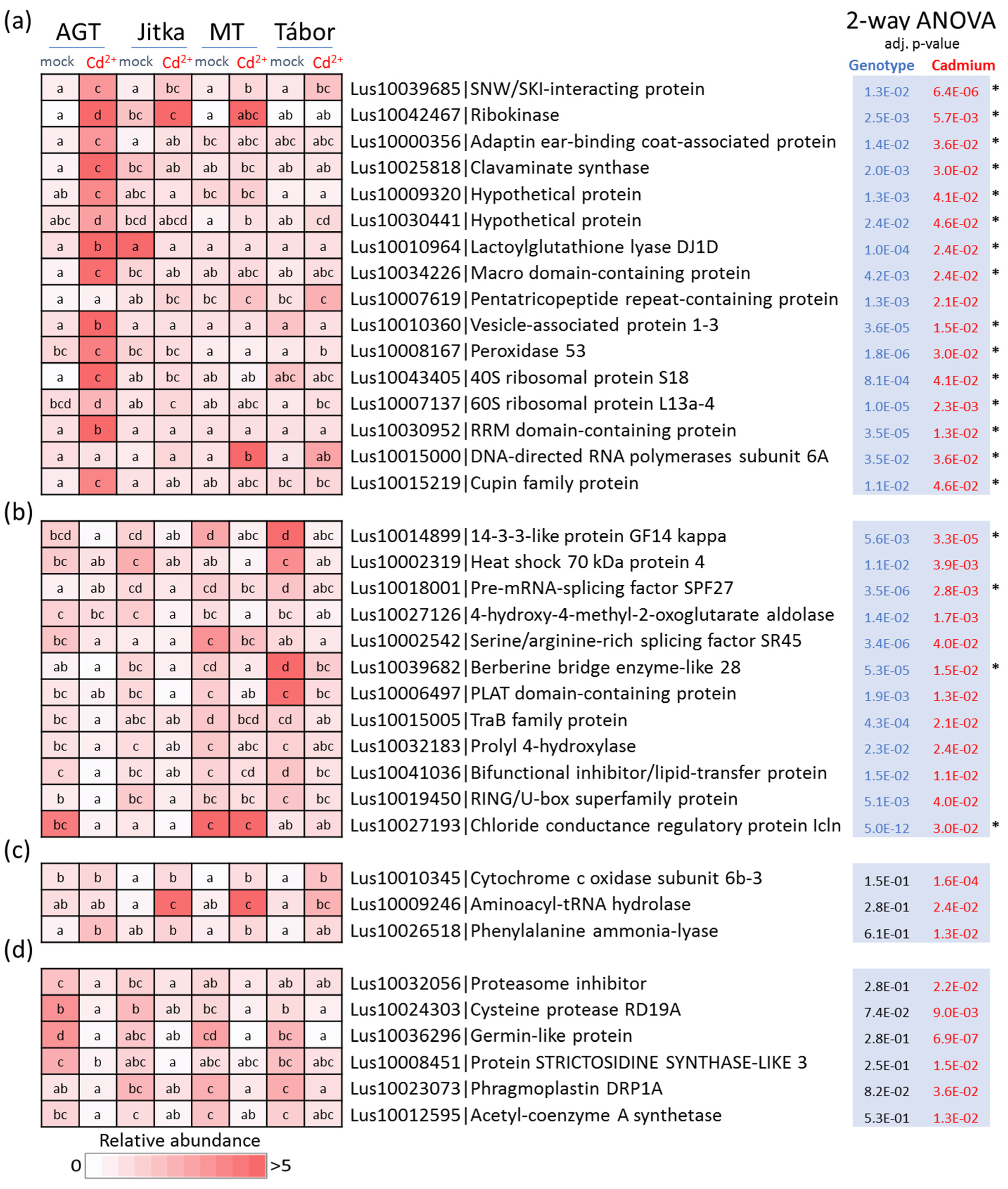
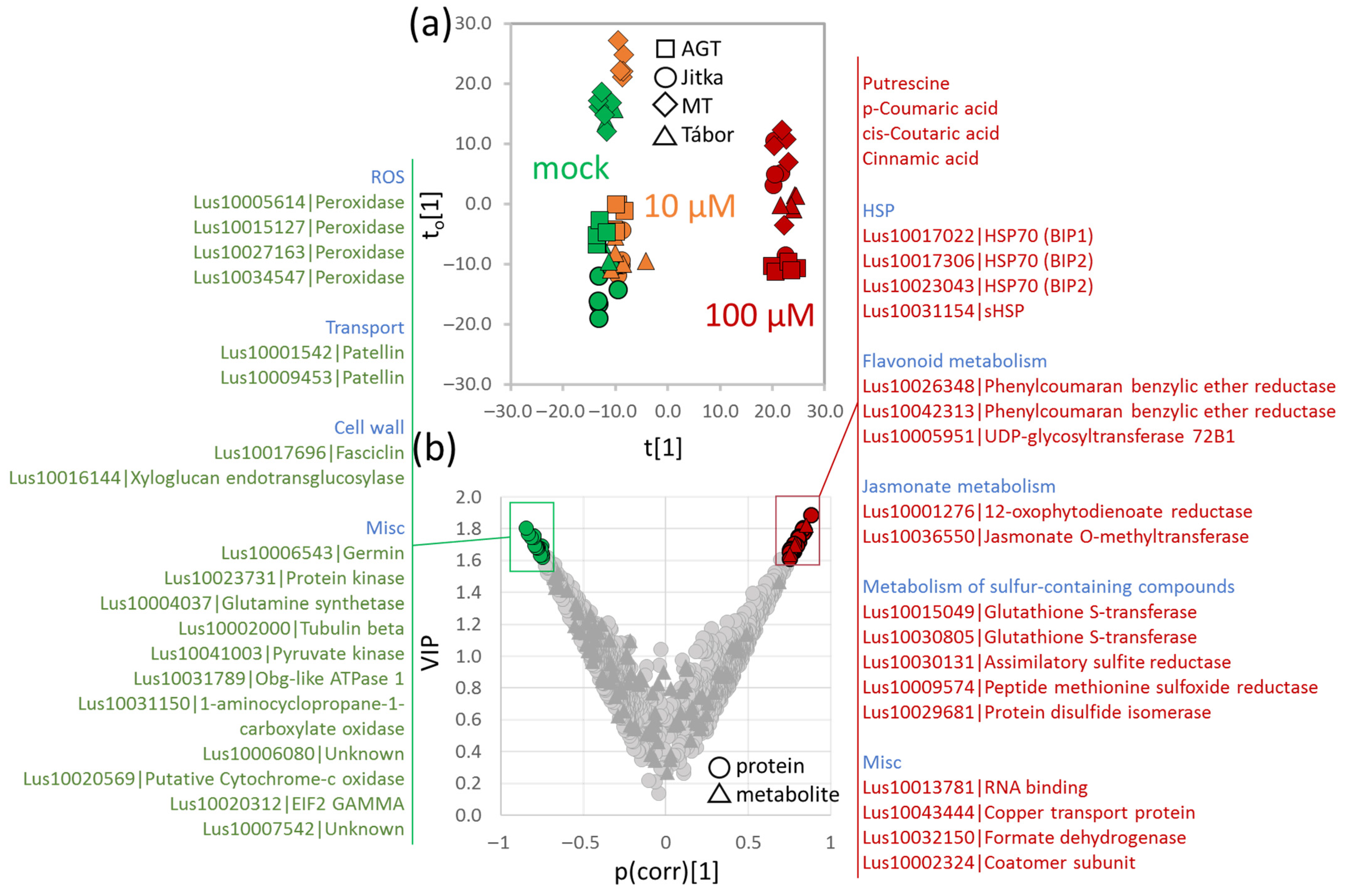
2.2. Proteome Analysis of Flax Suspension Cultures
2.3. Flax Root Proteome in Response to Cadmium
2.4. Comparison of Suspension Culture and Root Tissue Response to Cadmium
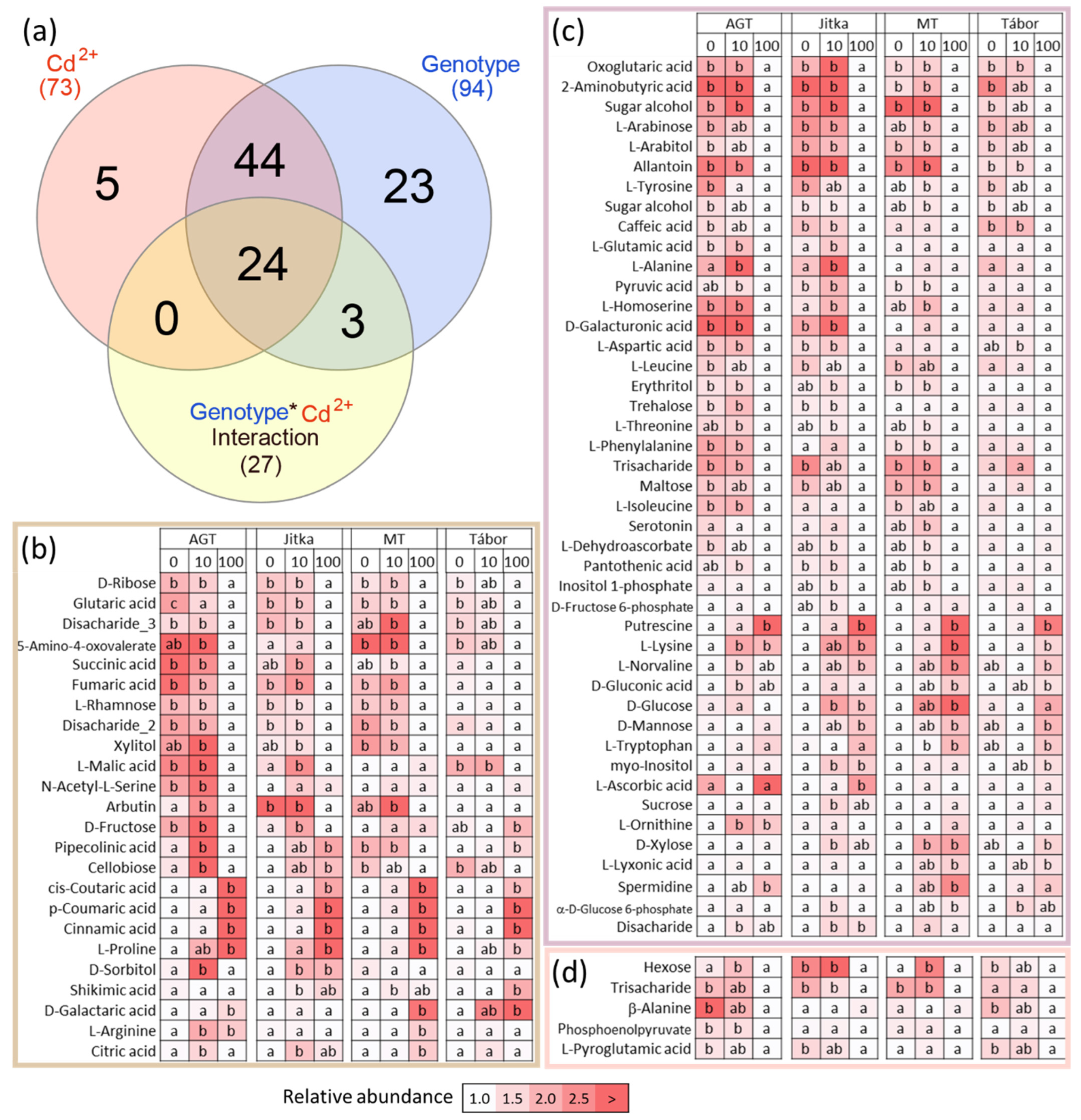
2.5. Metabolome Profiling
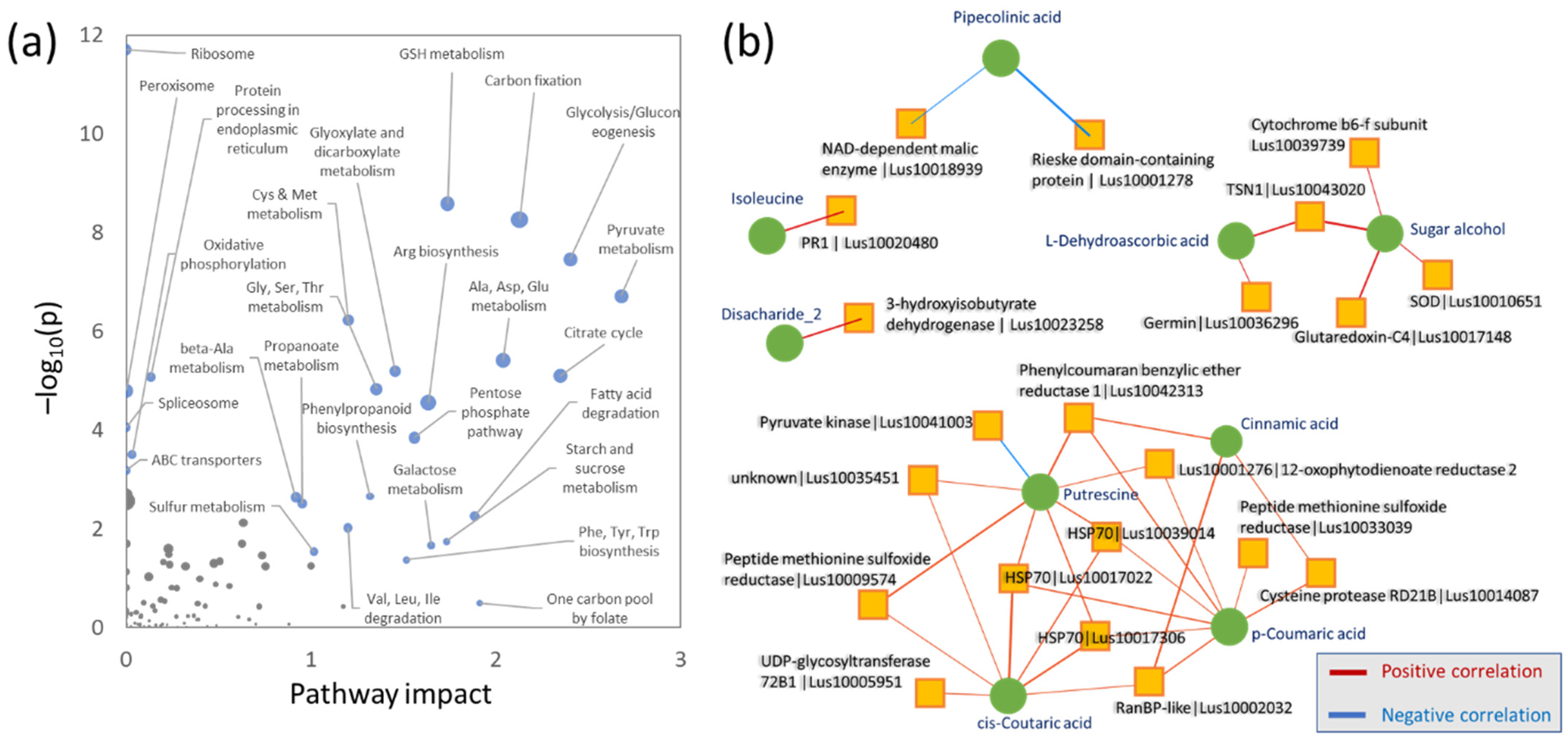
2.6. Integrative Analysis of Proteome and Metabolome
3. Discussion
3.1. Cadmium Response in Four Different Genotypes
3.2. Comparison of Cadmium-Responsive Proteins in Flax and Model Plant Arabidopsis Thaliana
3.3. Cadmium Response in Suspension Cultures Could Correlate with a Higher Cadmium Import in Cultivar Tábor
3.4. Polyamines, Jasmonic Acid, and Cytokinin Promoted Tolerance to Cadmium
3.5. HSP70 Proteins–Integral Components of Cadmium Tolerance?
3.6. Observed Decrease in Germin Proteins Could Indicate Reallocation of Resources to Abiotic Stress Response
3.7. Reactive Oxygen Species and Phenolics in Response to Cadmium
4. Materials and Methods
4.1. Plant Material
4.2. Cadmium Determination
4.3. Proteome Analysis
4.4. Metabolome Analysis
4.5. Statistics
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 2021, 211, 111887. [Google Scholar] [CrossRef] [PubMed]
- Hashem, A.; Abd_Allah, E.F.; Alqarawi, A.A.; Al Huqail, A.A.; Egamberdieva, D.; Wirth, S. Alleviation of cadmium stress in Solanum lycopersicum L. by arbuscular mycorrhizal fungi via induction of acquired systemic tolerance. Saudi J. Biol. Sci. 2016, 23, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Cao, Q.; Zheng, Y.M.; Huang, Y.Z.; Zhu, Y.G. Health risks of heavy metals in contaminated soils and food crops irrigated with wastewater in Beijing, China. Environ. Pollut. 2008, 152, 686–692. [Google Scholar] [CrossRef]
- Zhang, M.-K.; Liu, Z.-Y.; Wang, H. Use of Single Extraction Methods to Predict Bioavailability of Heavy Metals in Polluted Soils to Rice. Commun. Soil Sci. Plant Anal. 2010, 41, 820–831. [Google Scholar] [CrossRef]
- Salt, D.E.; Prince, R.C.; Pickering, I.J.; Raskin, I. Mechanisms of Cadmium Mobility and Accumulation in Indian Mustard. Plant Physiol. 1995, 109, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Gallego, S.M.; Pena, L.B.; Barcia, R.A.; Azpilicueta, C.E.; Iannone, M.F.; Rosales, E.P.; Zawoznik, M.S.; Groppa, M.D.; Benavides, M.P. Unravelling cadmium toxicity and tolerance in plants: Insight into regulatory mechanisms. Environ. Exp. Bot. 2012, 83, 33–46. [Google Scholar] [CrossRef]
- Schaefer, H.R.; Dennis, S.; Fitzpatrick, S. Cadmium: Mitigation strategies to reduce dietary exposure. J. Food Sci. 2020, 85, 260–267. [Google Scholar] [CrossRef]
- Nazar, R.; Iqbal, N.; Masood, A.; Khan, M.I.R.; Syeed, S.; Khan, N.A. Cadmium Toxicity in Plants and Role of Mineral Nutrients in Its Alleviation. Am. J. Plant Sci. 2012, 3, 1476–1489. [Google Scholar] [CrossRef]
- Ismael, M.A.; Elyamine, A.M.; Moussa, M.G.; Cai, M.; Zhao, X.; Hu, C. Cadmium in plants: Uptake, toxicity, and its interactions with selenium fertilizers. Metallomics 2019, 11, 255–277. [Google Scholar] [CrossRef]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef]
- Abbas, T.; Rizwan, M.; Ali, S.; Adrees, M.; Zia-ur-Rehman, M.; Qayyum, M.F.; Ok, Y.S.; Murtaza, G. Effect of biochar on alleviation of cadmium toxicity in wheat (Triticum aestivum L.) grown on Cd-contaminated saline soil. Environ. Sci. Pollut. Res. 2018, 25, 25668–25680. [Google Scholar] [CrossRef] [PubMed]
- Gratão, P.L.; Monteiro, C.C.; Rossi, M.L.; Martinelli, A.P.; Peres, L.E.P.; Medici, L.O.; Lea, P.J.; Azevedo, R.A. Differential ultrastructural changes in tomato hormonal mutants exposed to cadmium. Environ. Exp. Bot. 2009, 67, 387–394. [Google Scholar] [CrossRef]
- Xue, Z.-C.; Gao, H.-Y.; Zhang, L.-T. Effects of cadmium on growth, photosynthetic rate and chlorophyll content in leaves of soybean seedlings. Biol. Plant. 2013, 57, 587–590. [Google Scholar] [CrossRef]
- Jhala, A.J.; Hall, L.M. Flax (Linum usitatissimum L.): Current uses and future applications. Aust. J. Basic Appl. Sci. 2010, 4, 4304–4312. [Google Scholar]
- Fucassi, F.; Heikal, A.; Mikhalovska, L.I.; Standen, G.; Allan, I.U.; Mikhalovsky, S.V.; Cragg, P.J. Metal chelation by a plant lignan, secoisolariciresinol diglucoside. J. Incl. Phenom. Macrocycl. Chem. 2014, 80, 345–351. [Google Scholar] [CrossRef]
- Angelova, V.; Ivanova, R.; Delibaltova, V.; Ivanov, K. Bio-accumulation and distribution of heavy metals in fibre crops (flax, cotton and hemp). Ind. Crops Prod. 2004, 19, 197–205. [Google Scholar] [CrossRef]
- Smykalova, I.; Vrbova, M.; Tejklova, E.; Vetrovcova, M.; Griga, M. Large scale screening of heavy metal tolerance in flax/linseed (Linum usitatissimum L.) tested in vitro. Ind. Crops Prod. 2010, 32, 527–533. [Google Scholar] [CrossRef]
- Bjelková, M.; Genčurová, V.; Griga, M. Accumulation of cadmium by flax and linseed cultivars in field-simulated conditions: A potential for phytoremediation of Cd-contaminated soils. Ind. Crops Prod. 2011, 33, 761–774. [Google Scholar] [CrossRef]
- Ueno, D.; Milner, M.J.; Yamaji, N.; Yokosho, K.; Koyama, E.; Clemencia Zambrano, M.; Kaskie, M.; Ebbs, S.; Kochian, L.V.; Ma, J.F. Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011, 66, 852–862. [Google Scholar] [CrossRef]
- Hradilová, J.; Řehulka, P.; Řehulková, H.; Vrbová, M.; Griga, M.; Brzobohatý, B. Comparative analysis of proteomic changes in contrasting flax cultivars upon cadmium exposure. Electrophoresis 2010, 31, 421–431. [Google Scholar] [CrossRef]
- Höckner, M.; Piechnik, C.A.; Fiechtner, B.; Weinberger, B.; Tomanek, L. Cadmium-Related Effects on Cellular Immunity Comprises Altered Metabolism in Earthworm Coelomocytes. Int. J. Mol. Sci. 2020, 21, 599. [Google Scholar] [CrossRef] [PubMed]
- Daniel, B.; Wallner, S.; Steiner, B.; Oberdorfer, G.; Kumar, P.; van der Graaff, E.; Roitsch, T.; Sensen, C.W.; Gruber, K.; Macheroux, P. Structure of a Berberine Bridge Enzyme-Like Enzyme with an Active Site Specific to the Plant Family Brassicaceae. PLoS ONE 2016, 11, e0156892. [Google Scholar] [CrossRef] [PubMed]
- Hyun, T.K.; van der Graaff, E.; Albacete, A.; Eom, S.H.; Großkinsky, D.K.; Böhm, H.; Janschek, U.; Rim, Y.; Ali, W.W.; Kim, S.Y.; et al. The Arabidopsis PLAT Domain Protein1 Is Critically Involved in Abiotic Stress Tolerance. PLoS ONE 2014, 9, e112946. [Google Scholar] [CrossRef] [PubMed]
- Krapivinsky, G.B.; Ackerman, M.J.; Gordon, E.A.; Krapivinsky, L.D.; Clapham, D.E. Molecular characterization of a swelling-induced chloride conductance regulatory protein, plCIn. Cell 1994, 76, 439–448. [Google Scholar] [CrossRef]
- Collings, D.A.; Gebbie, L.K.; Howles, P.A.; Hurley, U.A.; Birch, R.J.; Cork, A.H.; Hocart, C.H.; Arioli, T.; Williamson, R.E. Arabidopsis dynamin-like protein DRP1A: A null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. J. Exp. Bot. 2008, 59, 361–376. [Google Scholar] [CrossRef]
- Gutierrez-Beltran, E.; Moschou, P.N.; Smertenko, A.P.; Bozhkov, P.V. Tudor Staphylococcal Nuclease Links Formation of Stress Granules and Processing Bodies with mRNA Catabolism in Arabidopsis. Plant Cell 2015, 27, 926–943. [Google Scholar] [CrossRef]
- Dutilleul, C.; Jourdain, A.; Bourguignon, J.; Hugouvieux, V. The Arabidopsis Putative Selenium-Binding Protein Family: Expression Study and Characterization of SBP1 as a Potential New Player in Cadmium Detoxification Processes. Plant Physiol. 2008, 147, 239–251. [Google Scholar] [CrossRef]
- Amaral dos Reis, R.; Hendrix, S.; Mourato, M.P.; Louro Martins, L.; Vangronsveld, J.; Cuypers, A. Efficient regulation of copper homeostasis underlies accession-specific sensitivities to excess copper and cadmium in roots of Arabidopsis thaliana. J. Plant Physiol. 2021, 261, 153434. [Google Scholar] [CrossRef]
- Heo, D.-H.; Baek, I.-J.; Kang, H.-J.; Kim, J.-H.; Chang, M.; Jeong, M.-Y.; Kim, T.-H.; Choi, I.-D.; Yun, C.-W. Cadmium regulates copper homoeostasis by inhibiting the activity of Mac1, a transcriptional activator of the copper regulon, in Saccharomyces cerevisiae. Biochem. J. 2010, 431, 257–265. [Google Scholar] [CrossRef][Green Version]
- Griga, M.; Bjelkova, M.; Tejklová, E. Potential of flax (Linum usitatissimum L.) for heavy metal phytoextraction and industrial processing of contaminated biomass—A review. In Risk Assessment and Sustainable Land Management Using Plants in Trace Elements-Contaminated Soils; Mench, M., Mocquot, B., Eds.; Institut National de la Recherche Agronomique: Villenave d’Ornon, France, 2003; pp. 173–179. [Google Scholar]
- Soudek, P.; Katrušáková, A.; Sedláček, L.; Petrová, Š.; Kočí, V.; Maršík, P.; Griga, M.; Vaněk, T. Effect of Heavy Metals on Inhibition of Root Elongation in 23 Cultivars of Flax (Linum usitatissimum L.). Arch. Environ. Contam. Toxicol. 2010, 59, 194–203. [Google Scholar] [CrossRef]
- Son, J.-H.; Park, K.-C.; Lee, S.-I.; Kim, H.-H.; Kim, J.-H.; Kim, S.-H.; Kim, N.-S. Isolation of cold-responsive genes from garlic, Allium sativum. Genes Genom. 2012, 34, 93–101. [Google Scholar] [CrossRef]
- Bernsdorff, F.; Döring, A.-C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic Acid Orchestrates Plant Systemic Acquired Resistance and Defense Priming via Salicylic Acid-Dependent and -Independent Pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef] [PubMed]
- Seifikalhor, M.; Aliniaeifard, S.; Bernard, F.; Seif, M.; Latifi, M.; Hassani, B.; Didaran, F.; Bosacchi, M.; Rezadoost, H.; Li, T. γ-Aminobutyric acid confers cadmium tolerance in maize plants by concerted regulation of polyamine metabolism and antioxidant defense systems. Sci. Rep. 2020, 10, 3356. [Google Scholar] [CrossRef]
- Groppa, M.D.; Tomaro, M.L.; Benavides, M.P. Polyamines as protectors against cadmium or copper-induced oxidative damage in sunflower leaf discs. Plant Sci. 2001, 161, 481–488. [Google Scholar] [CrossRef]
- Groppa, M.D.; Tomaro, M.L.; Benavides, M.P. Polyamines and heavy metal stress: The antioxidant behavior of spermine in cadmium- and copper-treated wheat leaves. BioMetals 2007, 20, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.T.; Kao, C.H. Cadmium-induced oxidative damage in rice leaves is reduced by polyamines. Plant Soil 2007, 291, 27–37. [Google Scholar] [CrossRef]
- Lei, G.J.; Sun, L.; Sun, Y.; Zhu, X.F.; Li, G.X.; Zheng, S.J. Jasmonic acid alleviates cadmium toxicity in Arabidopsis via suppression of cadmium uptake and translocation. J. Integr. Plant Biol. 2020, 62, 218–227. [Google Scholar] [CrossRef]
- Černý, M.; Novák, J.; Habánová, H.; Cerna, H.; Brzobohatý, B. Role of the proteome in phytohormonal signaling. Biochim. Biophys. Acta-Proteins Proteom. 2016, 1864, 1003–1015. [Google Scholar] [CrossRef]
- Lu, Q.; Chen, S.; Li, Y.; Zheng, F.; He, B.; Gu, M. Exogenous abscisic acid (ABA) promotes cadmium (Cd) accumulation in Sedum alfredii Hance by regulating the expression of Cd stress response genes. Environ. Sci. Pollut. Res. 2020, 27, 8719–8731. [Google Scholar] [CrossRef]
- Chen, K.; Chen, P.; Qiu, X.; Chen, J.; Gao, G.; Wang, X.; Zhu, A.; Yu, C. Regulating role of abscisic acid on cadmium enrichment in ramie (Boehmeria nivea L.). Sci. Rep. 2021, 11, 22045. [Google Scholar] [CrossRef]
- Pavlů, J.; Novák, J.; Koukalová, V.; Luklová, M.; Brzobohatý, B.; Černý, M. Cytokinin at the Crossroads of Abiotic Stress Signalling Pathways. Int. J. Mol. Sci. 2018, 19, 2450. [Google Scholar] [CrossRef] [PubMed]
- Brenner, W.G.; Schmülling, T. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biol. 2012, 12, 112. [Google Scholar] [CrossRef] [PubMed]
- Pavlů, J.; Kerchev, P.; Černý, M.; Novák, J.; Berka, M.; Jobe, T.O.; López-Ramos, J.M.; Saiz-Fernández, I.; Rashotte, A.M.; Kopriva, S.; et al. Cytokinin modulates sulfur and glutathione metabolic network. J. Exp. Bot. 2022. in print. [Google Scholar] [CrossRef] [PubMed]
- Berka, M.; Kopecká, R.; Berková, V.; Brzobohatý, B.; Černý, M. Regulation of heat shock proteins 70 and their role in plant immunity. J. Exp. Bot. 2022, 73, 1894–1909. [Google Scholar] [CrossRef] [PubMed]
- Sriram, M.; Osipiuk, J.; Freeman, B.C.; Morimoto, R.I.; Joachimiak, A. Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 1997, 5, 403–414. [Google Scholar] [CrossRef]
- Begum, N.; Hu, Z.; Cai, Q.; Lou, L. Influence of PGPB Inoculation on HSP70 and HMA3 Gene Expression in Switchgrass under Cadmium Stress. Plants 2019, 8, 504. [Google Scholar] [CrossRef]
- Li, X.; Zheng, H.; Shi, L.; Liu, Z.; He, L.; Gao, J. Stress-seventy subfamily A 4, A member of HSP70, confers yeast cadmium tolerance in the loss of mitochondria pyruvate carrier 1. Plant Signal. Behav. 2020, 15, 1719312. [Google Scholar] [CrossRef]
- Guan, C.; Jin, C.; Ji, J.; Wang, G.; Li, X. LcBiP, a endoplasmic reticulum chaperone binding protein gene from Lycium chinense, confers cadmium tolerance in transgenic tobacco. Biotechnol. Prog. 2015, 31, 358–368. [Google Scholar] [CrossRef]
- Wang, T.; Yuan, Y.; Zou, H.; Yang, J.; Zhao, S.; Ma, Y.; Wang, Y.; Bian, J.; Liu, X.; Gu, J.; et al. The ER stress regulator Bip mediates cadmium-induced autophagy and neuronal senescence. Sci. Rep. 2016, 6, 38091. [Google Scholar] [CrossRef]
- De Benedictis, M.; Gallo, A.; Migoni, D.; Papadia, P.; Roversi, P.; Santino, A. Cadmium treatment induces endoplasmic reticulum stress and unfolded protein response in Arabidopsis thaliana. bioRxiv 2022. [Google Scholar] [CrossRef]
- Yuan, B.; Yang, Y.; Fan, P.; Liu, J.; Xing, H.; Liu, Y.; Feng, D. Genome-Wide Identification and Characterization of Germin and Germin-Like Proteins (GLPs) and Their Response Under Powdery Mildew Stress in Wheat (Triticum aestivum L.). Plant Mol. Biol. Report. 2021, 39, 821–832. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Chang, X.; Sun, M.; Zhang, Y.; Li, W.; Li, Y. Overexpression of germin-like protein GmGLP10 enhances resistance to Sclerotinia sclerotiorum in transgenic tobacco. Biochem. Biophys. Res. Commun. 2018, 497, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Li, X.; Zhu, Y.; Ge, X.; Sun, Y.; Liu, N.; Jia, Y.; Li, F.; Hou, Y. GhABP19, a Novel Germin-Like Protein From Gossypium hirsutum, Plays an Important Role in the Regulation of Resistance to Verticillium and Fusarium Wilt Pathogens. Front. Plant Sci. 2019, 10, 583. [Google Scholar] [CrossRef] [PubMed]
- Cerny, M.; Berka, M.; Dvořák, M.; Milenković, I.; Saiz-Fernández, I.; Brzobohatý, B.; Ďurkovič, J. Defense mechanisms promoting tolerance to aggressive Phytophthora species in hybrid poplar. Front. Plant Sci. 2022, 13, 1018272. [Google Scholar] [CrossRef]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Saifullah; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, S.; Zhao, J.; Wang, F.; Du, Y.; Zou, S.; Li, H.; Wen, D.; Huang, Y. Comparative responses to silicon and selenium in relation to antioxidant enzyme system and the glutathione-ascorbate cycle in flowering Chinese cabbage (Brassica campestris L. ssp. chinensis var. utilis) under cadmium stress. Environ. Exp. Bot. 2017, 133, 1–11. [Google Scholar] [CrossRef]
- Qin, S.; Liu, H.; Nie, Z.; Gao, W.; Li, C.; Lin, Y.; Zhao, P. AsA–GSH Cycle and Antioxidant Enzymes Play Important Roles in Cd Tolerance of Wheat. Bull. Environ. Contam. Toxicol. 2018, 101, 684–690. [Google Scholar] [CrossRef]
- Chen, S.; Lin, R.; Lu, H.; Wang, Q.; Yang, J.; Liu, J.; Yan, C. Effects of phenolic acids on free radical scavenging and heavy metal bioavailability in Kandelia obovata under cadmium and zinc stress. Chemosphere 2020, 249, 126341. [Google Scholar] [CrossRef]
- Vrbová, M.; Kotrba, P.; Horáček, J.; Smýkal, P.; Švábová, L.; Větrovcová, M.; Smýkalová, I.; Griga, M. Enhanced accumulation of cadmium in Linum usitatissimum L. plants due to overproduction of metallothionein α-domain as a fusion to β-glucuronidase protein. Plant Cell Tissue Organ Cult. 2013, 112, 321–330. [Google Scholar] [CrossRef]
- Cvečková, M.; Smýkalová, I.; Větrovcová, M.; Vrbová, M. Testing of Heavy Metals Toxicity with Suspension Cultures of Hemp (Cannabis sativa ssp. sativa L.); Agritec: Šumperk, Czech Republic, 2015; pp. 1–20. [Google Scholar]
- Yaru, B.; Bainok, D.; Day, G. Determination of Cd, Cu, Pb, and Zn in biological tissues using Zeeman graphite furnace AAS after microwave digestion in non-pressurized, semi-closed vessel. At. Spectrosc. 1999, 20, 33–38. [Google Scholar]
- Berka, M.; Greplová, M.; Saiz-Fernández, I.; Novák, J.; Luklová, M.; Zelená, P.; Tomšovský, M.; Brzobohatý, B.; Černý, M. Peptide-based identification of Phytophthora isolates and Phytophthora detection in planta. Int. J. Mol. Sci. 2020, 21, 9463. [Google Scholar] [CrossRef] [PubMed]
- Saiz-Fernández, I.; Milenković, I.; Berka, M.; Černý, M.; Tomšovský, M.; Brzobohatý, B.; Kerchev, P. Integrated Proteomic and Metabolomic Profiling of Phytophthora cinnamomi Attack on Sweet Chestnut (Castanea sativa) Reveals Distinct Molecular Reprogramming Proximal to the Infection Site and Away from It. Int. J. Mol. Sci. 2020, 21, 8525. [Google Scholar] [CrossRef] [PubMed]
- Dufková, H.; Berka, M.; Greplová, M.; Shejbalová, Š.; Hampejsová, R.; Luklová, M.; Domkářová, J.; Novák, J.; Kopačka, V.; Brzobohatý, B.; et al. The Omics Hunt for Novel Molecular Markers of Resistance to Phytophthora infestans. Plants 2022, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hobson, N.; Galindo, L.; Zhu, S.; Shi, D.; McDill, J.; Yang, L.; Hawkins, S.; Neutelings, G.; Datla, R.; et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 2012, 72, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Berka, M.; Luklová, M.; Dufková, H.; Berková, V.; Novák, J.; Saiz-Fernández, I.; Rashotte, A.M.; Brzobohaty, B.; Cerny, M. Barley root proteome and metabolome in response to cytokinin and abiotic stimuli. Front. Plant Sci. 2020, 11, 590337. [Google Scholar] [CrossRef]
- Hampejsová, R.; Berka, M.; Berková, V.; Jersáková, J.; Domkářová, J.; von Rundstedt, F.; Frary, A.; Saiz-Fernández, I.; Brzobohatý, B.; Černý, M. Interaction with Fungi Promotes the Accumulation of Specific Defense Molecules in Orchid Tubers and May Increase the Value of Tubers for Biotechnological and Medicinal Applications: The Case Study of Interaction Between Dactylorhiza sp. and Tulasnella calospora. Front. Plant Sci. 2022, 13, 757852. [Google Scholar] [CrossRef]
- Pino, L.K.; Searle, B.C.; Bollinger, J.G.; Nunn, B.; MacLean, B.; MacCoss, M.J. The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev. 2020, 39, 229–244. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Zhou, G.; Ewald, J.; Xia, J. OmicsAnalyst: A comprehensive web-based platform for visual analytics of multi-omics data. Nucleic Acids Res. 2021, 49, W476–W482. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berková, V.; Berka, M.; Griga, M.; Kopecká, R.; Prokopová, M.; Luklová, M.; Horáček, J.; Smýkalová, I.; Čičmanec, P.; Novák, J.; et al. Molecular Mechanisms Underlying Flax (Linum usitatissimum L.) Tolerance to Cadmium: A Case Study of Proteome and Metabolome of Four Different Flax Genotypes. Plants 2022, 11, 2931. https://doi.org/10.3390/plants11212931
Berková V, Berka M, Griga M, Kopecká R, Prokopová M, Luklová M, Horáček J, Smýkalová I, Čičmanec P, Novák J, et al. Molecular Mechanisms Underlying Flax (Linum usitatissimum L.) Tolerance to Cadmium: A Case Study of Proteome and Metabolome of Four Different Flax Genotypes. Plants. 2022; 11(21):2931. https://doi.org/10.3390/plants11212931
Chicago/Turabian StyleBerková, Veronika, Miroslav Berka, Miroslav Griga, Romana Kopecká, Miroslava Prokopová, Markéta Luklová, Jiří Horáček, Iva Smýkalová, Petr Čičmanec, Jan Novák, and et al. 2022. "Molecular Mechanisms Underlying Flax (Linum usitatissimum L.) Tolerance to Cadmium: A Case Study of Proteome and Metabolome of Four Different Flax Genotypes" Plants 11, no. 21: 2931. https://doi.org/10.3390/plants11212931
APA StyleBerková, V., Berka, M., Griga, M., Kopecká, R., Prokopová, M., Luklová, M., Horáček, J., Smýkalová, I., Čičmanec, P., Novák, J., Brzobohatý, B., & Černý, M. (2022). Molecular Mechanisms Underlying Flax (Linum usitatissimum L.) Tolerance to Cadmium: A Case Study of Proteome and Metabolome of Four Different Flax Genotypes. Plants, 11(21), 2931. https://doi.org/10.3390/plants11212931






