Integrated Metabolites and Transcriptomics at Different Growth Stages Reveal Polysaccharide and Flavonoid Biosynthesis in Cynomorium songaricum
Abstract
:1. Introduction
2. Results
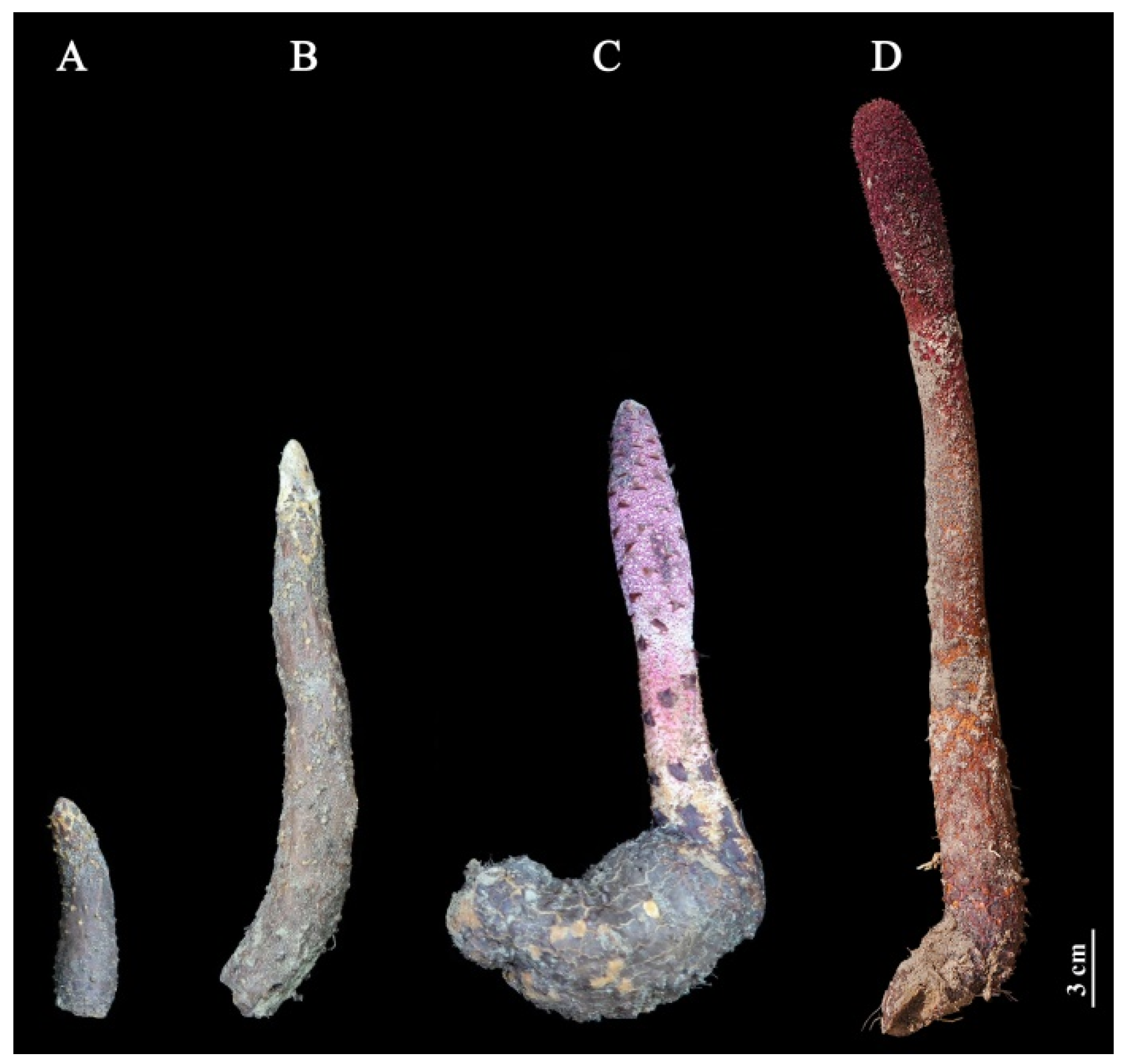
2.1. Changes of Fresh Weight at Different Growth Stages
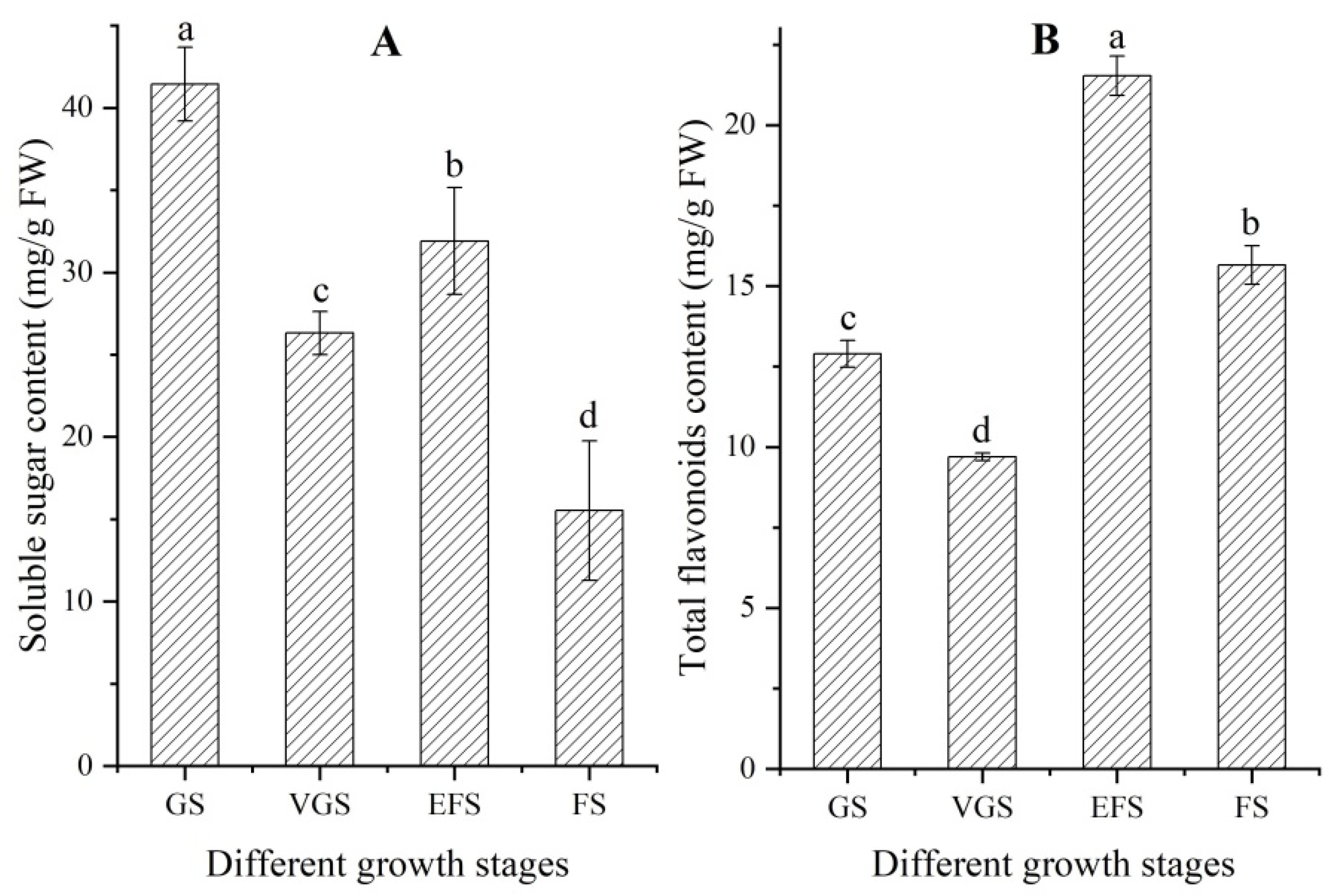
2.2. Changes of Soluble Sugar and Total Flavonoids Contents at Different Growth Stages
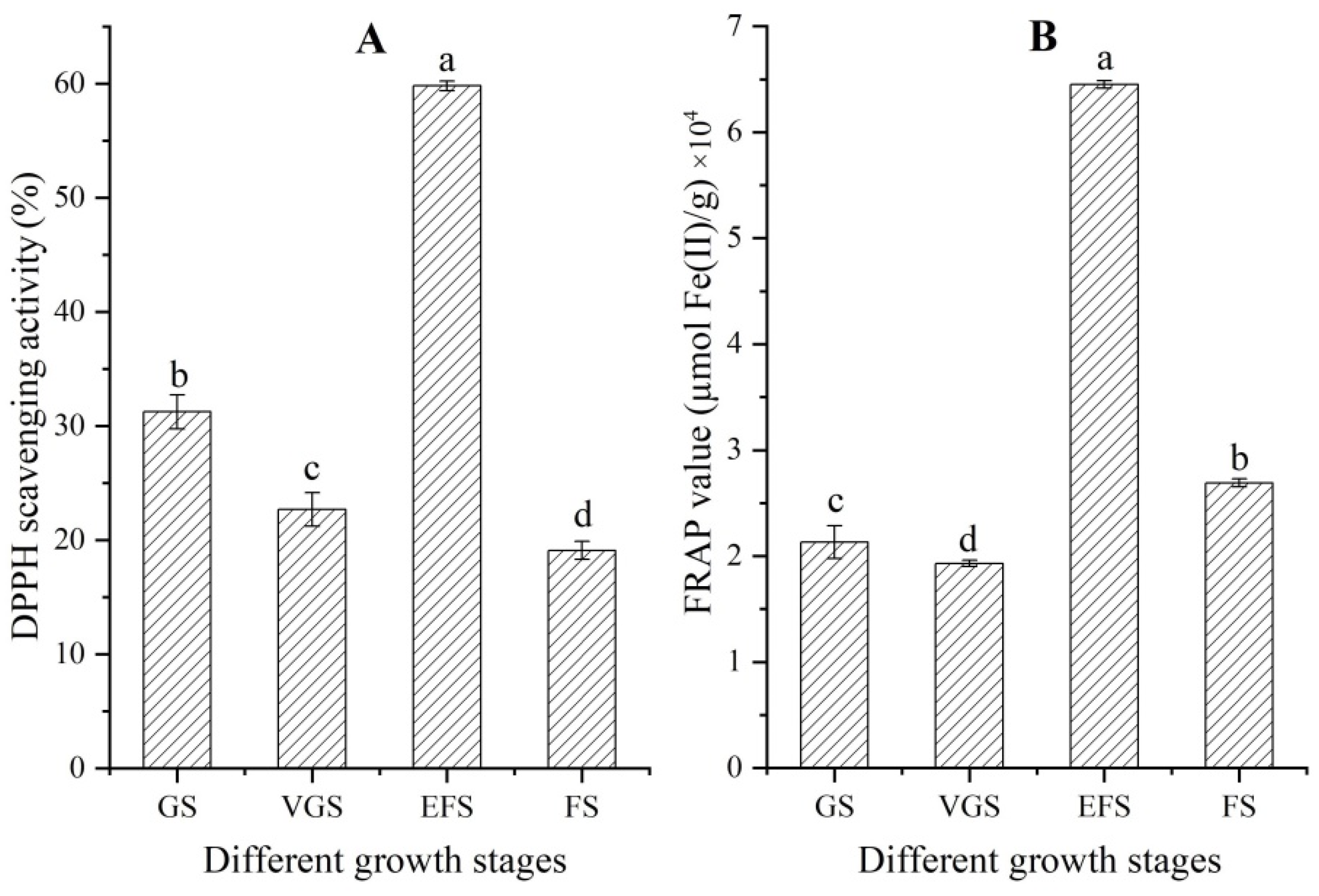
2.3. Changes of Antioxidant Capacity at Different Growth Stages
2.4. Transcriptomics Analysis at Different Growth Stages
2.4.1. Global Gene Analysis
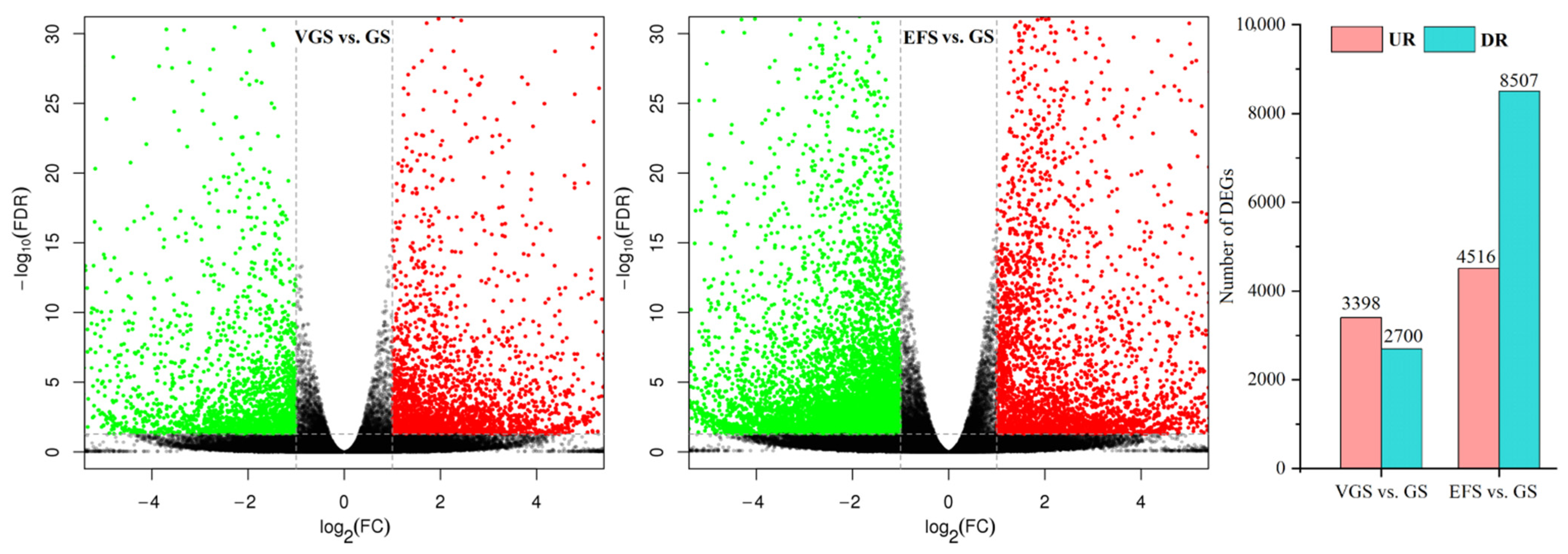
2.4.2. Identification of Differentially Expressed Genes (DEGs)
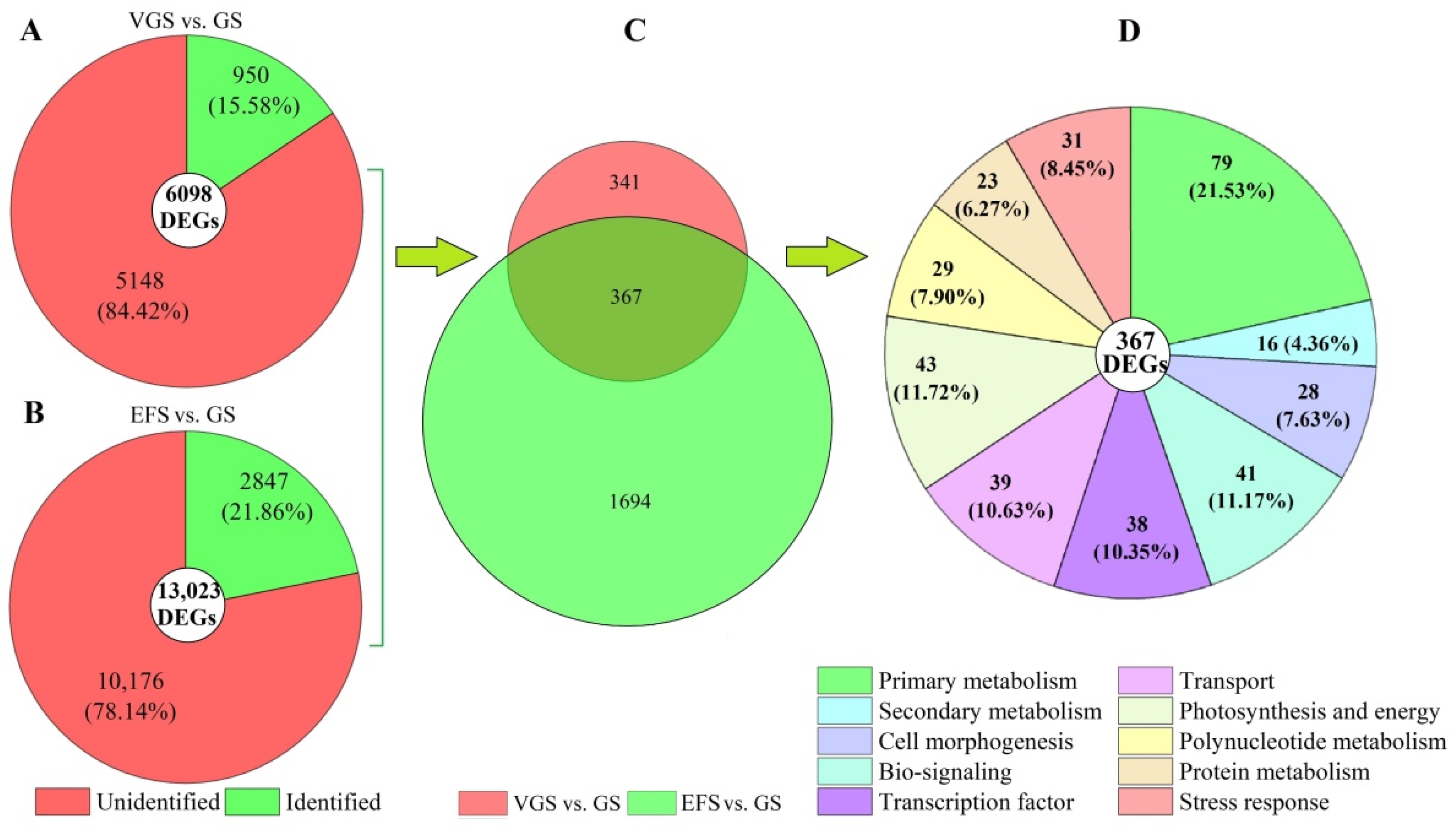
2.4.3. Distribution and Classification of DEGs
2.5. Functional Classification of DEGs
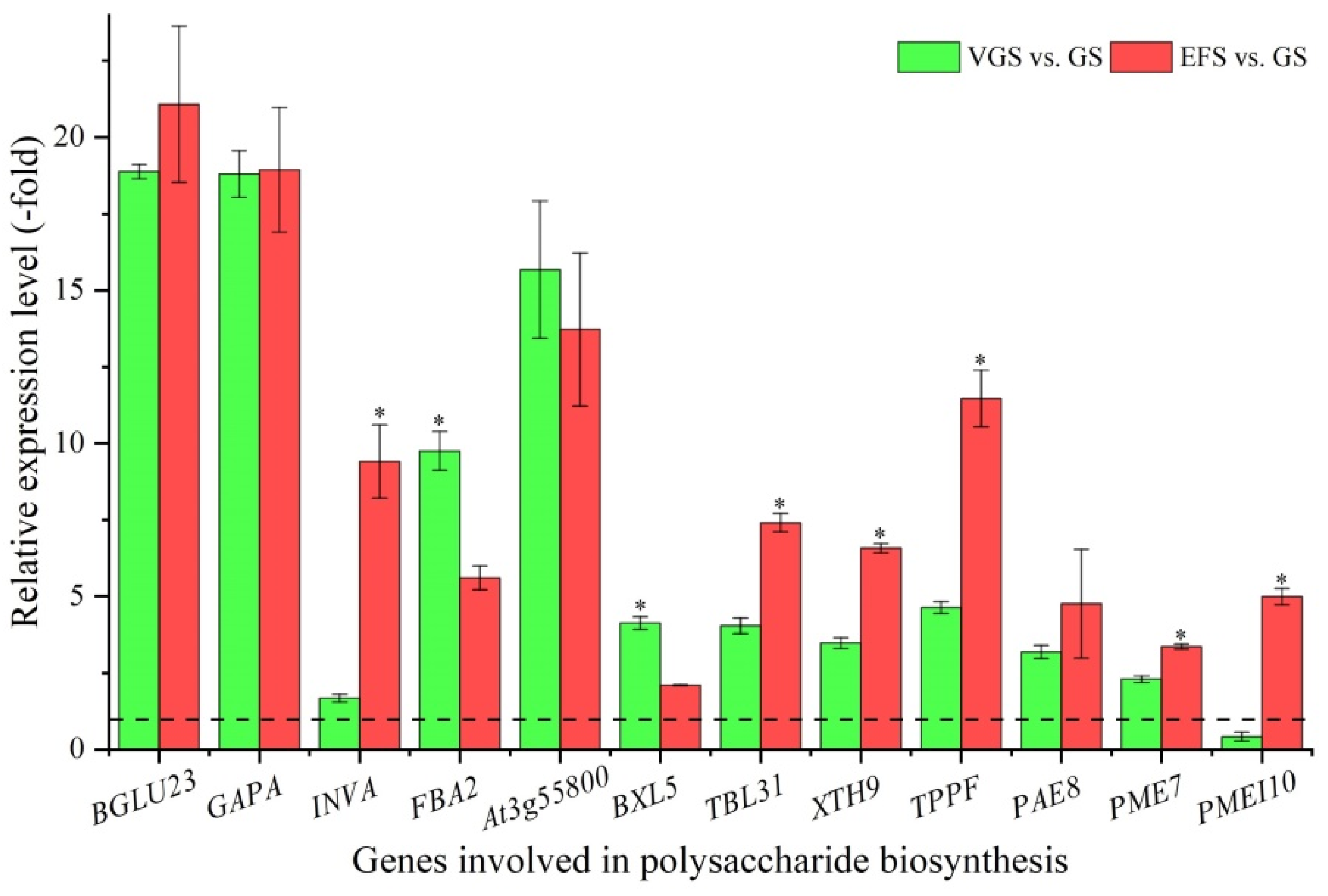
2.5.1. DEGs Involved in Polysaccharide Biosynthesis
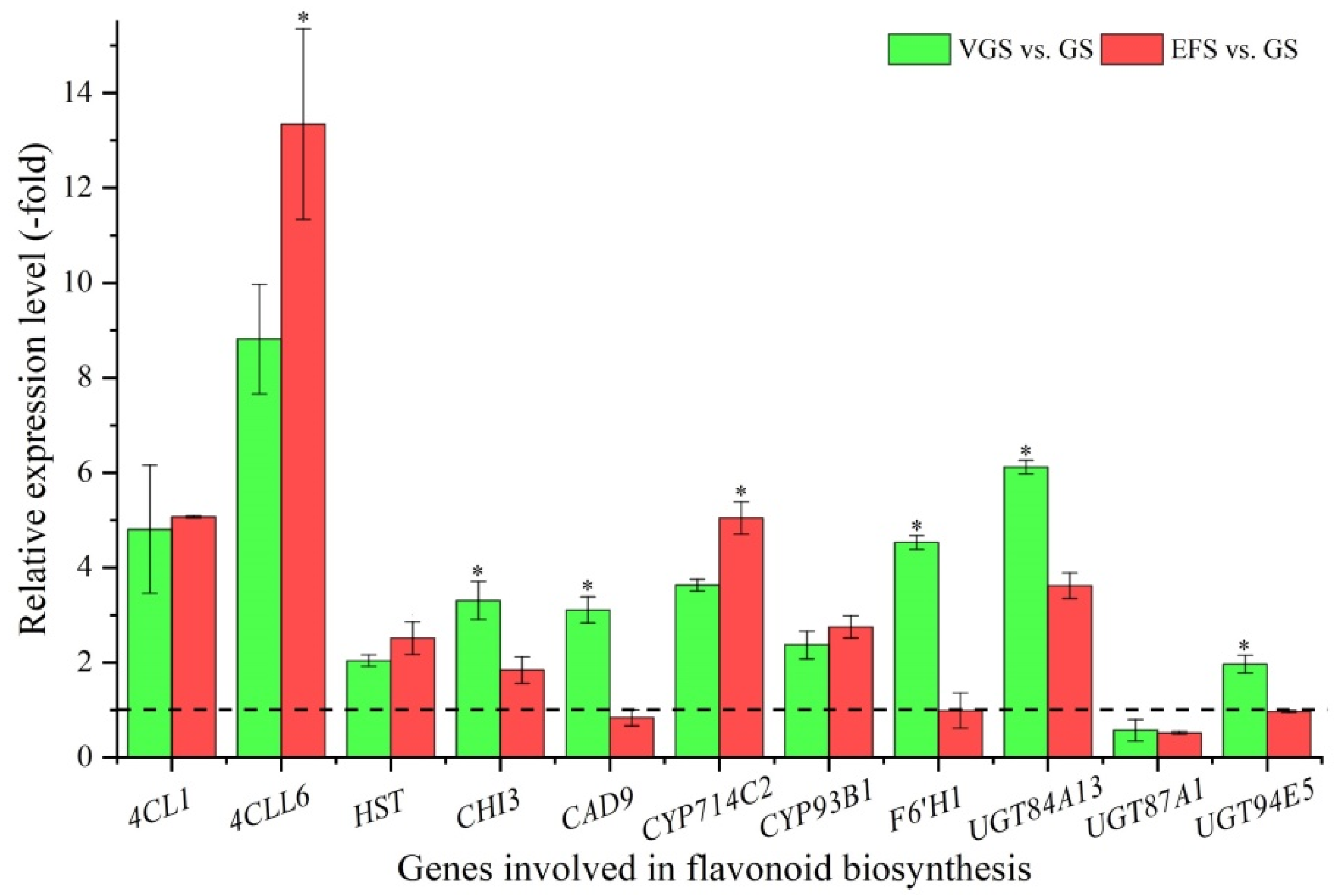
2.5.2. DEGs Involved in Flavonoid Biosynthesis
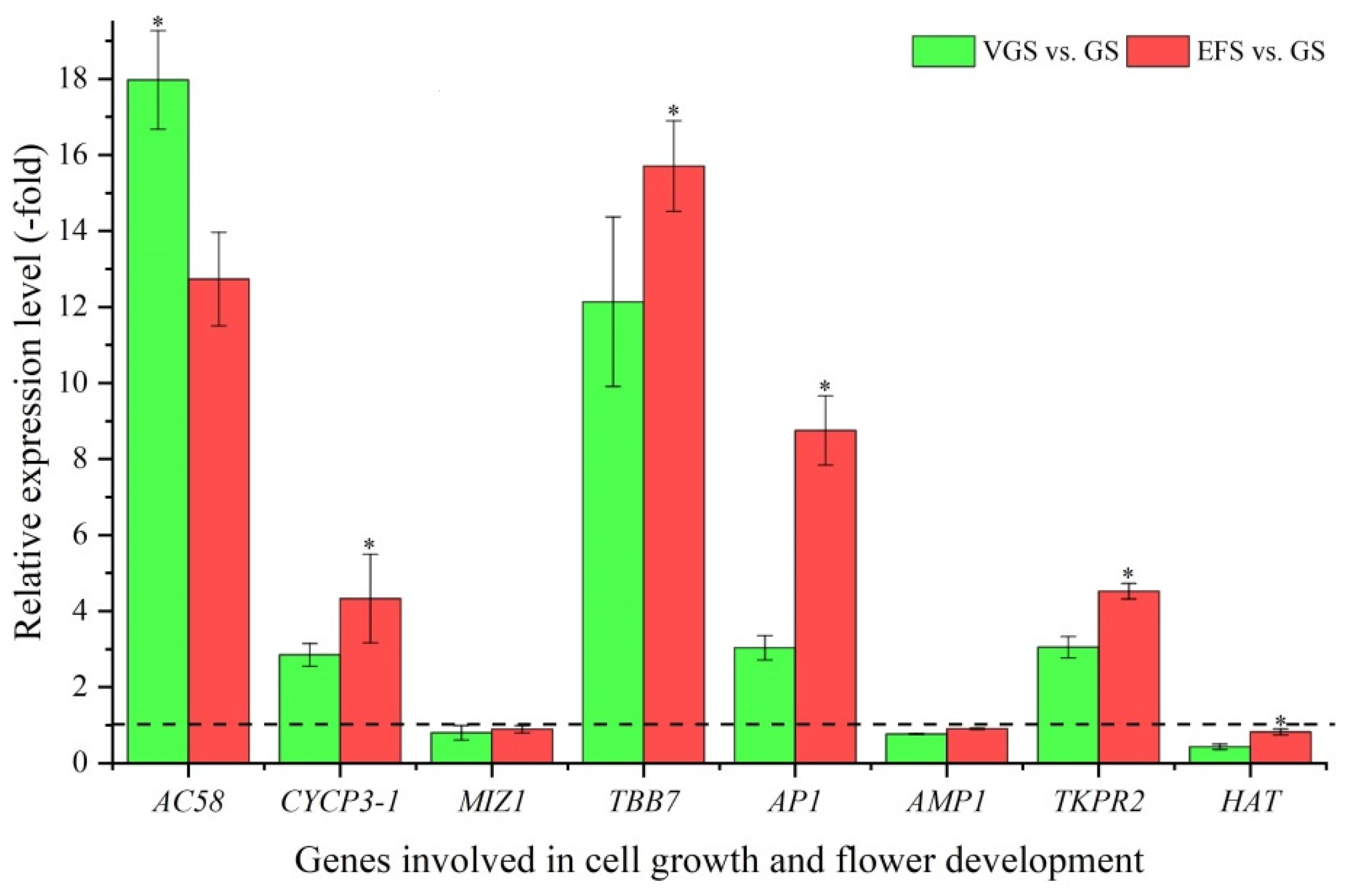
2.5.3. DEGs Involved in Cell Growth and Flower Development
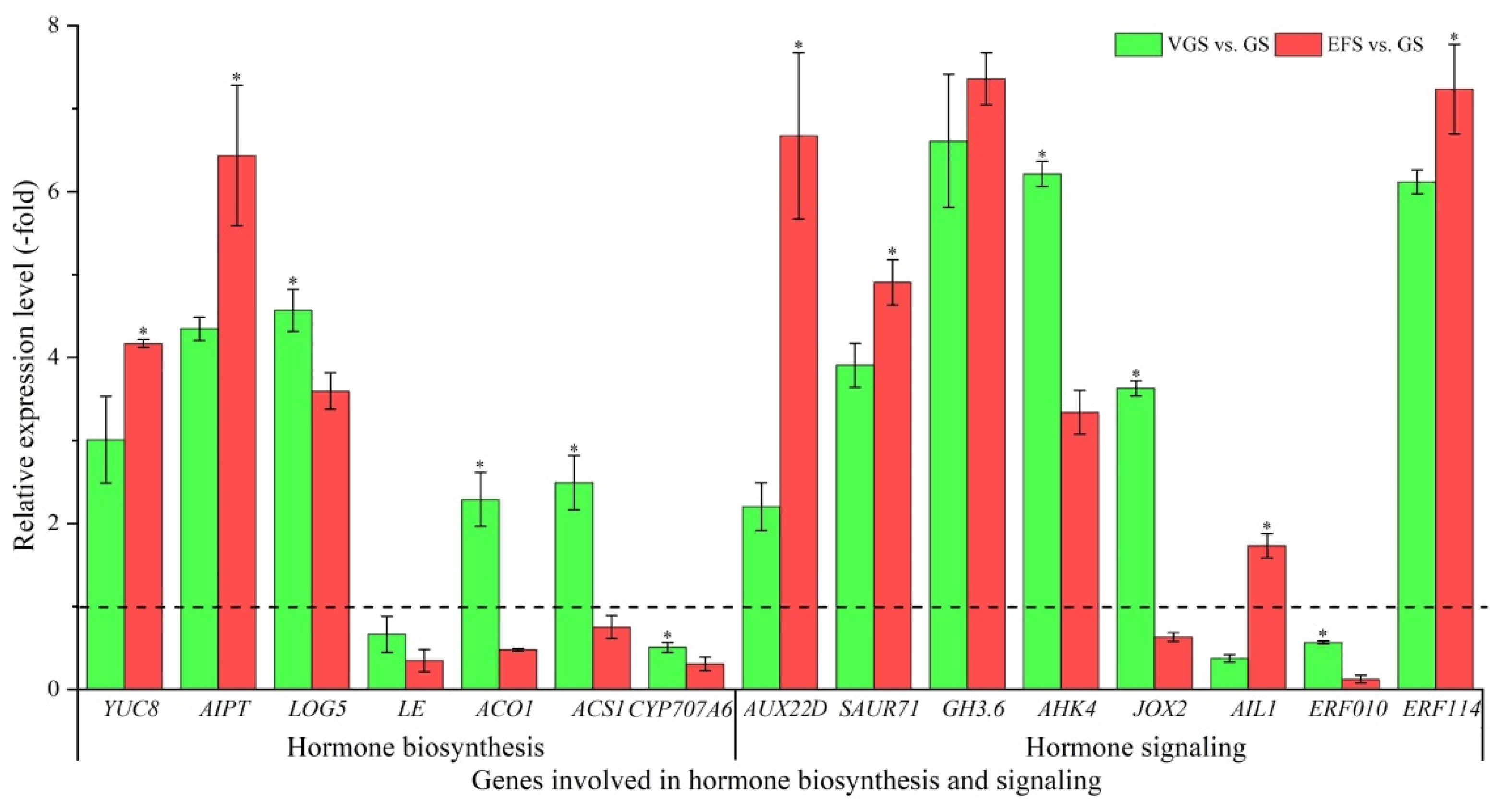
2.5.4. DEGs Involved in Hormone Biosynthesis and Signaling
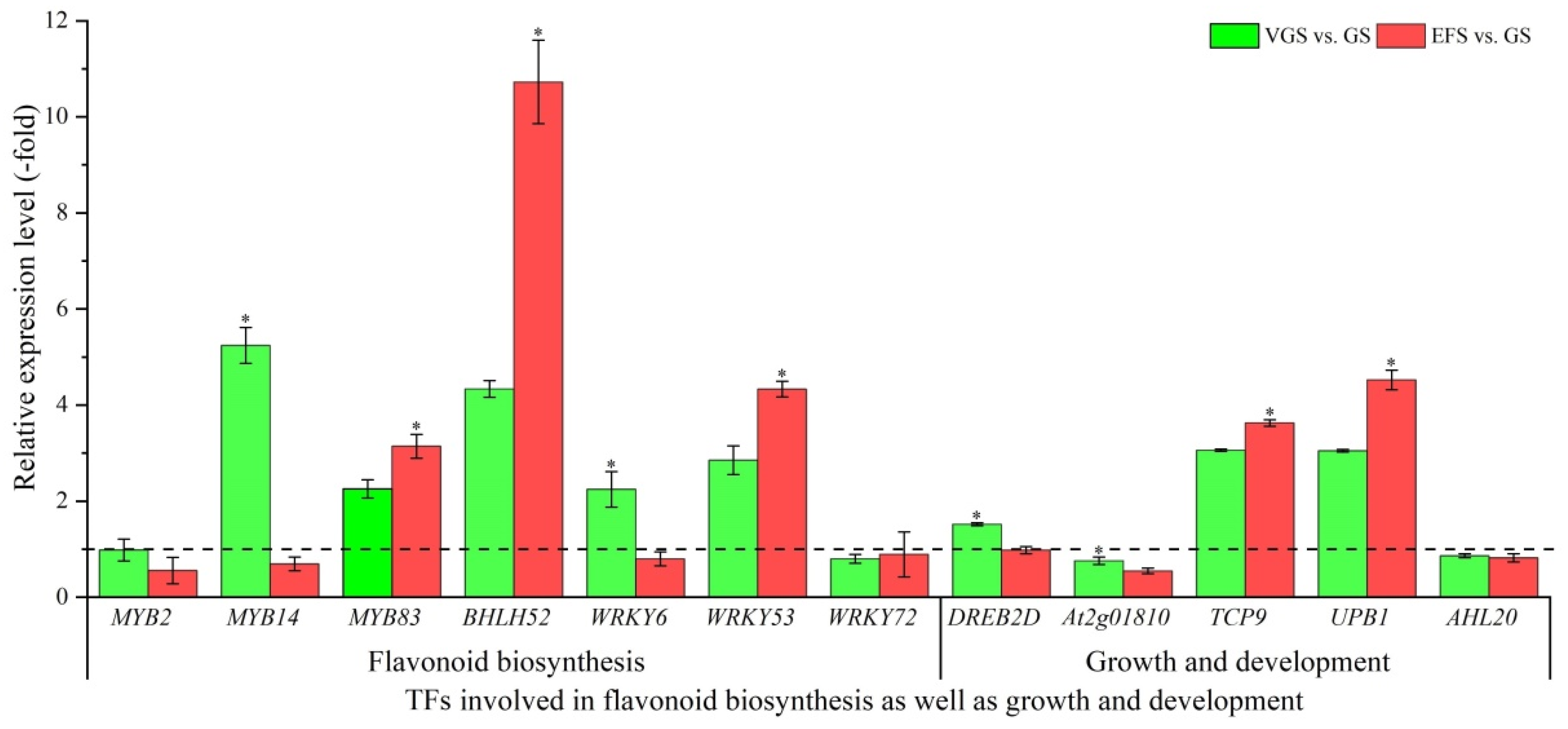
2.5.5. TFs Involved in Flavonoid Biosynthesis as Well as Growth and Development
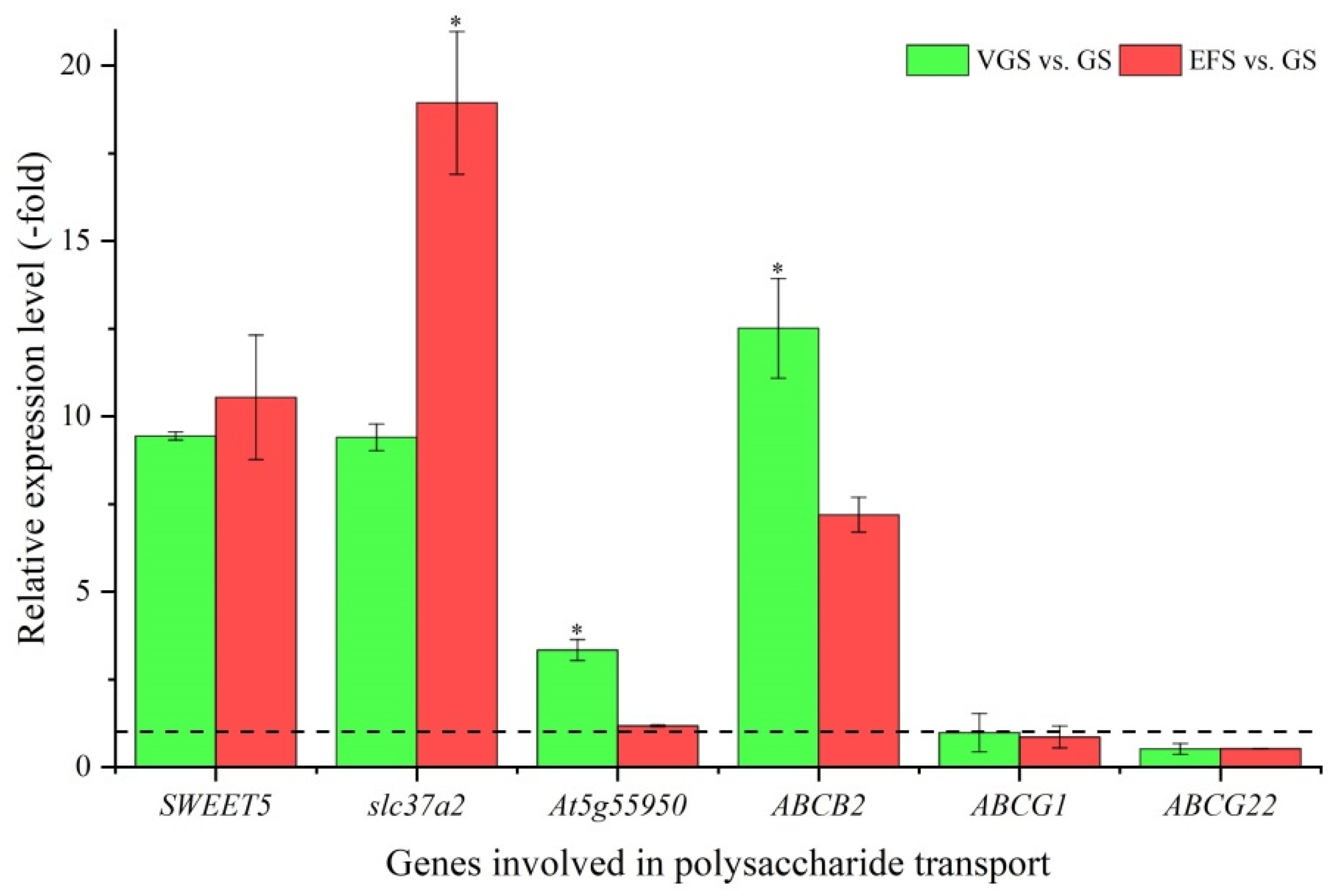
2.5.6. DEGs Involved in Polysaccharide Transport
2.5.7. DEGs Involved in Other Biological Functions
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Stem Biomass
4.3. Determination of Soluble Sugar and Total Flavonoids Contents as Well as Antioxidant Capacity
4.3.1. Extracts Preparation
4.3.2. Determination of Soluble Sugar Content
4.3.3. Determination of Flavonoids Content
4.3.4. Determination of Antioxidant Capacity
4.4. Transcriptomic Analysis
4.4.1. RNA Extraction and Illumina Sequencing
4.4.2. Reads Filtration, Assembly, Unigene Expression Analysis, and Basic Annotation
4.5. qRT-PCR Validation
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chinese Academy of Sciences, China Flora Editorial Committee. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 2000; Volume 53, pp. 152–154. [Google Scholar]
- Committee for the Pharmacopoeia of PR China. Pharmacopoeia of PR China; Chinese Medical Science and Technology Press: Beijing, China, 2020; Volume 1, p. 346.
- Ma, C.M.; Sato, N.; Li, X.Y.; Nakamura, N.; Hattori, M. Flavan-3-ol contents, anti-oxidative and α-glucosidase inhibitory activities of Cynomorium songaricum. Food Chem. 2010, 118, 116–119. [Google Scholar] [CrossRef]
- Chen, J.H.; Wong, H.S.; Leung, H.Y.; Leong, P.K.; Chan, W.M.; Ko, K.M. An ursolic acid-enriched Cynomorium songarium extract attenuates high fat diet-induced obesity in mice possibly through mitochondrial uncoupling. J. Funct. Foods 2014, 9, 211–224. [Google Scholar] [CrossRef]
- Tuvaanjav, S.; Han, S.Q.; Komata, M.; Ma, C.J.; Kanamoto, T.; Nakashima, H.; Yoshida, T. Isolation and antiviral activity of water-soluble Cynomorium songaricum Rupr. polysaccharides. J. Asian Nat. Prod. Res. 2016, 18, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Miao, M.S.; Li, Y.; Li, M.; Zhang, Y.; Tian, S. Effect of cynomorium favonoids on morphology of perimenopausal depression mice model. Saudi Pharm. J. 2016, 24, 322–328. [Google Scholar] [CrossRef]
- Guo, Y.H.; Lin, H.M.; Lin, Z.J. Study on identification characteristics of Cynomorium of the Nitraia siberian. Gansu Agric. Sci. Technol. 2010, 3, 15–17. [Google Scholar]
- Huang, L.; Xie, C.; Chen, S.; Duan, B.; Sun, C. Regional suitability evaluation on desert herbal Cynomorii. Chin. Bull. Bot. 2010, 45, 205–211. [Google Scholar]
- Chen, Y.; Han, D.H.; Gao, H.; Luo, G.H.; Wang, J. Distribution and utilization on germplasm resources of host plants of Cynomorium songaricum. Chin. Wild Plant Resour. 2013, 32, 45–47. [Google Scholar]
- Wang, J.; Su, H.Y.; Han, H.P.; Wang, W.S.; Li, M.C.; Zhou, Y.B.; Li, Y.; Li, M.F. Transcriptomics reveals host-dependent differences of polysaccharides biosynthesis in Cynomorium songaricum. Molecules 2022, 27, 44. [Google Scholar] [CrossRef]
- Zhou, Y.B.; Ye, R.R.; Lu, X.F.; Lin, P.C.; Yang, S.B.; Yue, P.P.; Zhang, C.X.; Peng, M. GC-MS analysis of liposoluble constituents from the stems of Cynomorium songaricum. J. Pharm. Biomed. Anal. 2009, 49, 1097–1100. [Google Scholar] [CrossRef]
- Shekarchi, M.; Kondori, B.; Hajimehdipoor, H.; Abdi, L.; Naseri, M.; Pourfarzib, M.; Amin, G. Finger printing and quantitative analysis of Cuscuta chinensis flavonoid contents from different hosts by RP-HPLC. Food Nutr. Sci. 2014, 5, 914–921. [Google Scholar]
- Chang, Y.X.; Li, J.; Su, G.E.; Wang, Y.C. Study on the dynamics trends of polysaccharide content of Cynomorium songaricum Rupr. in different growth stages. J. Inner Mongolia Univ. 2007, 38, 237–240. [Google Scholar]
- Wang, Q.; Wei, Q.H. Study on dynamics trends of protocatechuic acid content of the Cynomorium songaricum Rupr. in different growth stages. Chin. J. Exp. Tradit. Med. Form. 2010, 16, 54–56. [Google Scholar]
- Li, M.L.; Cui, X.W.; Jin, L.; Li, M.F.; Wei, J.H. Bolting reduces ferulic acid and flavonoid biosynthesis and induces root lignifi- cation in Angelica sinensis. Plant Physiol. Biochem. 2022, 170, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Kang, T.L.; Xing, H.; Zhang, Z.; Yang, D.L.; Zhang, J.L.; Pare, P.W.; Li, M.F. Phytochemical changes in aerial parts of Hypericum perforatum at different harvest stages. Rec. Nat. Prod. 2019, 13, 1–9. [Google Scholar] [CrossRef]
- Zhang, W.J.; Dong, C.F.; Ding, C.L.; Xu, N.X.; Cheng, Y.H. Effects of harvest stage on nutrients and silage quality of different cultivars of Italian ryegrass (Lolium multiflorum). Chin. J. Grassl. 2016, 38, 32–37. [Google Scholar]
- Feng, X.Y. Transcriptome Sequenceing of Cynomorium songaricum Rupr. and Analysis of Genes Involved in Catechin Biosynthesis. Master’s Thesis, Inner Mongolia University, Inner Mongolia, China, 2016. [Google Scholar]
- Jiang, Q.; Lv, Y.; Dai, W.; Miao, X.; Zhong, D. Extraction and bioactivity of polygonatum polysaccharides. Int. J. Biol. Macromol. 2013, 54, 131–135. [Google Scholar] [CrossRef]
- Wu, M.Q.; Xia, W.; Xu, Z.Z.; Song, J.X.; Chen, L.X.; Zhang, W.Q. Review on isolation and purification, structural elucidation and biological activity of botanical polysaccharides. Chem. World 2019, 60, 737–747. [Google Scholar]
- Li, X.G.; Zhang, X.; Yu, J.; Wang, X.F.; Dai, H.Y.; Chen, J.W.; Xu, J.B.; Cao, G.H.; He, S. Research progress on biosynthesis pathway of quality marker polysaccharide and involved key enzymes for medicinal plants. Chin. Tradit. Herb. Drugs 2021, 52, 4752–4762. [Google Scholar]
- Niu, J.F.; Zhao, G.M.; Mi, Z.Y.; Chen, L.J.; Liu, S.A.; Wang, S.Q.; Wang, D.H.; Wang, Z.Z. De novo sequencing of Bletilla striata (Orchidaceae) transcriptome and identification of genes involved in polysaccharide biosynthesis. Genet. Mol. Biol. 2020, 43, e20190417. [Google Scholar] [CrossRef]
- Wang, S.Q.; Wang, B.; Hua, W.P.; Niu, J.F.; Dang, K.K.; Qiang, Y.; Wang, Z.Z. De novo assembly and analysis of Polygonatum sibiricum transcriptome and identification of genes involved in polysaccharide biosynthesis. Int. J. Mol. Sci. 2017, 18, 1950. [Google Scholar] [CrossRef]
- He, C.; Teixeira da Silva, J.A.; Wang, H.; Si, C.; Zhang, M.; Zhang, X.; Li, M.; Tan, J.; Duan, J. Mining MYB transcription factors from the genomes of orchids (Phalaenopsis and Dendrobium) and characterization of an orchid R2R3-MYB gene involved in water-soluble polysaccharide biosynthesis. Sci. Rep. 2019, 9, 13818. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E.J.; Kandaswami, C.; Theoharides, T.C. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar] [PubMed]
- Tohge, T.; De Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef]
- Kim, J.I.; Hidalgo-Shrestha, C.; Bonawitz, N.D.; Franke, R.B.; Chapple, C. Spatio-temporal control of phenylpropanoid biosynthesis by inducible complementation of a cinnamate 4-hydroxylase mutant. J. Exp. Bot. 2021, 72, 3061–3073. [Google Scholar] [CrossRef]
- Santín, O.; Moncalián, G. Loading of malonyl-CoA onto tandem acyl carrier protein domains of polyunsaturated fatty acid synthases. J. Biol. Chem. 2018, 293, 12491–12501. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef]
- Rauf, A.; Imran, M.; Abu-Lzneid, T.; Lahtisham-Ul-Haq; Patel, S.; Pan, X.P.; Naz, S.; Silva, A.S.; Saeed, F.; Suleria, H.A.R. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Kleindt, C.K.; Stracke, R.; Mehrtens, F.; Weisshaar, B. Expression analysis of flavonoid biosynthesis genes during Arabidopsis thaliana silique and seed development with a primary focus on the proanthocyanidin biosynthetic pathway. BMC Res. Notes 2010, 3, 255. [Google Scholar] [CrossRef]
- Ayabe, S.I.; Akashi, T. Cytochrome P450s in flavonoid metabolism. Phytochem. Rev. 2006, 5, 271–282. [Google Scholar] [CrossRef]
- Winkel, B.S.J. Metabolic channeling in plants. Annu Rev. Plant Biol. 2004, 55, 85–107. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.T.; Zhang, M.H.; Su, H.Y.; Li, M.L.; Wang, Y.Y.; Jin, L.; Li, M.F. Integrated metabolomic and transcriptomic analysis reveals differential mechanism of flavonoid biosynthesis in two cultivars of Angelica sinensis. Molecules 2022, 27, 306. [Google Scholar] [CrossRef] [PubMed]
- Li, M.F.; Pare, P.W.; Zhang, J.L.; Kang, T.L.; Zhang, Z.; Yang, D.L.; Wang, K.P.; Xing, H. Antioxidant capacity connection with phenolic and flavonoid content in Chinese medicinal herbs. Rec. Nat. Prod. 2018, 12, 239–250. [Google Scholar] [CrossRef]
- Fu, L.; Xu, B.T.; Xu, X.R.; Gan, R.Y.; Zhang, Y.; Xia, E.Q.; Li, H.B. Antioxidant capacities and total phenolic contents of 62 fruits. Food Chem. 2011, 129, 345–350. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.P.; Jin, L.X.; Gao, R.J.; Bao, J.Q.; Cui, B.W. Preparation, characterization and antioxidant activity of polysaccharide from Fallopia multiflora (Thunb.) Harald. Int. J. Biol. Macromol. 2018, 108, 259–262. [Google Scholar] [CrossRef]
- Nakano, R.T.; Pislewska-Bednarek, M.; Yamada, K.; Edger, P.P.; Miyahara, M.; Kondo, M.; Bottcher, C.; Mori, M.; Nishimura, M.; Schulze-Lefert, P.; et al. PYK10 myrosinase reveals a functional coordination between endoplasmic reticulum bodies and glucosinolates in Arabidopsis thaliana. Plant J. 2017, 89, 204–220. [Google Scholar] [CrossRef]
- Arai, M.; Mori, H.; Imaseki, H. Cloning and sequence of cDNAs for an intracellular acid invertase from etiolated hypocotyls of mung bean and expression of the gene during growth of seedlings. Plant Cell Physiol. 1992, 33, 245–252. [Google Scholar] [CrossRef]
- Mininno, M.; Brugiere, S.; Pautre, V.; Gilgen, A.; Ma, S.; Ferro, M.; Tardif, M.; Alban, C.; Ravanel, S. Characterization of chloroplastic fructose 1,6-bisphosphate aldolases as lysine-methylated proteins in plants. J. Biol. Chem. 2012, 287, 21034–21044. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, S.; Lawson, T.; Zakhleniuk, O.V.; Lloyd, J.C.; Raines, C.A.; Fryer, M. Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 2005, 138, 451–460. [Google Scholar] [CrossRef]
- Yuan, Y.; Teng, Q.; Zhong, R.; Ye, Z.H. TBL3 and TBL31, Two Arabidopsis DUF231 domain proteins, are required for 3-O-monoacetylation of xylan. Plant Cell Physiol. 2016, 57, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Hyodo, H.; Yamakawa, S.; Takeda, Y.; Tsuduki, M.; Yokota, A.; Nishitani, K.; Kohchi, T. Active gene expression of a xyloglucan endotransglucosylase/hydrolase gene, XTH9, in inflorescence apices is related to cell elongation in Arabidopsis thaliana. Plant Mol. Biol. 2003, 52, 473–482. [Google Scholar] [CrossRef] [PubMed]
- De Souza, A.; Hull, P.A.; Gille, S.; Pauly, M. Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants. Planta 2014, 240, 1123–1138. [Google Scholar] [CrossRef]
- Louvet, R.; Cavel, E.; Gutierrez, L.; Guenin, S.; Roger, D.; Gillet, F.; Guerineau, F.; Pelloux, J. Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 2006, 224, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Fabri, E.; De Caroli, M.; Hansen, A.R.; Willats, W.G.; Piro, G.; Bellincampi, D. Three pectin methyl esterase inhibitors protect cell wall integrity for immunity to Botrytis. Plant Physiol. 2017, 173, 1844–1863. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.J.; Chen, S.Y.; Jun, H.S.; Lin, S.R.; Mansfield, B.C.; Chou, J.Y. SLC37A1 and SLC37A2 are phosphate-linked, glucose-6-phosphate antiporters. PLoS ONE 2011, 6, e23157. [Google Scholar] [CrossRef]
- Szulc, B.; Sosicka, P.; Maszczak-Seneczko, D.; Skurska, E.; Shauchuk, A.; Olczak, T.; Freeze, H.H.; Olczak, M. Biosynthesis of GlcNAc-rich N- and O-glycans in the Golgi apparatus does not require the nucleotide sugar transporter SLC35A3. J. Biol. Chem. 2020, 295, 16445–16463. [Google Scholar] [CrossRef]
- Ahmad, N.; Liu, J.Y.; Xu, T.; Noman, M.; Jameel, A.; Yao, N.; Dong, Y.Y.; Wang, N.; Li, X.W.; Wang, F.W.; et al. Overexpression of a novel Cytochrome P450 promotes flavonoid biosynthesis and osmotic stress tolerance in transgenic Arabidopsis. Genes 2019, 10, 756. [Google Scholar] [CrossRef] [Green Version]
- Besseau, S.; Hoffmann, L.; Geoffroy, P.; Lapierre, C.; Pollet, B.; Legrand, M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 2007, 19, 148–162. [Google Scholar] [CrossRef]
- Eudes, A.; Pollet, B.; Sibout, R.; Do, C.T.; Seguin, A.; Lapierre, C.; Jouanin, L. Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 2006, 225, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Frangne, N.; Eggmann, T.; Koblischke, C.; Weissenbock, G.; Martinoia, E.; Klein, M. Flavone glucoside uptake into barley mesophyll and Arabidopsis cell culture vacuoles. energization occurs by H+-antiport and ATP-binding cassette-type mechanisms. Plant Physiol. 2002, 128, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhou, P.; Gui, C.; Da, G.Z.; Gong, L.; Zhang, X.Q. Comparative transcriptome analysis of Ampelopsis megalophylla for identifying genes involved in flavonoid biosynthesis and accumulation during different seasons. Molecules 2019, 24, 1267. [Google Scholar] [CrossRef] [PubMed]
- Drouin, G.; Dover, G.A. Independent gene evolution in the potato actin gene family demonstrated by phylogenetic procedures for resolving gene conversions and the phylogeny of angiosperm actin genes. J. Mol. Evol. 1990, 31, 132–150. [Google Scholar] [CrossRef]
- Chen, Y.X.; Sun, S.Y.; Wang, X.L. The epidermis specific cyclin CYCP3;1 is involved in the excess BR signaling inhibited root meristem cell division. J. Integr. Plant Biol. 2020, 62, 14. [Google Scholar] [CrossRef]
- Yamazaki, T.; Miyazawa, Y.; Kobayashi, A.; Moriwaki, T.; Fujii, N.; Takahashi, H. MIZ1, an essential protein for root hydrotropism, is associated with the cytoplasmic face of the endoplasmic reticulum membrane in Arabidopsis root cells. FEBS Lett. 2012, 586, 398–402. [Google Scholar] [CrossRef]
- Pelaz, S.; Gustafson-Brown, C.; Kohalmi, S.E.; Crosby, W.L.; Yanofsky, M.F. APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 2001, 26, 385–394. [Google Scholar] [CrossRef]
- Vidaurre, D.P.; Ploense, S.; Krogan, N.T.; Berleth, T. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 2007, 134, 2561–2567. [Google Scholar] [CrossRef]
- Grienenberger, E.; Kim, S.S.; Lallemand, B.; Geoffroy, P.; Heintz, D.; de Azevedo Souza, C.; Heitz, T.; Douglas, C.J.; Legrand, M. Analysis of TETRAKETIDE?-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 2010, 22, 4067–4083. [Google Scholar] [CrossRef] [Green Version]
- Bundock, P.; Hooykaas, P. An Arabidopsis hAT-like transposase is essential for plant development. Nature 2005, 436, 282–284. [Google Scholar] [CrossRef]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef] [PubMed]
- Sakano, Y.; Okada, Y.; Matsunaga, A.; Suwama, T.; Kaneko, T.; Ito, K.; Noguchi, H.; Abe, I. Molecular cloning, expression, and characterization of adenylate isopentenyltransferase from hop (Humulus lupulus L.). Phytochemistry 2004, 65, 2439–2446. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.N.; Proebsting, W.M.; Hedden, P. Mendel’s dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 8907–8911. [Google Scholar] [CrossRef] [PubMed]
- Barry, S.C.; Blume, B.; Bouzayen, M.; Cooper, W.; Hamilton, A.J.; Grierson, D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 1996, 9, 525–535. [Google Scholar] [CrossRef]
- Rottmann, W.H.; Peter, G.F.; Oeller, P.W.; Keller, J.A.; Shen, N.F.; Nagy, B.P.; Taylor, L.P.; Campbell, A.D.; Theologis, A. 1-Aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J. Mol. Biol. 1991, 222, 937–961. [Google Scholar] [CrossRef]
- Yang, S.H.; Choi, D. Characterization of genes encoding ABA 8’-hydroxylase in ethylene-induced stem growth of deepwater rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 2006, 350, 685–690. [Google Scholar] [CrossRef]
- Xu, G.; Huang, J.; Yang, Y.; Yao, Y.A. Transcriptome analysis of flower sex differentiation in Jatropha curcas L. using RNA sequencing. PLoS ONE 2016, 11, 014561. [Google Scholar] [CrossRef]
- Kong, Y.; Zhu, Y.; Gao, C.; She, W.; Lin, W.; Chen, Y.; Han, N.; Bian, H.; Zhu, M.; Wang, J. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 2013, 54, 609–621. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef]
- Ueguchi, C.; Sato, S.; Kato, T.; Tabata, S. The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.J.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Mehrnia, M.; Balazadeh, S.; Zanor, M.I.; Mueller-Roeber, B. EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol. 2013, 162, 842–857. [Google Scholar] [CrossRef]
- Ciarmiello, L.F.; Woodrow, P.; Piccirillo, P.; Luca, A.D.; Carillo, P. Chapter 3-Transcription Factors and Environmental Stresses in Plants. In Emerging Technologies and Management of Crop Stress Tolerance; Ahmad, P., Rasool, S., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 57–78. [Google Scholar]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Afrin, S.; Nuruzzaman, M.; Zhu, J.; Luo, Z.Y. Combinatorial interactions of MYB and bHLH in flavonoid biosynthesis and their function in plants. J. Plant Biol. 2014, 3, 65–77. [Google Scholar]
- Wang, N.; Liu, W.; Zhang, T.; Jiang, S.; Xu, H.; Wang, Y.; Zhang, Z.; Wang, C.; Chen, X. Transcriptomic analysis of red-fleshed apples reveals the novel role of MdWRKY11 in flavonoid and anthocyanin biosynthesis. J. Agric. Food Chem. 2018, 66, 7076–7086. [Google Scholar] [CrossRef] [PubMed]
- Danisman, S.; van der Wal, F.; Dhondt, S.; Waites, R.; de Folter, S.; Bimbo, A.; van Dijk, A.D.; Muino, J.M.; Cutri, L.; Dornelas, M.C.; et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martinez, J.A.; Poza-Carrion, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Busch, W.; Benfey, P.N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 2010, 143, 606–616. [Google Scholar] [CrossRef]
- Tayengwa, R.; Sharma Koirala, P.; Pierce, C.F.; Werner, B.E.; Neff, M.M. Overexpression of AtAHL20 causes delayed flowering in Arabidopsis via repression of FT expression. BMC Plant Biol. 2020, 20, 559. [Google Scholar] [CrossRef]
- Yin, H.; Cai, B.; Li, C.H.; Chen, F. Genome- wide analysis of bHLH transcription factor family in grape. Acta Agric. Jiangxi 2013, 25, 1–6. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Lay, M.M.; Karsani, S.A.; Mohajer, S.; Abd Malek, S.N. Phytochemical constituents, nutritional values, phenolics, flavonols, flavonoids, antioxidant and cytotoxicity studies on Phaleria macrocarpa (Scheff.) Boerl fruits. BMC Complement. Altern. Med. 2014, 14, 152. [Google Scholar]
- Dong, H.; Li, M.L.; Jin, L.; Xie, X.R.; Li, M.F.; Wei, J.H. Cool temperature enhances growth, ferulic acid and flavonoid biosynthesis while inhibiting polysaccharide biosynthesis in Angelica sinensis. Molecules 2022, 27, 320. [Google Scholar] [CrossRef]
- Arnao, M.B. Some methodological problems in the determination of antioxidant activity using chromogen radicals: A practical case. Trends Food Sci. Technol. 2000, 11, 419–421. [Google Scholar] [CrossRef]
- Nencini, C.; Menchiari, A.; Franchi, G.G.; Micheli, L. In vitro antioxidant activity of aged extracts of some Italian Allium species. Plant Foods Hum. Nutr. 2011, 66, 11–16. [Google Scholar] [CrossRef]
- Li, M.F.; Li, T.; Li, W.; Yang, D.L. Changes in antioxidant capacity, levels of soluble sugar, total polyphenol, organosulfur compound and constituents in garlic clove during storage. Ind. Crop. Prod. 2015, 69, 137–142. [Google Scholar]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of antioxidant power: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Li, M.F.; Sun, P.; Kang, T.L.; Xing, H.; Yang, D.L.; Zhang, J.L.; Paré, P.W. Mapping podophyllotoxin biosynthesis and growth related transcripts with high elevation in Sinopodophyllum hexandrum. Ind. Crop. Prod. 2018, 124, 510–518. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNAseq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef] [PubMed]

| GS | VGS | EFS | |
|---|---|---|---|
| Filtered data | |||
| Data of reads number (million) | 50.22 ± 5.50 | 51.24 ± 3.29 | 52.69 ± 3.95 |
| Data of reads number × read length (million) | 7533 ± 824.75 | 7686 ± 493.36 | 7904 ± 592.2 |
| Q20 (%) | 97.16 ± 0.19 | 96.83 ± 0.47 | 97.05 ± 0.27 |
| Q30 (%) | 92.21 ± 0.42 | 91.54 ± 0.92 | 92.00 ± 0.54 |
| Mapped data | |||
| Data of unique mapped reads (million) | 41.17 ± 4.37 | 42.46 ± 2.79 | 43.59 ± 3.30 |
| Data of multiple mapped reads (million) | 1.62 ± 0.18 | 1.61 ± 0.13 | 1.62 ± 0.12 |
| Mapping ratio (%) | 85.21 ± 3.68 | 86.01 ± 4.22 | 85.80 ± 4.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Su, H.; Wu, Z.; Wang, W.; Zhou, Y.; Li, M. Integrated Metabolites and Transcriptomics at Different Growth Stages Reveal Polysaccharide and Flavonoid Biosynthesis in Cynomorium songaricum. Int. J. Mol. Sci. 2022, 23, 10675. https://doi.org/10.3390/ijms231810675
Wang J, Su H, Wu Z, Wang W, Zhou Y, Li M. Integrated Metabolites and Transcriptomics at Different Growth Stages Reveal Polysaccharide and Flavonoid Biosynthesis in Cynomorium songaricum. International Journal of Molecular Sciences. 2022; 23(18):10675. https://doi.org/10.3390/ijms231810675
Chicago/Turabian StyleWang, Jie, Hongyan Su, Zhibo Wu, Wenshu Wang, Yubi Zhou, and Mengfei Li. 2022. "Integrated Metabolites and Transcriptomics at Different Growth Stages Reveal Polysaccharide and Flavonoid Biosynthesis in Cynomorium songaricum" International Journal of Molecular Sciences 23, no. 18: 10675. https://doi.org/10.3390/ijms231810675
APA StyleWang, J., Su, H., Wu, Z., Wang, W., Zhou, Y., & Li, M. (2022). Integrated Metabolites and Transcriptomics at Different Growth Stages Reveal Polysaccharide and Flavonoid Biosynthesis in Cynomorium songaricum. International Journal of Molecular Sciences, 23(18), 10675. https://doi.org/10.3390/ijms231810675







