Full-Length Transcriptome Sequencing Combined with RNA-Seq to Analyze Genes Related to Terpenoid Biosynthesis in Cinnamomum burmannii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Chemical Compositions
2.3. RNA Extraction, cDNA Library Construction, and Sequencing
2.4. PacBio SMRTbell Library Construction and SMRT Sequencing
2.5. PacBio Sequence Data Processing
2.6. Illumina RNA-Sequencing Library Construction and Sequencing
2.7. Second-Generation Data Processing
2.8. Differential Gene Expression Analysis
2.9. Functional Annotation of DEGs
2.10. RT-qPCR Validation of DEGs
2.11. Phylogenetic Analysis of Terpenoid Synthases
3. Results
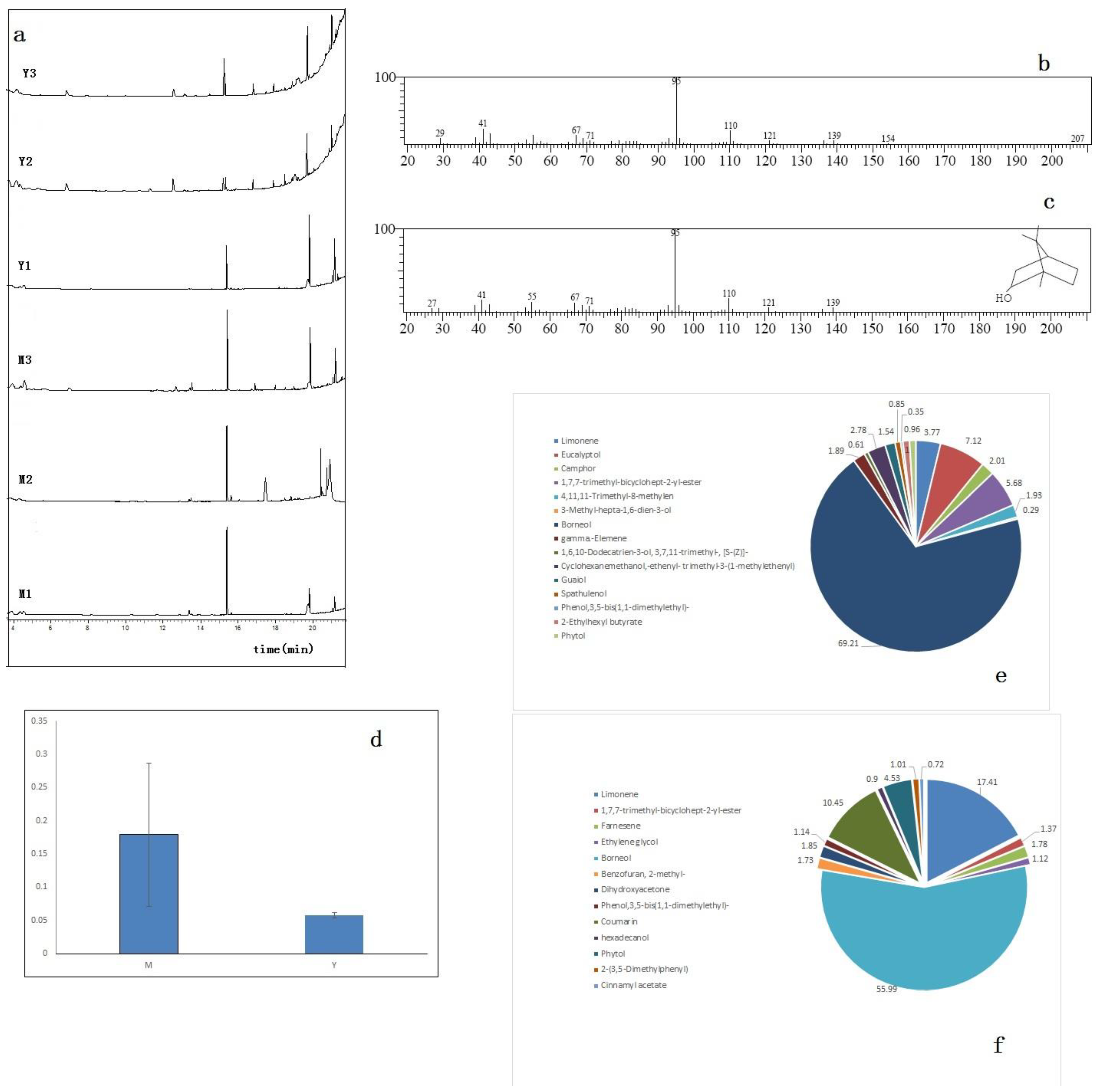
3.1. Determination of D-Borneol in C. Burmanni
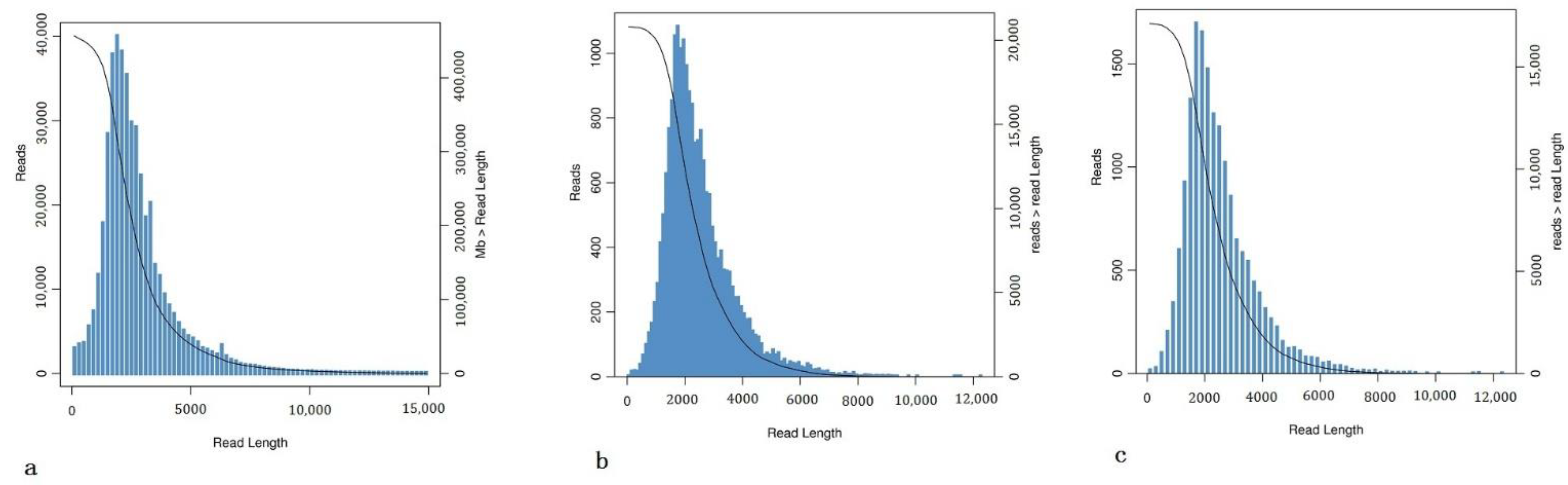
3.2. RNA Sequencing and Transcriptomic Assembly
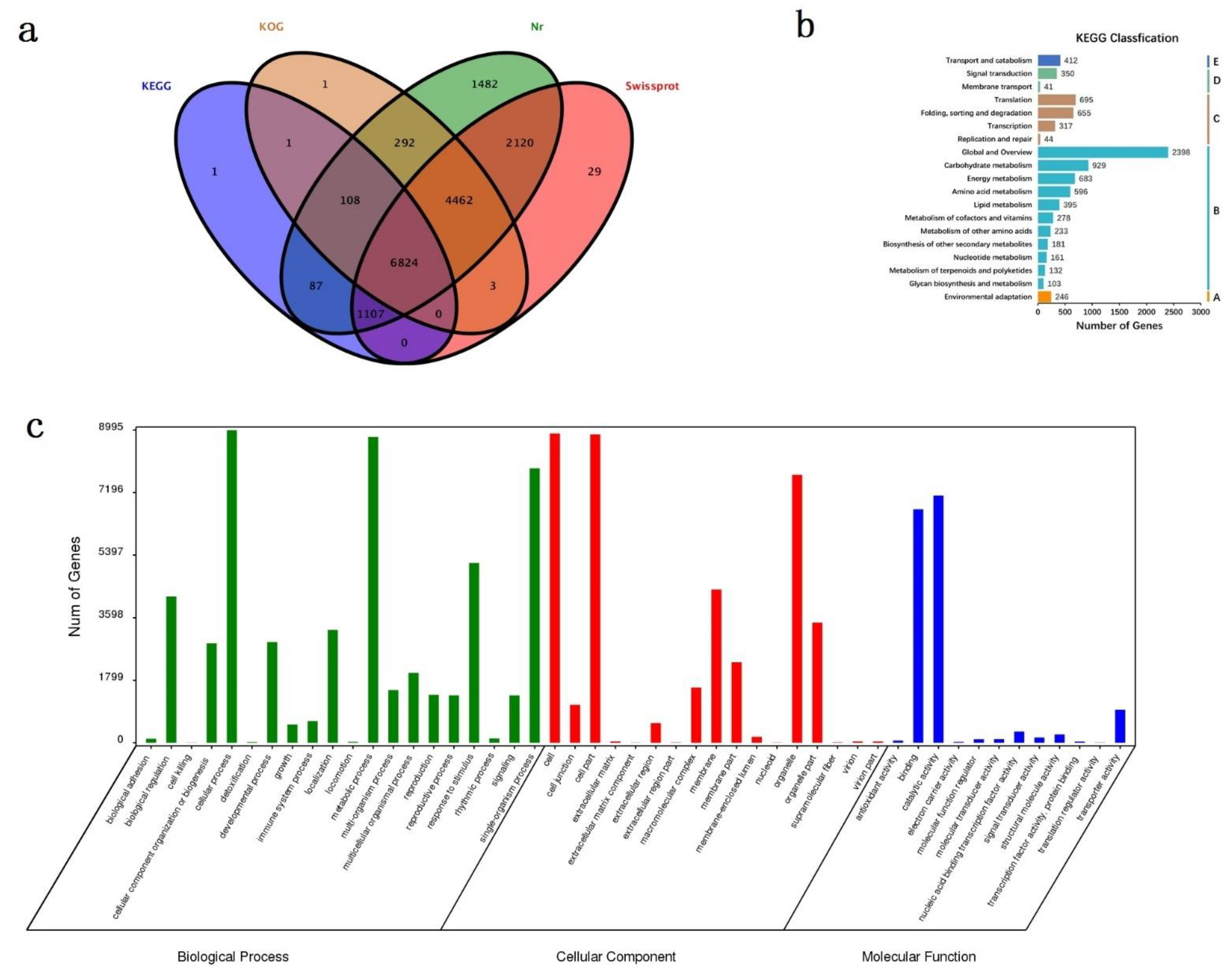
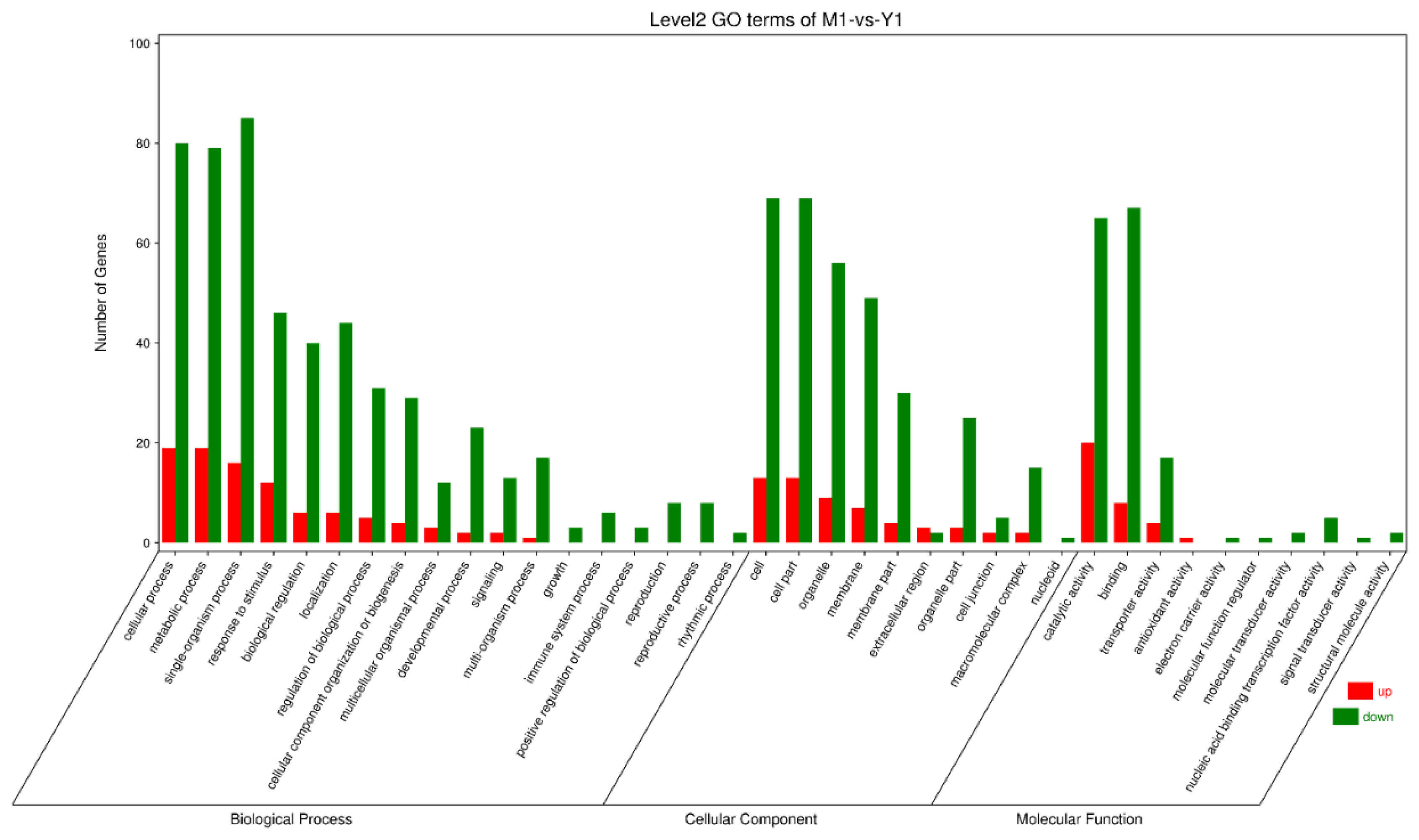
3.3. Gene Annotation and Functional Classification
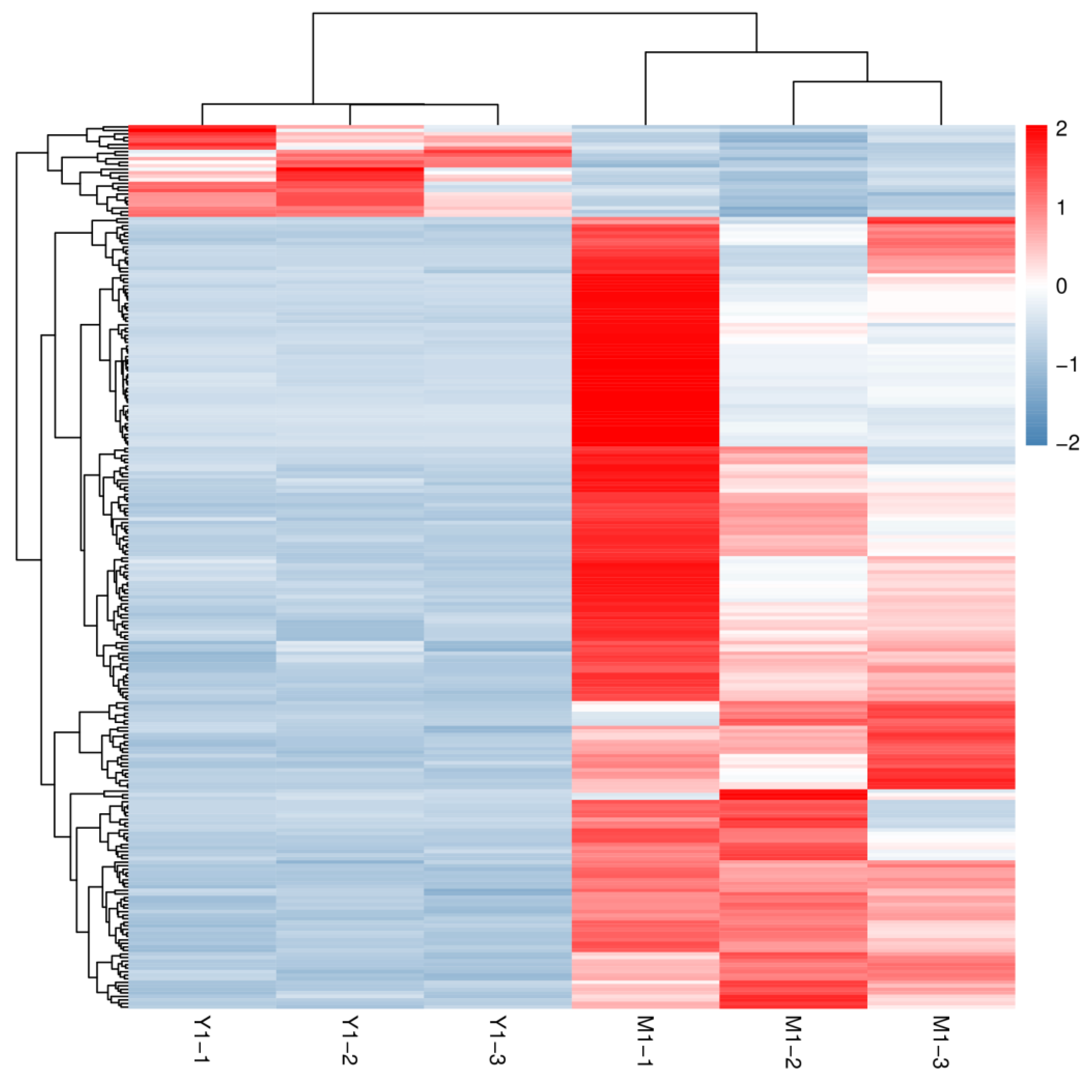
3.4. Analysis of DEGs between Different Groups
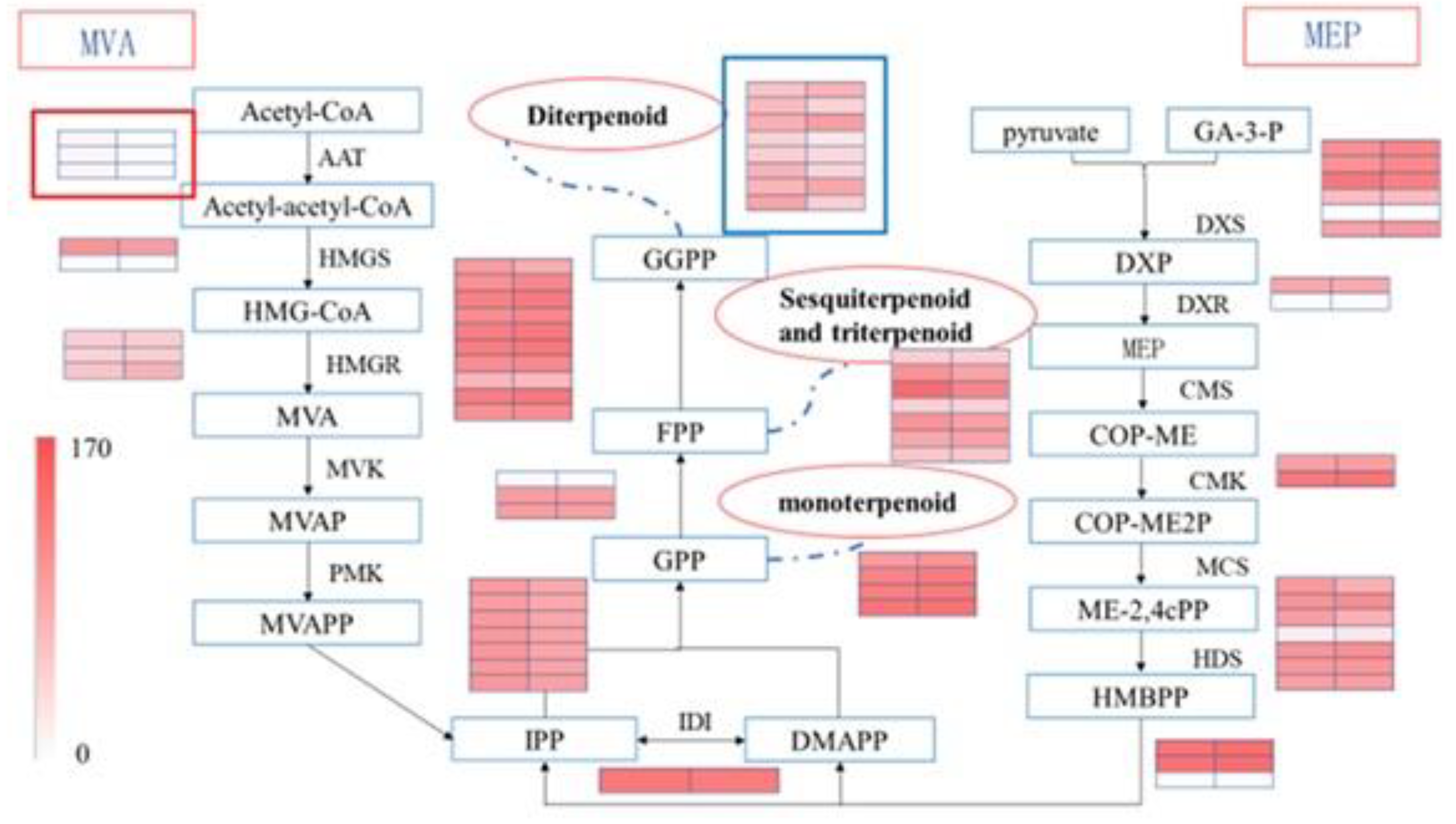
3.5. Screening of Genes Related to Terpenoid Synthesis
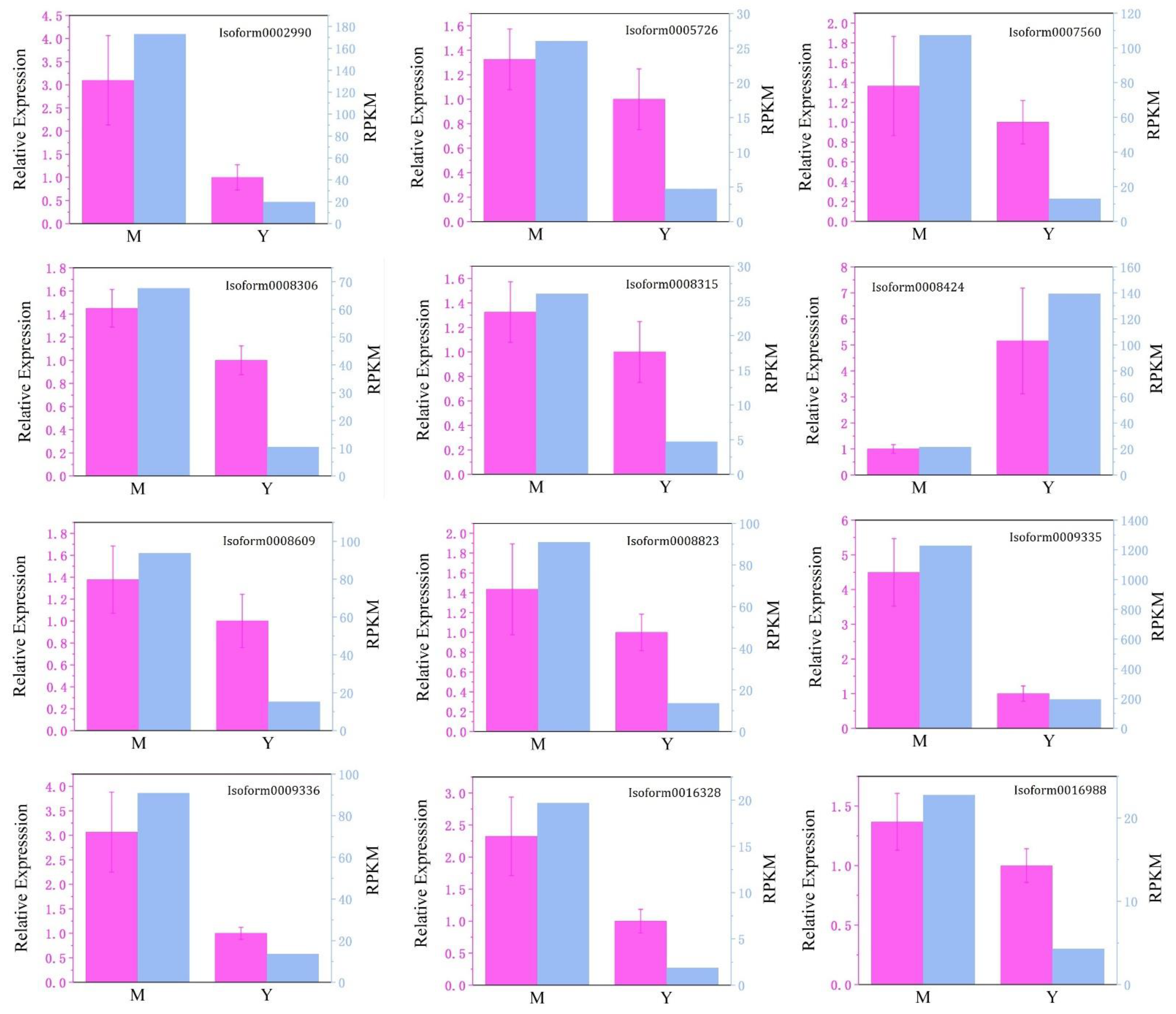
3.6. qRT-PCR Validation of DEGs from RNA-Seq Analysis
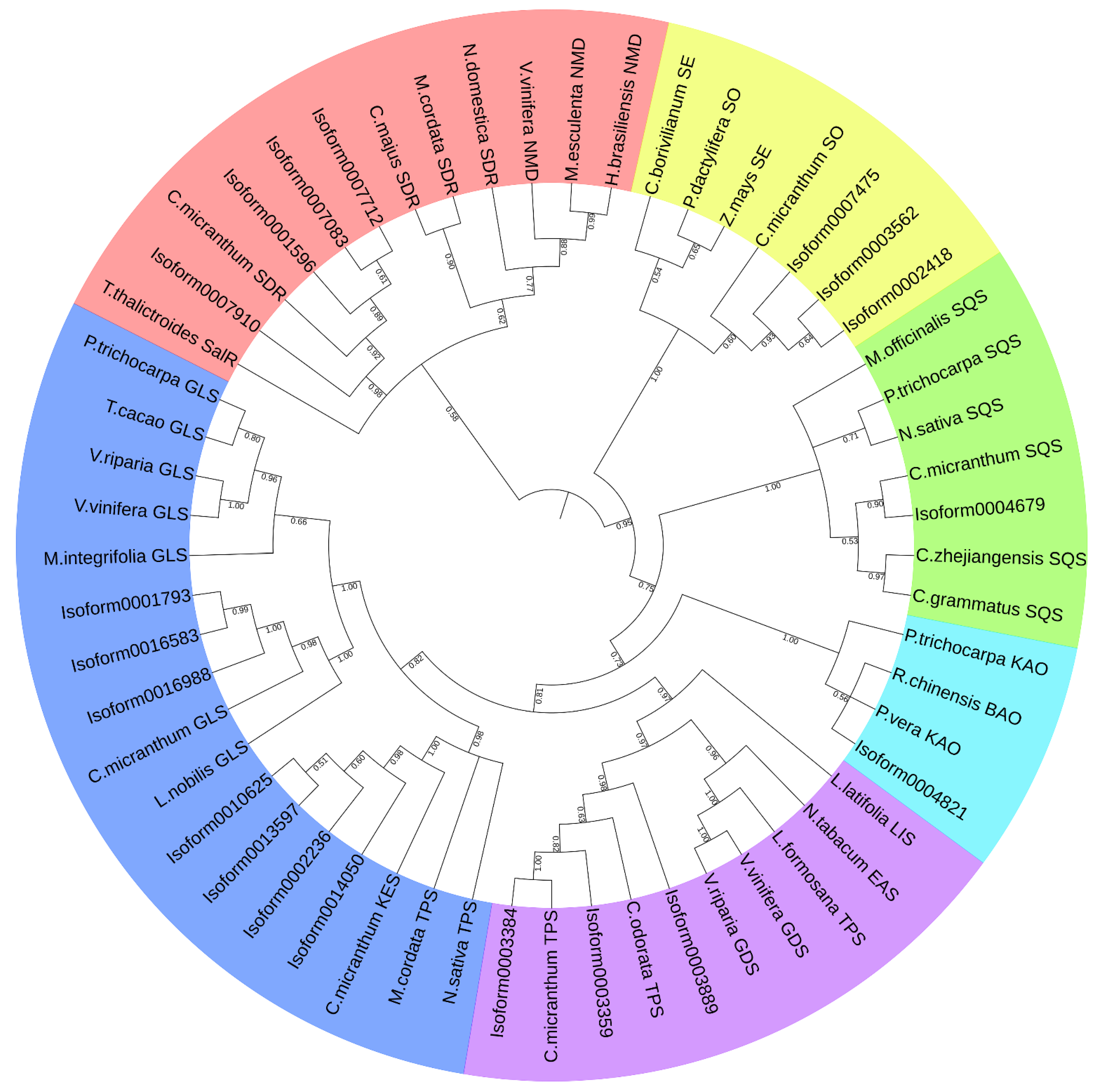
3.7. The Phylogenetic Analysis of Terpenoid Synthases
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, L.; Su, J.; Li, L.; Li, B.; Li, W. A new source of natural D-borneol and its characteristic. J. Med. Plants Res. 2010, 5, 3440–3447. [Google Scholar]
- Wang, R.; Wang, R.; Yang, B. Extraction of essential oils from five cinnamon leaves and identification of their volatile compound compositions. Innov. Food Sci. Emerg. 2009, 10, 289–292. [Google Scholar] [CrossRef]
- Daker, M.; Lin, V.Y.; Akowuah, G.A.; Yam, M.F.; Mariam, A. Inhibitory effects of Cinnamomum burmannii Blume stem bark extract and trans-cinnamaldehyde on nasopharyngeal carcinoma cells; synergism with cisplatin. Exp. Ther. Med. 2013, 5, 1701–1709. [Google Scholar] [CrossRef]
- Meng, X.; Dong, X.; Wang, W.; Yang, L.; Zhang, X.; Li, Y.; Chen, T.; Ma, H.; Qi, D.; Su, J. Natural borneol enhances paclitaxel-induced apoptosis of ESCC cells by inactivation of the PI3K/AKT. J. Food Sci. 2018, 83, 1436–1443. [Google Scholar] [CrossRef]
- Shan, B.; Cai, Y.; Brooks, J.; Corke, H. Antibacterial properties and major bioactive components of cinnamon stick (Cinnamomum burmannii): Activity against foodborne pathogenic bacteria. J. Agric. Food Chem. 2007, 55, 5484–5490. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, Z.; Xie, J.; Chen, G.; Zeng, L.; Lin, C.; Luo, W. The study on selection and breeding of the high D-borneol content Cinnamomum burmannii. For. Sci. Technol. 2017, 8, 17–19. [Google Scholar]
- Chen, D.; Ye, H.; Li, G.; Liu, Y. Advances in molecular biology of plant isoprenoid metabolic pathway. Acta Bot. Sin. 2005, 42, 551–558. [Google Scholar]
- Cheng, A.; Lou, Y.; Mao, Y.; Lu, S.; Wang, L.; Chen, X. Plant terpenoids: Biosynthesis and ecological functions. J. Integr. Plant Biol. 2007, 49, 179–186. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Wang, Y.H.; Shen, S.K. Transcriptome analysis of Cinnamomum chago: A revelation of candidate genes for abiotic stress response and terpenoid and fatty acid biosyntheses. Front. Genet. 2018, 9, 505. [Google Scholar]
- Kalra, S.; Puniya, B.L.; Kulshreshtha, D.; Kumar, S.; Kaur, J.; Ramachandran, S.; Singh, K. De novo transcriptome sequencing reveals important molecular networks and metabolic pathways of the plant, Chlorophytum borivilianum. PLoS ONE 2013, 8, e83336. [Google Scholar] [CrossRef]
- Nautiyal, A.K.; Gani, U.; Sharma, P.; Kundan, M.; Misra, P. Comprehensive transcriptome analysis provides insights into metabolic and gene regulatory networks in trichomes of Nicotiana tabacum. Plant Mol. Biol. 2020, 102, 625–644. [Google Scholar] [CrossRef]
- Wang, T.; Chen, Y.; Zhuang, W.; Zhang, F.; Yang, Q. Transcriptome Sequencing Reveals Regulatory Mechanisms of Taxol Synthesis in Taxus wallichiana var. Mairei. Int. J. Genom. 2019, 2019, 1596895. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Ge, G.; Xiao, P.; Zhang, Y.; Yang, L. The first insight into the tissue specific taxus transcriptome via Illumina second-generation sequencing. PLoS ONE 2011, 6, e21220. [Google Scholar] [CrossRef]
- Li, J.; Harata-Lee, Y.; Denton, M.D.; Feng, Q.; Rathjen, J.R.; Qu, Z.; Adelson, D.L. Long read reference genome-free reconstruction of a full-length transcriptome from Astragalus membranaceus reveals transcript variants involved in bioactive compound biosynthesis. Cell Discov. 2017, 3, 17031. [Google Scholar] [CrossRef] [Green Version]
- Fu, M.; Lu, Z.; Ma, X. Enhanced extraction efficiency of natural D-borneol from Mei Pian tree leaves pretreated with deep eutectic solvents. Food Sci. Nutr. 2020, 8, 3806–3813. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.J.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zhu, L. Recent advances in the research of essential oil resources of Cinnamomum B1. in China. J. Plant Resour. Environ. 1994, 3, 51–55. [Google Scholar]
- Chen, C.; Zheng, Y.; Zhong, Y.; Wu, Y.; Xu, M. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genom. 2018, 19, 550–564. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, J.; Zhang, B.; Jin, X.; Jin, Z. Transcriptional analysis of metabolic pathways and regulatory mechanisms of essential oil biosynthesis in the leaves of Cinnamomum camphora (L.) Presl. Front. Genet. 2020, 11, 598714. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wu, Y.; Xiao, F.; Xiong, Z.; Xu, H. Transcriptome analysis for leaves of five chemical types in Cinnamomum camphora. Yi Chuan 2014, 36, 58–68. [Google Scholar]
- Yan, K.; Wei, Q.; Feng, R.; Zhou, W.; Chen, F. Transcriptome analysis of Cinnamomum longepaniculatum by high-throughput sequencing. Electron. J. Biotechnol. 2017, 28, 58–66. [Google Scholar] [CrossRef]
- Yang, Z.; An, W.; Liu, S.; Huang, Y.; Xie, C.; Huang, S.; Zheng, X. Mining of candidate genes involved in the biosynthesis of dextrorotatory borneol in Cinnamomum burmannii by transcriptomic analysis on three chemotypes. Peer J. 2020, 8, e9311. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Wang, X.; Zheng, Y.; Wang, H.; Liu, X.; Su, X. Full-Length transcriptome sequencing and different chemotype expression profile analysis of genes related to monoterpenoid biosynthesis in Cinnamomum porrectum. Int. J. Mol. Sci. 2019, 20, 6230. [Google Scholar] [CrossRef]
- Bohlmann, J.; Meyer-Gauen, G.; Croteau, R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1998, 95, 4126–4133. [Google Scholar] [CrossRef]
- Trapp, S.C.; Croteau, R.B. Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 2001, 158, 811–832. [Google Scholar] [CrossRef]

| Item | Answer |
|---|---|
| Total base(bp) | 36,323,230,896 |
| average length | 2181 |
| N50 | 2610 |
| Number of reads | 458,065 |
| Number of CCS bases | 1,273,679,005 |
| CCS Read Length (mean) | 2780 |
| Number of Passes (mean) | 8 |
| Number of unpolished consensus isoforms | 20,796 |
| 2499.751 | Mean unpolished consensus isoforms read length |
| Number of polished high-quality isoforms | 20,582 |
| Number of polished low-quality isoforms | 199 |
| Total Number | Total Length (bp) | Maximum Length (bp) | Minimum Length (bp) | Average Length (bp) | N50 Length (bp) | GC Content (%) |
|---|---|---|---|---|---|---|
| 17,116 | 42,907,024 | 12,292 | 86 | 2506.84 | 2749 | 44.04 |
| Sample | Raw Data (bp) | Clean Data (bp) | Q20 | Q30 | GC Content (%) |
|---|---|---|---|---|---|
| M1 | 7,228,626,300 | 7,187,544,129 | 6,927,661,015 (96.38%) | 6,494,362,280 (90.36%) | 48.56 |
| M2 | 6,788,385,300 | 6,750,745,242 | 6,482,068,702 (96.02%) | 6,041,734,157 (89.50%) | 48.91 |
| M3 | 7,040,023,800 | 6,999,531,017 | 6,741,621,103 (96.32%) | 6,315,230,155 (90.22%) | 48.85 |
| Y1 | 6,238,760,400 | 6,201,129,378 | 5,953,219,753 (96.00%) | 5,556,042,709 (89.60%) | 48.46 |
| Y2 | 6,938,253,600 | 6,898,066,477 | 6,616,909,592 (95.92%) | 6,166,342,127 (89.39%) | 48.47 |
| Y3 | 6,646,860,900 | 6,620,075,722 | 6,377,116,047 (96.33%) | 5,976,061,626 (90.27%) | 49.17 |
| Gene Name | Primer Sequence | Amplification Efficiency (%) | R2 | Annealing Temperature (°C) |
|---|---|---|---|---|
| Isoform0005426 | F:AGACGGTTGCTGTTGGAGTT R: CACCATCCACCCCTTTGTCA | 98.6 | 0.999 | 56 |
| Isoform0002990 | F: GGCATCAAGCCATCAACT R:AGAATCGCTGGAGAATCAT | 101.7 | 0.997 | 56 |
| Isoform0005726 | F: GCATCCAAACTGGCAAGA R: CAATCGGAGAACCTCAAATA | 95.4 | 0.999 | 56 |
| Isoform0007560 | F: GAAAGCAAGGGAAGAGGT R: GTGGATACAATCGGAGAAC | 98.8 | 0.995 | 56 |
| Isoform0008306 | F:TAGTTCAGACGGCGAAGGAC R: GCTCAGATGCGGCAAGTG | 95.5 | 0.997 | 56 |
| Isoform0008315 | F: GTATTCCCGTTGTTGATG R: ATTTGACCTAACCGTGCC | 97.6 | 0.996 | 56 |
| Isoform0008424 | F: CGCCCTTCATCTCAACCT R: GCTCGTCCCACCAATACTT | 97.5 | 0.992 | 56 |
| Isoform0008609 | F: GAAAGCAAGGGAAGAGGT R: GTGGATACAATCGGAGAAC | 96.8 | 0.993 | 56 |
| Isoform0008823 | F: GAAAGCAAGGGAAGAGGT R: GTGGATACAATCGGAGAAC | 100.6 | 0.999 | 56 |
| Isoform0009335 | F:TCAATAATCAGGGCAACACT R:TCTCATCCTACGCAACACC | 99.8 | 0.996 | 56 |
| Isoform0009336 | F: ACTCCACCTTACTTTCATCT R: CTTATTAGCACGGTTTCC | 99.9 | 0.998 | 56 |
| Isoform0016328 | F:GAGCGATGACAGTGAAAGCG R: CCTTCATCGGCCAAATCCCT | 100.8 | 0.992 | 56 |
| Isoform0016988 | F: TCCACCCTTTTATCCCATC R: TGCTCTGACAAGCCCAAT | 103.2 | 0.999 | 56 |
| Species Name | Accession No. | Gene Name | Rename |
|---|---|---|---|
| Cinnamomum micranthum | RWR72162.1 | Short-chain dehydrogenase/reductase SDR | C.micranthum_SDR |
| Thalictrum thalictroides | KAF5191451.1 | Salutarine reductase | T.thalictroides_SalR |
| Chelidonium majus | ACN87274.1 | Short-chain dehydrogenase/reductase, partial | C.majus_SDR |
| Morella rubra | KAB1199009.1 | Salutarine reductase | M.rubra_SalR |
| Lavandula latifolia | ABD77417.1 | linalool synthase | L.latifolia_LIS |
| Nandina domestica | ACN87275.1 | Short-chain dehydrogenase/reductase | N.domestica_SDR |
| Hevea brasiliensis | XP_021653042.1 | (+)-neomenthol dehydrogenase-like | H.brasiliensis_NMD |
| Vitis vinifera | RVW27891.1 | (+)-neomenthol dehydrogenase | V.vinifera_NMD |
| Manihot esculenta | XP_021615637.1 | (+)-neomenthol dehydrogenase | M.esculenta_NMD |
| Nicotiana tabacum | AAA19216.1 | 5-epi-aristolochene synthase | N.tabacum_EAS |
| Cinnamomum micranthum | RWR82502.1 | squalene oxygenase-like protein isoform X2 | C.micranthum_SO |
| Cinnamomum micranthum | RWR82644.1 | squalene synthase | C.micranthum_SQS |
| Cinnamomum micranthum | RWR94586.1 | terpenoid synthase 1 | C.micranthum_TPS |
| Chlorophytum borivilianum | AFN61200.1 | squalene epoxidase | C.borivilianum_SE |
| Zea mays | ONL95392.1 | squalene epoxidase 1 | Z.mays_SE |
| Phoenix dactylifera | XP_038983584.1 | squalene oxygenase SE1 | P.dactylifera_SO |
| Chimonanthus grammatus | AYP73106.1 | squalene synthase | C.grammatus_SQS |
| Chimonanthus zhejiangensis | AYP73108.1 | squalene synthase | C.zhejiangensis_SQS |
| Magnolia officinalis | AMK48128.1 | squalene synthase | M.officinalis_SQS |
| Nigella sativa | AMA66327.1 | squalane synthase 1 | N.sativa_SQS |
| Populus trichocarpa | XP_002313765.1 | squalene synthase | P.trichocarpa_SQS |
| Cananga odorata | QMW48843.1 | terpenoid synthase 2 | C.odorata_TPS |
| Liquidambar formosana | AIO10964.1 | TPS01 | L.formosana_TPS |
| Vitis riparia | XP_034678035.1 | (−)-germacrene D synthase-like | V.riparia_GDS |
| Vitis vinifera | RVW94686.1 | (−)-germacrene D synthase | V.vinifera_GDS |
| Cinnamomum micranthum | RWR88021.1 | terpene geranyllinalool synthase | C.micranthum_GLS |
| Laurus nobilis | AKQ19359.1 | terpene geranyllinalool synthase | L.nobilis_GLS |
| Vitis riparia | XP_034698703.1 | (E,E)-geranyllinalool synthase-like | V.riparia_GLS |
| Macadamia integrifolia | XP_042496091.1 | (E,E)-geranyllinalool synthase | M.integrifolia_GLS |
| Vitis vinifera | NP_001268201.1 | (E,E)-geranyllinalool synthase-like | V.vinifera_GLS |
| Populus trichocarpa | XP_024454968.1 | (E,E)-geranyllinalool synthase | P.trichocarpa_GLS |
| Theobroma cacao | EOY28337.1 | P(E)-nerolidol/(E,E)-geranyl linalool synthase | T.cacao_GLS |
| Pistacia vera | XP_031276291.1 | ent-kaurenoic acid oxidase 1-like | P.vera_KAO |
| Populus trichocarpa | XP_002318613.3 | ent-kaurenoic acid oxidase 1 isoform X3 | P.trichocarpa_KAO |
| Rosa chinensis | XP_024174916.2 | beta-amyrin 11-oxidase | R.chinensis_BAO |
| Cinnamomum micranthum | RWR88499.1 | ent-kaur-16-ene synthase, chloroplastic isoform X1 | C.micranthum_KES |
| Macleaya cordata | OVA15215.1 | Terpenoid synthase | M.cordata_TPS |
| Nigella sativa | QLI42521.1 | terpenoid synthase-like 2 protein, partial | N.sativa_TPS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Liang, J.; Deng, Z.; Lu, Z.; Fu, M.; Su, J. Full-Length Transcriptome Sequencing Combined with RNA-Seq to Analyze Genes Related to Terpenoid Biosynthesis in Cinnamomum burmannii. Curr. Issues Mol. Biol. 2022, 44, 4197-4215. https://doi.org/10.3390/cimb44090288
Guo S, Liang J, Deng Z, Lu Z, Fu M, Su J. Full-Length Transcriptome Sequencing Combined with RNA-Seq to Analyze Genes Related to Terpenoid Biosynthesis in Cinnamomum burmannii. Current Issues in Molecular Biology. 2022; 44(9):4197-4215. https://doi.org/10.3390/cimb44090288
Chicago/Turabian StyleGuo, Siyuan, Jiahao Liang, Zhiwei Deng, Ziqing Lu, Minghui Fu, and Jianyu Su. 2022. "Full-Length Transcriptome Sequencing Combined with RNA-Seq to Analyze Genes Related to Terpenoid Biosynthesis in Cinnamomum burmannii" Current Issues in Molecular Biology 44, no. 9: 4197-4215. https://doi.org/10.3390/cimb44090288
APA StyleGuo, S., Liang, J., Deng, Z., Lu, Z., Fu, M., & Su, J. (2022). Full-Length Transcriptome Sequencing Combined with RNA-Seq to Analyze Genes Related to Terpenoid Biosynthesis in Cinnamomum burmannii. Current Issues in Molecular Biology, 44(9), 4197-4215. https://doi.org/10.3390/cimb44090288





