3.1. Paralog-Specific Phenotypes Are Observed in Multiple Pairs of Ribosome Paralog Deletions in Fission Yeast
We initially aimed at elucidating the genetic pathways that influence the epigenetic stability of the centromeric chromatin organization in fission yeast [
21]. To this end, we performed a genetic screen in an annotated gene deletion haploid strain collection, using an established centromere position-effect variegation (CEN-PEV) system as the readout [
22]. In this system, the variegated expression of the ade6 reporter gene inserted in Centromere 2 (cnt2::ade6) is visualized by the coloration of individual colonies grown with a low supply of adenine. The white or red color of the colony corresponds to the ON or OFF state of ade6 expression, respectively. We previously have shown that CENP-A/Cnp1 nucleosome occupancy on ade6 directly correlates with its silencing and that the CEN-PEV effect reflects the epigenetic stability of Cnp1 nucleosome positioning within the centromere throughout mitotic cell generations [
23]. We reasoned that any mutation altering CEN-PEV implies that the mutated gene is functionally involved in regulating centromere epigenetic stability directly or indirectly. Based on the altered coloration patterns of the colonies, almost half of the tested RP deletions (27/54) were found to change the colony coloration pattern.
In the pombe genome, 140 genes encode 78 ribosomal proteins (RPs) (
http://ribosome.med.miyazaki-u.ac.jp, accessed on 11 December 2007), with 56 RPs encoded by two or more paralogous genes. Among 21 paralog pairs tested in this study, 10 pairs exhibited a paralog-specific phenotype in CEN-PEV (
Table S1), suggesting that these paralogous genes differently impact centromere epigenetic stability. Such broad paralog specificity in RPs is in agreement with the findings in the budding yeast S. cerevisiae, in which various phenotype-based RP paralog specificities were identified and have led to the hypothesis of the “ribosome code” [
11].
To test whether the functional differences between pombe RP paralogs are in the context of ribosomes or not, we further characterized the phenotypes of the paralog deletions in cellular processes that are directly linked to ribosomal functions.
It is known that, in fission yeast, high-quality nitrogen sources such as NH
4+ inhibit the uptake of poor nitrogen sources such as amino acids [
24]. For auxotrophic strains leu1-32 and ura4D-18, which cannot synthesize leucine and uracil, respectively, excessive NH
4+ effectively causes leucine or uracil starvation. To compare their tolerance to starvation, we assessed the growth rates of the RP paralog deletions (under the proper auxotrophic genetic background) on YE media supplied with excess NH
4Cl. In comparison to wild-type cells, the mutants were categorized into three types by their responses to excess NH
4Cl: wild type-like, more resistant, or more sensitive to NH
4Cl than WT. Among 26 pairs tested in total, 13 pairs with sister paralog deletions were sorted into different categories (
Table S1), suggesting that, for these 13 RPs, the paralogs function differently in ribosomes so that their deletions exhibit different responses to amino acid starvation.
We also examined the growth of the RP paralog deletion mutants on YE media supplied with sublethal concentrations of cycloheximide, a potent protein synthesis inhibitor that blocks translational elongation by interfering with tRNA translocation on the ribosome. We found that, in 10 pairs, the paralog deletion mutants exhibited different levels of sensitivity (
Table S1, 16 pairs tested in total).
Noticeably, Rps8/eS8, Rps11/uS17, Rps23/uS12, Rps28/eS28, Rpl28/uL15, and Rpl43/eL43 exhibited significant paralog specificity in the phenotypes by all three surveys above. Thus, consistent with previous studies on the budding yeast, our genetic study in fission yeast revealed that RP paralog specificity broadly exists in multiple biological processes in this organism, probably in the context of ribosome functions.
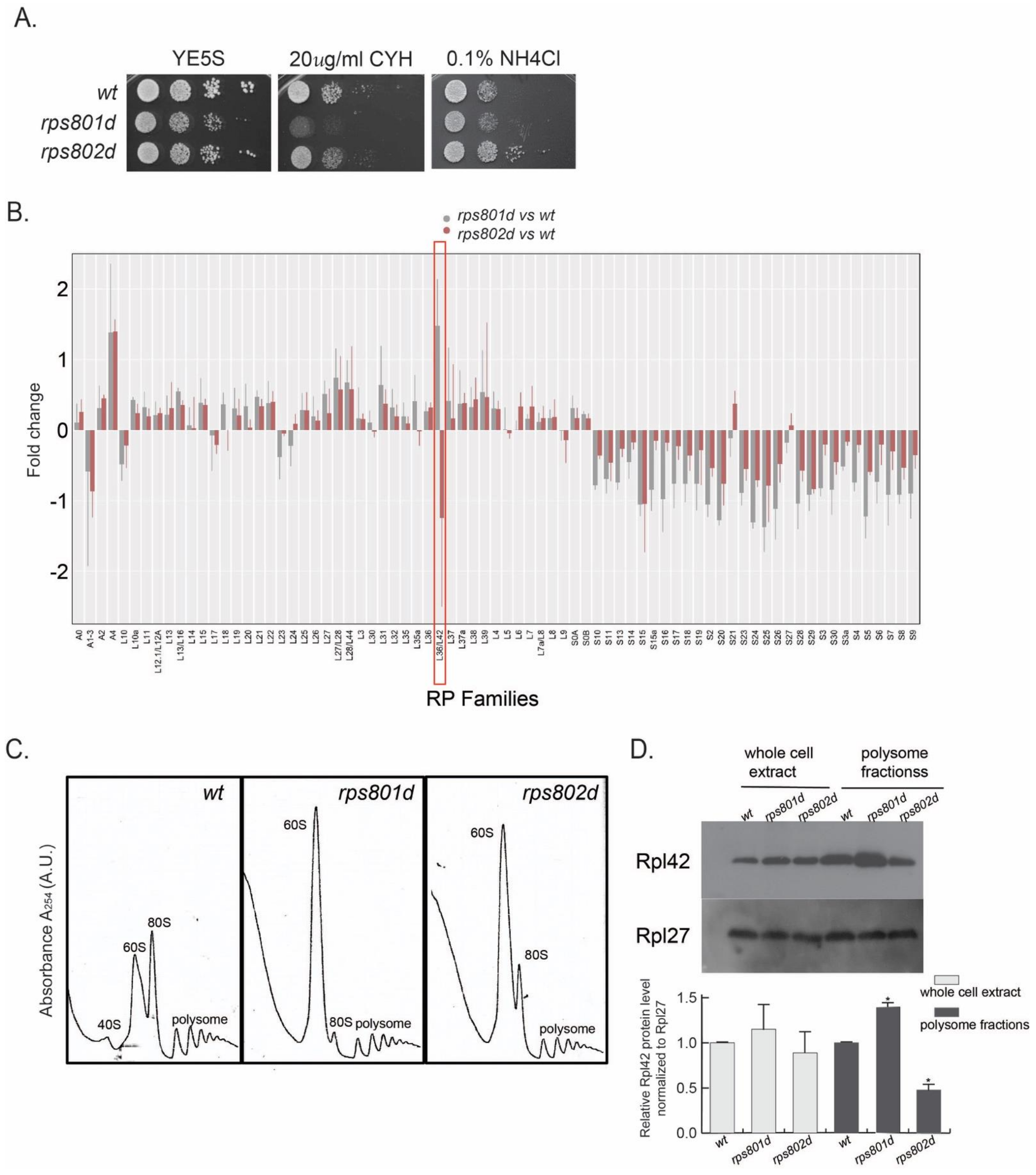
Among the tested RPs, the rps8 paralog deletions overall exhibited the highest contrast in phenotypical difference (
Figure 1A and
Figure S1). In the CEN-PEV assay, rps801d colonies showed a significant reduction in the frequency of color switching, while rps802d was wild type-like. Upon amino acid starvation, rps801d was moderately sensitive, while rps802d was resistant to amino acid starvation in comparison to the wild type. In addition, rps801d exhibited a higher sensitivity to cycloheximide than the wild type or rps802d; the latter two were indistinguishable under the tested conditions. We thus focused on rps8 paralogs to further investigate the molecular mechanisms underlying their phenotypic divergence.
3.2. Rps8 Deletions Impact on the Level of Large Subunit Protein Rpl42p/eL42 in Actively Translating Ribosome in a Paralog-Specific Manner
We hypothesized that the RP paralog deletions above, via differentially affecting the protein synthesis machinery, cause specific changes in the proteome, which eventually lead to the distinct phenotypes. We wished to identify the target proteins whose abundances were altered differentially due to rps8 paralog deletions. To do so, we quantified the relative abundance of individual proteins in whole-cell protein extracts, comparing between wild type and rps801d or rps802d, respectively, using quantitative mass spectrometry (LC-MS/MS; the details are described in
Section 2).
Proteins whose abundances were significantly altered (≥two-fold) in rps8 paralog deletion strains were shown (
Figure S2). Interestingly, the ribosomal proteins that were most frequently detected by mass spectrometry consistently exhibited significant alterations in abundance. Specifically, the abundances of all the small subunit proteins (RPSs) were consistently decreased in both rps8 paralog deletions compared to the wild type, whereas that of most large subunit proteins (RPLs) had no significant change (
Figure 1B). The sole exception of this trend was the large subunit component Rpl42p/eL42, whose abundance was increased significantly in rps801d but decreased in rps802d compared to the wild type.
In order to determine whether the differential Rpl42p/eL42 protein levels in rps8 paralog deletions reflected the differences in the ribosome compositions, cell lysates were subjected to a polysome profiling analysis by sucrose gradient ultracentrifugation, separating free RPs from those incorporated into ribosome subunits (40S and 60S) or 80S ribosomes and polysomes. Polysome profiling detected a diminished level of 40S small subunits but a prominent accumulation of 60S large subunits in both rps801d and rps802d compared to the wild type (
Figure 1C and
Figure S3). This is consistent with a recent report of the accumulation of RPLs in the budding yeast rps mutants, although the accumulation of 60S there does not seem as drastic [
25,
26]. Firstly, the Rpl42p/eL42 levels in each fraction separated by polysome profiling were determined by Western blotting using an antibody specifically against Rpl42p/eL42. The result showed that the Rpl42p/eL42 protein is present and accumulated in the 60S fraction, as well as 80S and polysome fractions in both rps8 paralog deletions (
Figure S3). Next, the Rpl42p/eL42 levels were measured in the whole-cell lysate or actively translating ribosomes. The result showed that the Rpl42p/eL42 level in the polysome fractions was increased in rps801d compared to that in the wild type but decreased in rps802d (
Figure 1D), suggesting the Rpl42p/eL42 level is varied in actively translating ribosomes between rps8 paralog deletions. Intriguingly, no significant difference in the total Rpl42p/eL42 level could be detected consistently in the whole-cell extracts of rps8 paralog deletions by Western blotting, which might be due to compensatory variations in free Rpl42p/eL42 in the cytoplasm and/or technical differences between mass spectrometry (
Figure 1B) and Western blotting.
Together, these results demonstrate that, while deletions of Rps8/eS8 paralogs indiscriminately affect the overall ribosome assembly at comparable levels, they impose opposite effects on the level of Rpl42/eL42 incorporation in intact ribosomes.
3.3. Differential Rpl42/eL42 Levels Contribute to Paralog-Specific Sensitivity to Amino Acid Starvation in rps8 Deletion Mutants
To test whether differential impacts on the Rpl42/eL42 level in ribosomes by rps8 paralog deletions may account for their phenotypical differences, we took a genetic approach to investigate the possible functional connection between Rpl42/eL42 and Rps8/eS8 paralogs.
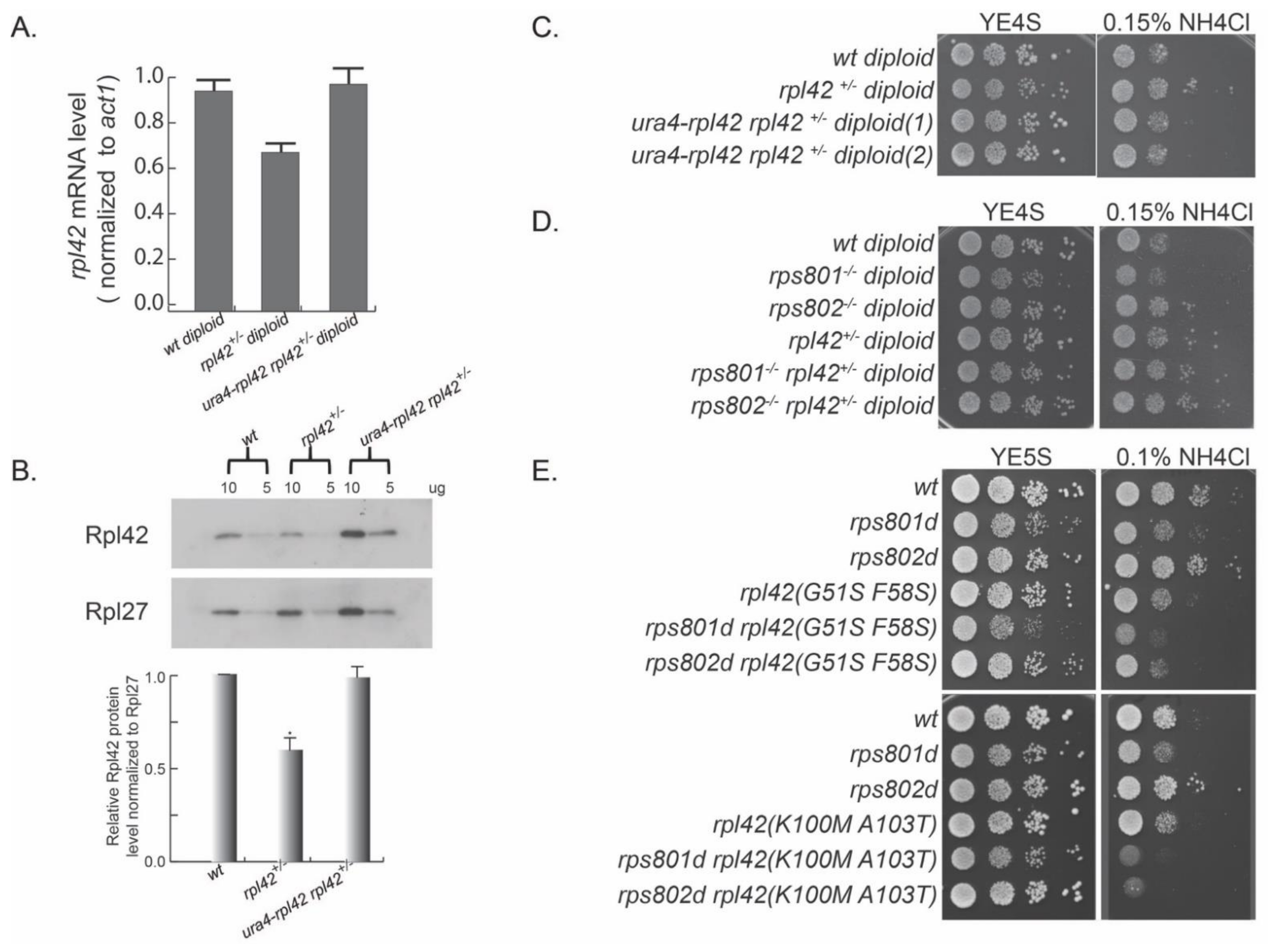
We first constructed a heterozygous rpl42 deletion (rpl42
+/−) diploid strain with the standard yeast molecular genetic methods. We induced meiosis in rpl42
+/− and performed a tetrad dissection for the meiosis progeny. The result showed that rpl42 gene deletion is lethal in haploids, demonstrating that rpl42 is an essential gene (
Figure S4A), in contrast to the result derived from systematic genome-wide gene deletion characterization [
27].
We speculated that different levels of Rpl42/eL42 in ribosomes may be the primary molecular alteration that eventually causes phenotypical differences in Rps8/eS8 paralog deletion mutants. To this end, we tested whether altering rpl42 expression was sufficient to mimic the phenotypes of rps8 paralog deletions or not. We first measured the Rpl42/eL42 mRNA level in rpl42
+/− diploid by qRT-PCR and found that it was decreased to about 70% of the wild-type diploid level (
Figure 2A). Consistently, the Rpl42/eL42 protein level measured in rpl42
+/− cell extracts by Western blotting was also decreased to a comparable degree (
Figure 2B). Growing in YE supplied with excess NH
4Cl, the rpl42
+/− heterozygous diploid strain was resistant to this stress, displaying enhanced growth compared to the wild-type diploid strain (
Figure 2C). Conversely, the ectopic expression of rpl42 by inserting an extra copy of rpl42 ORF with its native promoter at the ura4 site (ura4::rpl42) strain reversed the expression level of rpl42 in the heterozygous rpl42
+/− diploid to the wild-type level. In addition, cell growth was reversed to the level of wild-type diploid cells with excess NH
4Cl.
In order to test the genetic interaction between rps8 paralog deletions and rpl42, we deleted one copy of the rpl42 gene in rps801
−/− homozygous diploid and rps802
−/− homozygous diploid. Consistent with the rps8 paralog deletion haploid cells, the rps801
−/− diploid cells showed sensitivity to excess NH
4Cl, and rps802
−/− diploid cells showed resistance to excess NH
4Cl (
Figure 2D). A single deletion of the rpl42 gene (rpl42
+/−) reversed the growth of rps801
−/− diploid cells in YE supplied with excess NH
4Cl. Ribosomal proteins, such as Rpl36A (homolog of Rpl42/eL42), cannot be overproduced in budding yeast, because the excess proteins are rapidly degraded [
28]. Consistently, while an extra copy of rpl42(ura4::rpl42) in wild-type fission yeast haploid cells did cause increased rpl42 transcription (
Figure S4B), nonetheless, no increase was detected in the levels of the total Rpl42/eL42 protein or ribosome-integrated Rpl42/eL42 by Western blotting (
Figure S4C). This precluded the test on whether increased Rpl42/eL42 incorporation in ribosomes could mimic the phenotypes of rps801d.
Together, these results support the notion that changes in the Rpl42p/eL42 level in rps8 paralog deletions contribute mainly, if not solely, to the paralog-specific phenotypes in response to amino acid starvation.
To further explore the genetic interactions between rpl42 and rps8 paralogs, we constructed a rpl42 random mutation library by mutagenic PCR and replaced the endogenous rpl42 gene in rps801d or rps802d haploid cells, respectively, via the standard procedure of yeast transformation/homologous recombination. We then screened for rpl42 mutation alleles that were able to reverse the phenotypes or reduce the phenotypic contrast in amino acid starvation (excess NH4Cl in YE media) of rps801d or rps802d.
Among over 1000 transformants covered in each genetic screen, rpl42(G51S and, F58S) and rpl42(K100M and A103T) mutations were identified to specifically suppress the enhanced growth of rps802d cells under the condition of YE supplied with excess NH
4Cl (
Figure 2E). All of these rpl42 mutations by themselves exhibited a wild type-like growth rate on YE with excess NH
4Cl. Together, these results showed that ribosome small subunit protein Rps8/eS8 paralogs have a strong genetic interaction with the ribosome large subunit protein Rpl42/eL42 in response to amino acid starvation.
3.4. Differential Transcription Level of Rps8/eS8 Paralogs but Not Protein Sequence Variation Results in the Divergent Phenotypes of Rps8/eS8 Paralogs
Rps801p and Rps802p vary in the protein sequence by only two residues—130S in Rps801p vs. 130A in Rps802p and 133N in Rps801p vs. 133T in Rps802p (
Figure 3A and
Figure S5A). On the other hand, the mRNA expression levels of rps801 and rps802 differed (55% and 45%) in wild-type cells, measured by cDNA deep sequencing (see
Section 2,
Figure S5B), presumably due to the difference in strength of their promoters. Thus, deletions of rps801 and rps802 would impose at least two types of changes. First, the protein identity of Rps8p within ribosomes is changed—a mixture of Rps801p and Rps802p for wild-type cells and pure Rps802p and Rps801p for rps801d and rps802d cells, respectively. Second, the total expression of rps8 may be reduced to different levels by paralog deletions.
To determine whether either of the two changes accounted for the phenotypical differences in the rps8 paralog deletion strains, we stepwise constructed a series of haploid strains in which Rps8p/eS8 was produced only in the Rps801p or Rps802p form (
Figure 3B), with the gene dosage either maintained at the wild-type level in the “homogenic” rps8 strains or reduced in the “chimeric” rps8 strains (see below).
We replaced the two non-conserved residues (130aa and 133aa) in one paralog for the corresponding residues of the other by site-directed mutagenesis. The resulting mutation alleles rps801mm (S130A and N133T) and rps802mm (A130S and T133N) encode the protein sequences of Rps802p and Rps801p, respectively. These “homogenic” strains, rps801mm and rps802mm, each harbor two copies of the Rps802p- and Rps801p-coding sequences at both native loci.
On the other hand, for comparing paralogs at a single-copy dosage, we normalized the difference in the expression levels between the paralogs. To do so, the rps802 and rps801 ORFs were deleted at their native loci in the rps801mm and rps802mm homogenic strains to yield “chimeric” strains (rps801mm rps802d and rps802mm rps801d); each had only one type of Rps8p/eS8, Rps802p, and Rps801p protein expressed under the promoter of their paralogous sister (hence, chimeric), respectively (see
Table S2 for the summary of the above strains).
Firstly, the Rps8/eS8 mRNA level of the above strains was measured by qRT-PCR (
Figure 3C). The results showed that the Rps8/eS8 mRNA level was decreased in rps801d and rps802mm rps801d to about 40% of the wild type and, in rps802d and rps801mm rps802d, to about 60% of the wild type, respectively. It suggests that the deletion strains, as well as the chimeric strains, have a similar mRNA level to Rps8/eS8. In addition, the Rps8/eS8 mRNA level in the homogenic strains was comparable or slightly decreased (rps801mm in
Figure 3C) compared to the wild-type strain.
We then measured the colony size as a proxy of the growth rate (
Figure 3D). Both single rps8 paralog deletions and chimeric strains exhibited a decreased colony size compared with the wild type. No significant difference was detected between rps801d and rps801d rps802mm, rps802d, and rps802d rps801mm. Additionally, the colony sizes of rps802d and rps802d rps801mm were larger than those of rps801d and rps801d rps801mm. These data suggest that, in mutant cells harboring only one rps8 allele, the rps801 locus has a bigger impact than the rps802 locus on cell growth, regardless of the produced protein being Rps801p or Rps802p. For homogenic strains (rps801mm and rps802mm) with two copies of the same paralog, the colony size was indistinguishable from that of the wild type.
These findings indicate that the genetic loci (and thereby, presumably, the promoter strengths) of the rps8 paralogous genes, rather than the particular Rps8/eS8 protein paralogs, affect the cell growth rate.
We then measured the Rpl42/eL42 levels of the actively translating ribosome (polysome fractions from polysome profiling) in the Rps8/eS8 homogenic and chimeric strains by Western blotting (
Figure 3E,F). The mutants harboring only one rps8 allele consistently had diminished levels of the 40S subunits, decreased levels of 80S and the polysomes, and a drastic accumulation of 60S subunits relative to the wild type. On the other hand, the polysome profiles of the homogenic rps801mm and rps802mm strains were similar to that of the wild-type strain.
Western blotting showed that the level of Rpl42p/eL42 was indistinguishable between rps801mm, rps802mm and the wild type. The Rpl42p/eL42 protein level was more abundant in rps802mm rps801d (expressing Rps801p under the promoter of rps802) than the wild type and even more than rps801d, which expressed Rps802p under the promoter of rps802. In addition, the Rpl42p/eL42 protein level was less abundant in rps801mm rps802d (expressing Rps802p under the promoter of rps801) than the wild type and was indistinguishable from rps802d, which expressed Rps801p under the promoter of rps801 (
Figure 3F). Thus, a reduction in rps8 expression to variegated levels caused the different levels in the Rpl42p/eL42 protein incorporation in the ribosomes among these strains.
In order to explore whether the differential expression level of Rps8 in the above mutants causes differential rpl42 transcription, which ultimately leads to different levels in the Rpl42p/eL42 protein incorporation into ribosomes, we measured the Rpl42/eL42 mRNA level in the above strains by qRT-PCR (
Figure S5C). However, no significant change in the rpl42 transcription level was detected in these mutants in comparison to the wild type.
Lastly, we determined whether the varied Rpl42p/eL42 protein levels caused by differential Rps8/eS8 transcription levels would also affect the responses to amino acid starvation. We examined the growth of these strains on YE media supplied with excess NH
4Cl (
Figure 3G) and found that the growth of both rps801mm and rps802mm homogenic strains were comparable to that of the wild type. rps802mm rps801d displayed the highest sensitivity to amino acid starvation among the tested strains, since the Rpl42p/eL42 protein level was the most abundant in this strain by the previous Western blotting results (
Figure 3F). In contrast, rps801mm rps802d was resistant to amino acid starvation, similar to rps802d. Thus, the differential Rps8/eS8 transcription level, but not the specific Rps8p/eS8 protein identity, contributed to the divergent phenotypes between rps8 paralog deletions and chimeric strains.
Altogether, our results demonstrated that the cause for divergent phenotypes of rps8 paralog deletions resides in the differential rps8 expression rather than the variations in the Rps8/eS8 paralog protein sequence.
3.5. Additional 40S Ribosomal Protein Paralog Gene Deletions also Cause Differential Levels of Rpl42/eL42
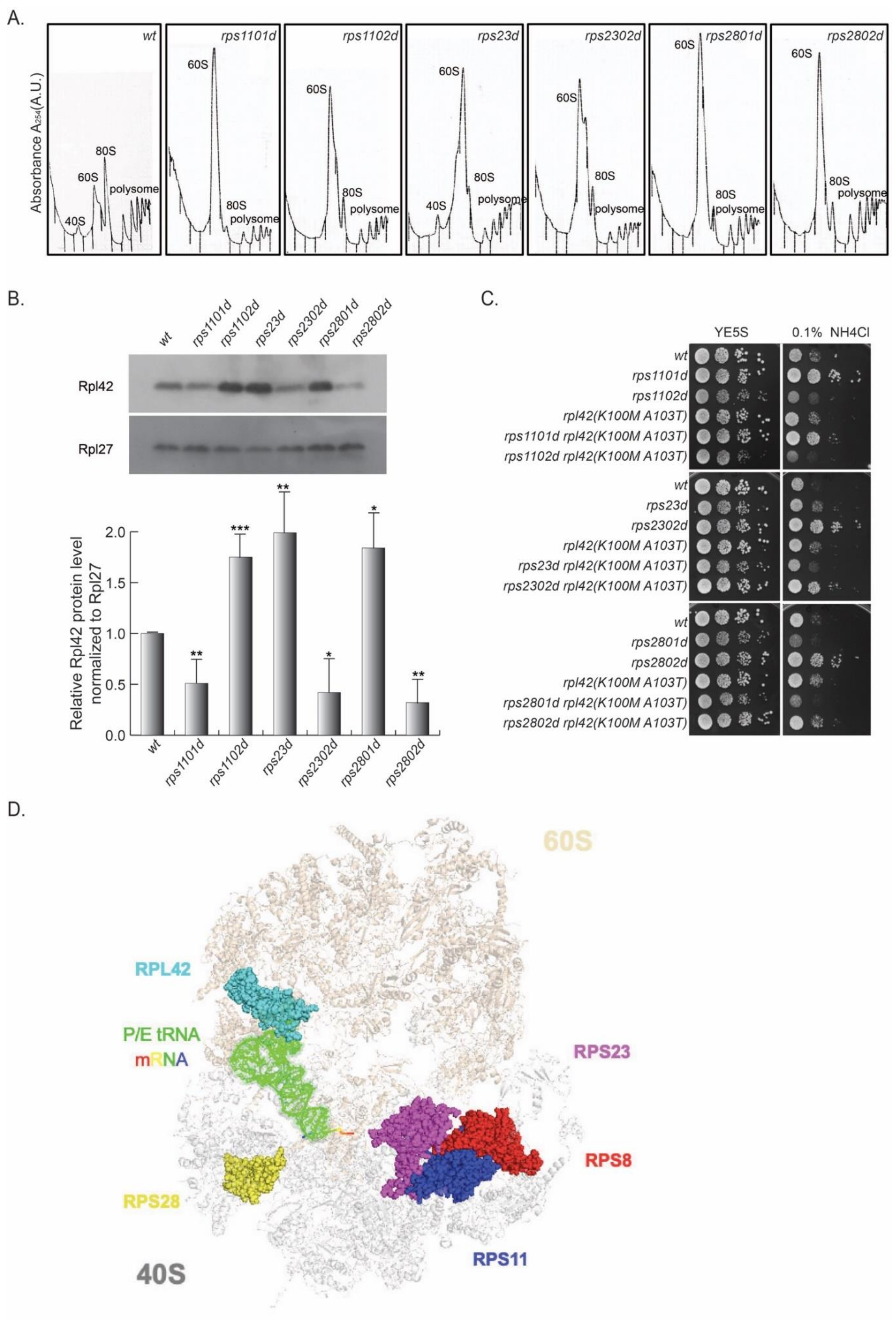
We wished to investigate whether the variations in Rpl42/eL42 level were broadly related to paralog-specific phenotypes in other RPs or not in addition to Rps8/eS8. Since the genetic interaction between Rpl42/eL42 and Rps8/eS8 was clearly reflected by the cell growth phenotype in the presence of excessive NH4Cl, we reasoned that this phenotype might serve as an indicator for other RPSs with paralog-specific effects on the Rpl42p/eL42 level.
In this vein, a collection of RP paralog deletions (
Table S1) was surveyed for differential growth rates at the presence of excess NH
4Cl. Compared to the wild type, rps1102d, rps2301d, and rps2801d were sensitive to excess NH
4Cl, whereas their corresponding paralog deletions (rps1101d, rps2302d, and rps2802d) were resistant (
Figure 4).
Next, the cell lysates of these strains were subjected to a polysome profiling analysis by sucrose gradient ultracentrifugation. Similar to rps8 paralog deletions, the abundance of 40S small subunits was diminished, whereas the 60S large subunits accumulated drastically in the rps11, rps23, and rps28 paralog deletions compared to the wild type (
Figure 4A). The actively translating ribosomes (the polysome fractions) of all six deletion strains, as well as wild type, were collected, and Rpl42p/eL42 levels were assessed by Western blotting. Higher levels of Rpl42p/eL42 were detected in rps1102d, rps23d, and rps2801d cells than in rps1101d, rps2302d, and rps2802d, respectively (
Figure 4B).
Furthermore, by introducing the rpl42(K100M and A103T) mutation in the rps11, rps23, and rps28 paralog deletion strains, the phenotype of the enhanced growth of rps1101d, rps2302d, and rps2802d in the excess of NH
4Cl was reversed, similar to that observed in rps802d. This genetic evidence suggests that the variegated Rpl42p/eL42 levels in the paralog deletions of these RPSs are responsible for the paralog-specific phenotypes, similar to that in rps8 (
Figure 4C).
According to the crystal structure of the yeast 80S ribosome [
29], Rps8/eS8, Rps11/uS17, Rps23/uS12, and Rps28/eS28 are all located at the interface of the 40S and 60S subunits (
Figure 4D). In order to test whether these RP paralog deletions are epistatic to the rps8 paralog deletions, we tested the growth of double mutants rps801d rps2801d, and rps802d rps2802d under the condition of YE supplied with excess NH
4Cl. The results showed that the double mutants did not exhibit a greater sensitivity or resistance to excess NH
4Cl than single mutants (
Figure S6A), suggesting that rps801d, rps2801d, rps802d, and rps2802d function in the same genetic pathway in response to amino acid starvation.
Rps11/uS17, Rps23/uS12, and Rps28/eS28 paralogous gene pairs encode the RP paralogs with identical sequences. This strongly suggests that the paralogs are unlikely to carry distinct biochemical activities to account for the differential incorporation of Rpl42/eL42. Rather, the variegated Rpl42/eL42 protein levels observed in these rps paralog deletions may be caused by differential RPS expression levels. Consistent with this, the single deletion of paralogous genes impacts the total expression level of the RP differently. By measuring the mRNA levels of each paralog upon deletions of its paralog partner in comparison to the wild-type cells (
Figure S6B), we found that the Rps11/uS17 mRNA level in rps1101d was decreased to about 90% of the wild type and, in rps1102d, to about 70% of the wild type, respectively. The Rps23/uS12 mRNA level in rps23d was decreased to about 50% of the wild type but increased in rps2302d to about 120% of the wild type, suggesting that the deletion of rps2302 may alter the expression of rps23.
Together, these results demonstrate that a variation in the Rpl42p/eL42 level is not restricted to rps8 paralog deletions but, rather, broadly underscores paralog-specific phenotypes of multiple RPS paralog deletions.
3.6. 60S Subunits Accumulation Is Independent to Varied Rpl42/eL42 Level in rps8 Paralogs
60S subunit accumulation was observed in rps deletion mutants, including rps8, rps11, rps23, and rps28 paralog deletion mutants (
Figure 1,
Figure 4 and
Figure S7). This accumulation might result either from a block in the maturation of 60S subunits or from the cellular tolerance of super-stoichiometric mature large subunits relative to small subunits. Ribosome assembly follows a discrete order in which RPs are integrated into precursors of large and small subunits at specific stages in conjunction with the precursors migrating from the nucleolus to the nucleoplasm and, eventually, to the cytoplasm in the cell. Rpl42p/eL42 is integrated into pre-60S at a late stage, immediately before or after pre-60Ss are exported to the cytoplasm [
30]. We wished to test whether 60S accumulation in the cytoplasm might contribute to differential Rpl42/eL42 incorporation in rps8 paralog deletions or not, perhaps by providing the time window necessary for creating such a difference.
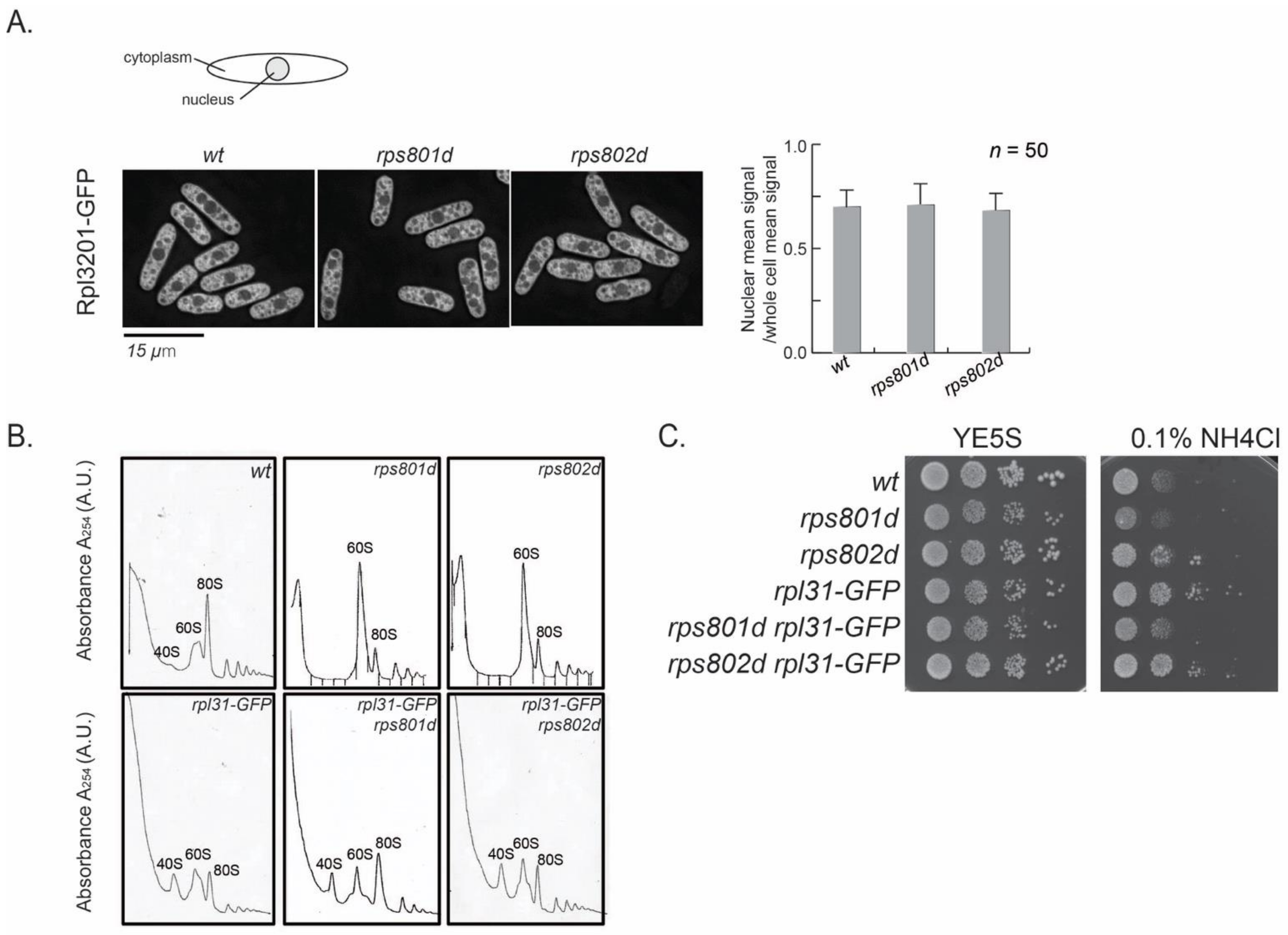
First, the process of ribosome assembly was examined microscopically. The Rpl3201-GFP fusion protein, which is integrated into pre-60S at an early step in the nucleus [
30], was used as an indicator for ribosome subunit precursors. In wild-type cells during exponential growth, nuclear GFP signal density represents the intermediates in the process of 60S large subunit assembly and is approximately half of the total Rpl3201-GFP in the cell (
Figure 5A). If 60S maturation were blocked in rps8 mutants, the accumulation of Rpl3201-GFP in the nucleus could be observed. The results showed that the subcellular distribution of Rpl3201-GFP was not accumulated in the nucleus in rps8 paralog deletions. The Rpl3201-GFP signals in the nucleus vs. whole cell were indistinguishable between rps8 paralog deletions and wild-type cells (
Figure 5A). It suggests that the accumulated 60S subunits detected by polysome profiling in the mutants were mature (or late-stage) large subunits and were exported into the cytoplasm. This is consistent with the mass spectrometry result in which the protein level of nearly all RPLs (except for Rpl42p/eL42) uniformly increased (
Figure 1B).
We then tested whether alleviating the 60S subunits accumulation in rps8 paralog deletions would suppress their differential Rpl42p incorporation phenotypes or not. Here, a Rpl31/eL31-GFP fusion construct was tested, because Rpl31/eL31 in fission yeast is encoded by a single gene and that GFP tagging partially compromises the functions of Rpl31 and, thus, alters the kinetics of 60S assembly (
Figure 5B, the reduced 80S peak of Rpl31-GFP in comparison to the wild type). The polysome profiling analysis showed that Rpl31/eL31-GFP indeed alleviates 60S subunit accumulation in both the rps801d and rps802d strains (
Figure 5B). However, rpl31-GFP did not affect the growth of rps801d or rps802d at the tested conditions (
Figure 5C). These results suggest that the suppression of 60S subunits accumulation is independent of the differential growth rates of rps8 paralog deletions in YE media supplied with excess NH
4Cl and, by extension, may be independent of the varied Rpl42/eL42 protein levels, the primary determinant to the differential growth rates in these strains (
Figure 1, above).