Association of High-Density Lipoprotein Cholesterol, Renal Function, and Metabolic Syndrome: An Assessment of the 2013–2018 National Health and Nutrition Examination Surveys
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sample
2.2. Demographic and Variable Information
2.3. Statistical Analyses
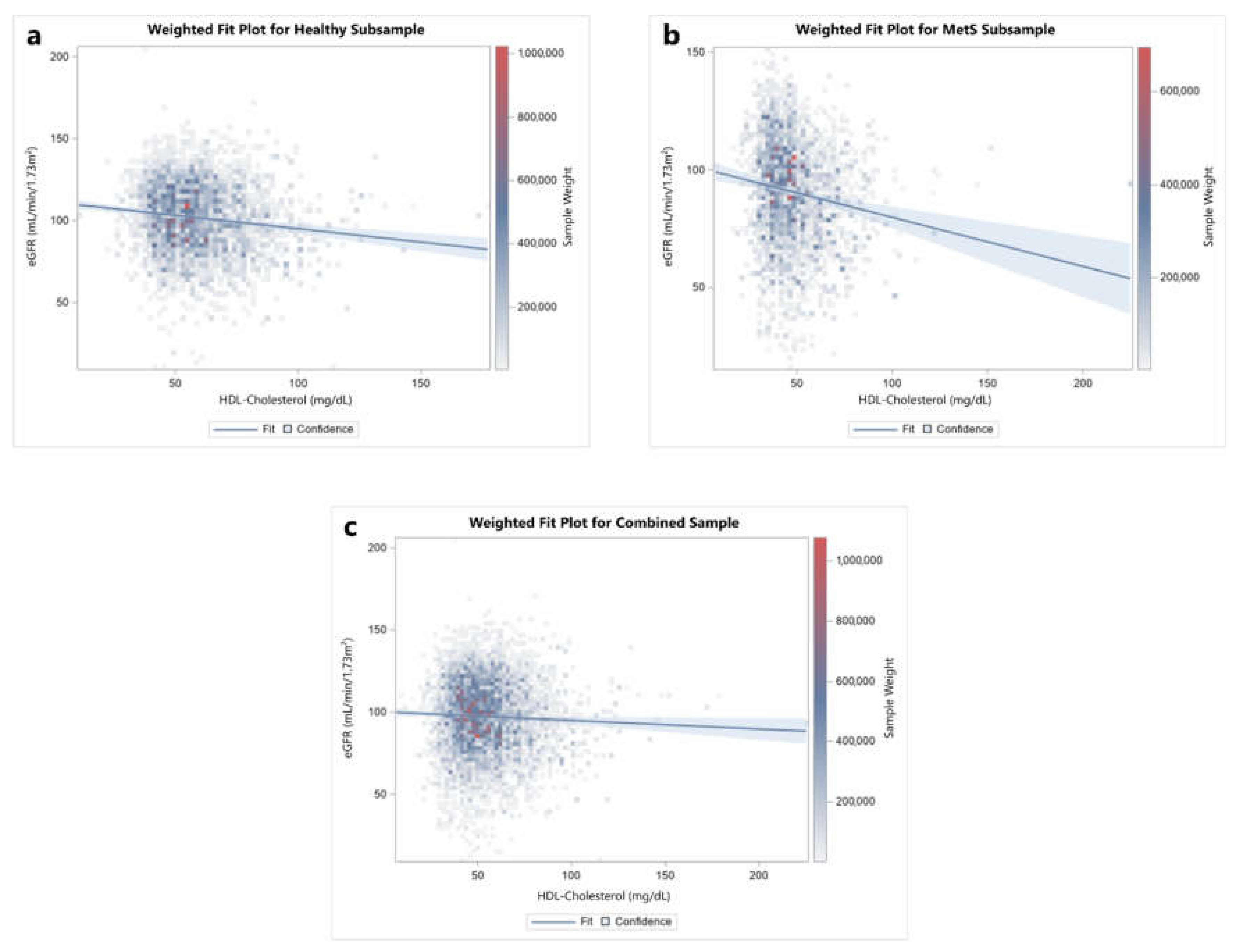
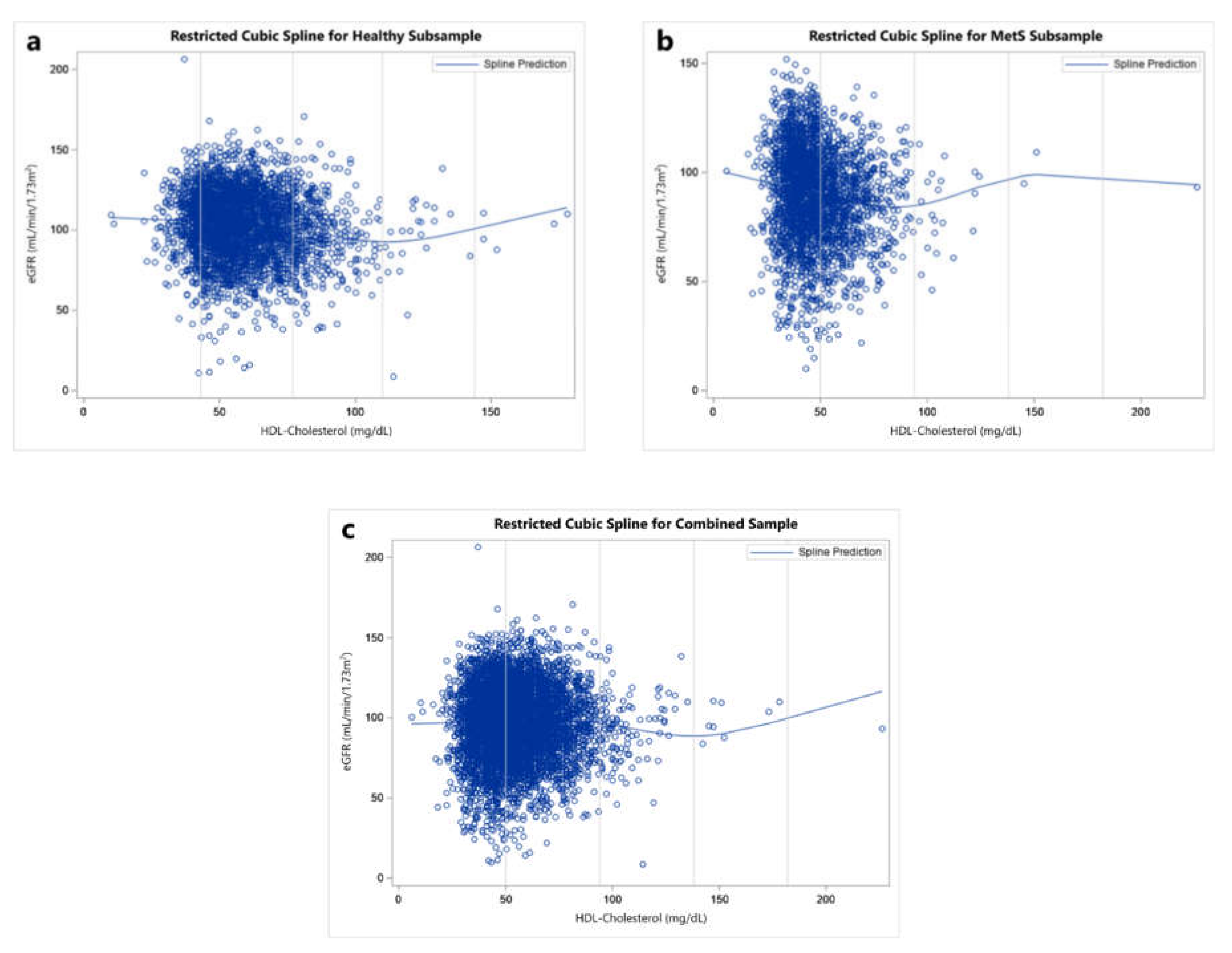
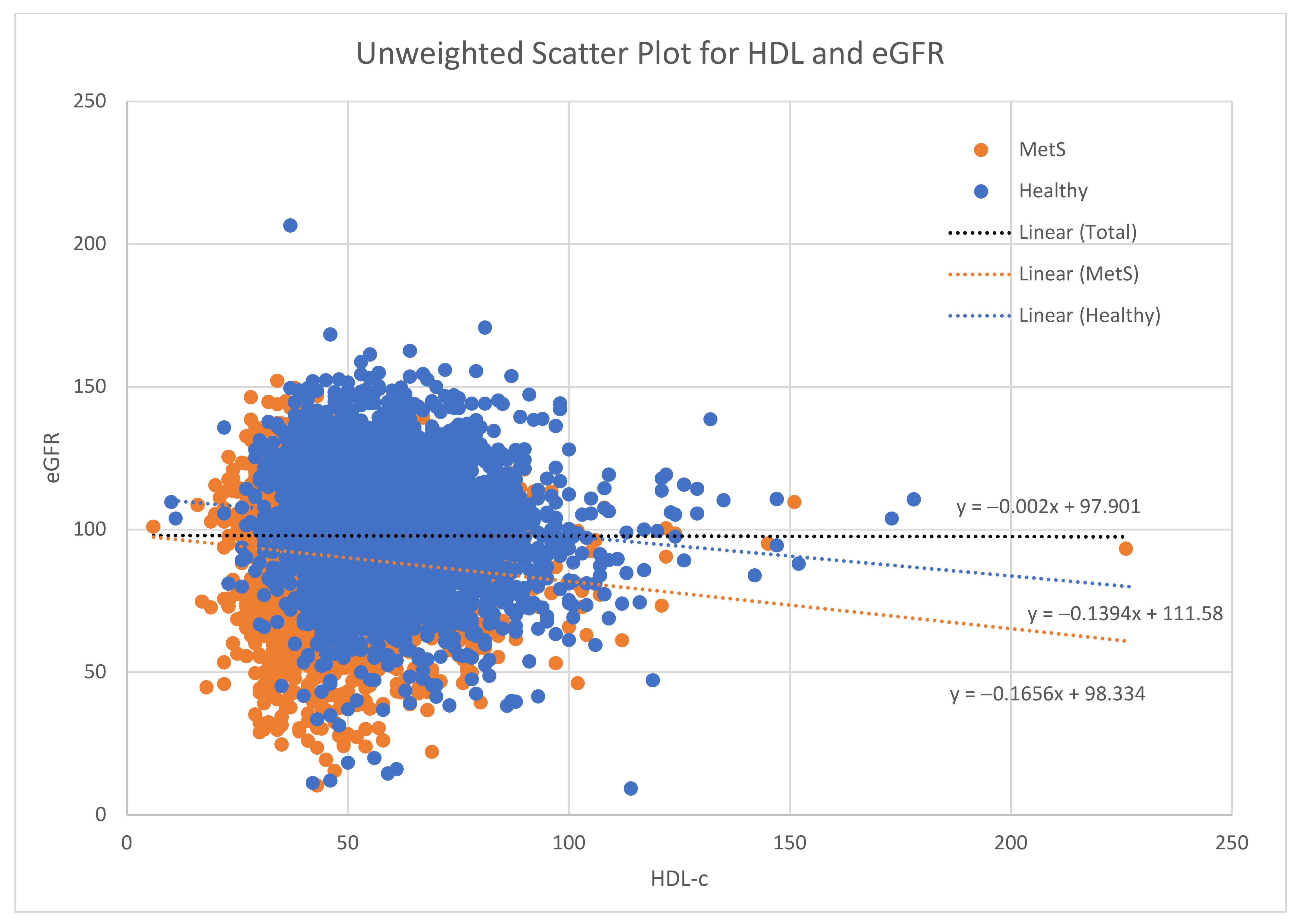
3. Results
4. Discussion
Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Chronic Kidney Disease in the United States, 2019; US Department of Health and Human Services, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Dunaief, D.M.; Fuhrman, J.; Dunaief, J.L.; Ying, G. Glycemic and cardiovascular parameters improved in type 2 diabetes with the high nutrient density (HND) diet. Open J. Prev. Med. 2012, 2, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Lim, E.L.; Hollingsworth, K.G.; Aribisala, B.S.; Chen, M.J.; Mathers, J.C.; Taylor, R. Reversal of type 2 diabetes: Normalisation of beta cell function in association with decreased pancreas and liver triacylglycerol. Diabetologia 2011, 54, 2506–2514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDougall, J.; Thomas, L.E.; McDougall, C.; Moloney, G.; Saul, B.; Finnell, J.S.; Richardson, K.; Petersen, K.M. Effects of 7 days on an ad libitum low-fat vegan diet: The McDougall Program cohort. Nutr. J. 2014, 13, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnard, N.D.; Cohen, J.; Jenkins, D.J.; Turner-McGrievy, G.; Gloede, L.; Green, A.; Ferdowsian, H. A low-fat vegan diet and a conventional diabetes diet in the treatment of type 2 diabetes: A randomized, controlled, 74-wk clinical trial. Am. J. Clin. Nutr. 2009, 89, 1588S–1596S. [Google Scholar] [CrossRef]
- Alberti, K.G.M.M.; Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z.; Cleeman, J.I.; Donato, K.A.; Fruchart, J.-C.; James, W.P.T.; Loria, C.M.; Smith, S.C. Harmonizing the Metabolic Syndrome. Circulation 2009, 120, 1640–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundy, S.M.; Cleeman, J.I.; Daniels, S.R.; Donato, K.A.; Eckel, R.H.; Franklin, B.A.; Gordon, D.J.; Krauss, R.M.; Savage, P.J.; Smith, S.C., Jr.; et al. Diagnosis and Management of the Metabolic Syndrome. Circulation 2005, 112, 2735–2752. [Google Scholar] [CrossRef] [Green Version]
- National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (adult Treatment Panel III): Final Report. Circulation 2002, 106, 3143–3421. [Google Scholar] [CrossRef]
- Voight, B.F.; Peloso, G.M.; Orho-Melander, M.; Frikke-Schmidt, R.; Barbalic, M.; Jensen, M.K.; Hindy, G.; Hólm, H.; Ding, E.L.; Johnson, T.; et al. Plasma HDL cholesterol and risk of myocardial infarction: A mendelian randomisation study. Lancet 2012, 380, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Keene, D.; Price, C.; Shun-Shin, M.J.; Francis, D.P. Effect on cardiovascular risk of high density lipoprotein targeted drug treatments niacin, fibrates, and CETP inhibitors: Meta-analysis of randomised controlled trials including 117 411 patients. BMJ 2014, 349, g4379. [Google Scholar] [CrossRef] [Green Version]
- Thomas, G.; Sehgal, A.R.; Kashyap, S.R.; Srinivas, T.R.; Kirwan, J.P.; Navaneethan, S.D. Metabolic Syndrome and Kidney Disease: A Systematic Review and Meta-analysis. Clin. J. Am. Soc. Nephrol. 2011, 6, 2364–2373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-B.; Sheng, L.-T.; Wei, W.; Guo, H.; Yang, H.; Min, X.; Guo, K.; Yang, K.; Zhang, X.; He, M.; et al. Association of blood lipid profile with incident chronic kidney disease: A Mendelian randomization study. Atherosclerosis 2020, 300, 19–25. [Google Scholar] [CrossRef]
- Lanktree, M.B.; Thériault, S.; Walsh, M.; Paré, G. HDL Cholesterol, LDL Cholesterol, and Triglycerides as Risk Factors for CKD: A Mendelian Randomization Study. Am. J. Kidney Dis. 2018, 71, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Lo, J.C.; Go, A.S.; Chandra, M.; Fan, D.; Kaysen, G.A. GFR, Body Mass Index, and Low High-Density Lipoprotein Concentration in Adults With and Without CKD. Am. J. Kidney Dis. 2007, 50, 552–558. [Google Scholar] [CrossRef]
- Navaneethan, S.D.; Schold, J.D.; Walther, C.P.; Arrigain, S.; Jolly, S.E.; Virani, S.S.; Winkelmayer, W.C.; Nally, J.V. High-density lipoprotein cholesterol and causes of death in chronic kidney disease. J. Clin. Lipidol. 2018, 12, 1061–1071.e7. [Google Scholar] [CrossRef] [PubMed]
- Bowe, B.; Xie, Y.; Xian, H.; Balasubramanian, S.; Al-Aly, Z. Low levels of high-density lipoprotein cholesterol increase the risk of incident kidney disease and its progression. Kidney Int. 2016, 89, 886–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowe, B.; Xie, Y.; Xian, H.; Balasubramanian, S.; Zayed, M.A.; Al-Aly, Z. High Density Lipoprotein Cholesterol and the Risk of All-Cause Mortality among U.S. Veterans. Clin. J. Am. Soc. Nephrol. 2016, 11, 1784–1793. [Google Scholar] [CrossRef] [Green Version]
- Krikken, J.A.; Gansevoort, R.T.; Dullaart, R.P.F. Lower HDL-C and apolipoprotein A-I are related to higher glomerular filtration rate in subjects without kidney disease. J. Lipid Res. 2010, 51, 1982–1990. [Google Scholar] [CrossRef] [Green Version]
- Melsom, T.; Norvik, J.V.; Enoksen, I.T.; Stefansson, V.; Rismo, R.; Jenssen, T.; Solbu, M.D.; Eriksen, B.O. Association of High-Density Lipoprotein Cholesterol With GFR Decline in a General Nondiabetic Population. Kidney Int. Rep. 2021, 6, 2084–2094. [Google Scholar] [CrossRef]
- Zanoni, P.; Khetarpal, S.A.; Larach, D.B.; Hancock-Cerutti, W.F.; Millar, J.S.; Cuchel, M.; DerOhannessian, S.; Kontush, A.; Surendran, P.; Saleheen, D.; et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science 2016, 351, 1166–1171. [Google Scholar] [CrossRef] [Green Version]
- Kwiterovich, P.O. The metabolic pathways of high-density lipoprotein, low-density lipoprotein, and triglycerides: A current review. Am. J. Cardiol. 2000, 86, 5–10. [Google Scholar] [CrossRef]
- Rosenson, R.S.; Brewer, H.B.; Ansell, B.J.; Barter, P.; Chapman, M.J.; Heinecke, J.W.; Kontush, A.; Tall, A.R.; Webb, N.R. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat. Rev. Cardiol. 2016, 13, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Ansell, B.J.; Navab, M.; Hama, S.; Kamranpour, N.; Fonarow, G.; Hough, G.; Rahmani, S.; Mottahedeh, R.; Dave, R.; Reddy, S.T.; et al. Inflammatory/Antiinflammatory Properties of High-Density Lipoprotein Distinguish Patients From Control Subjects Better Than High-Density Lipoprotein Cholesterol Levels and Are Favorably Affected by Simvastatin Treatment. Circulation 2003, 108, 2751–2756. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-H.; Cho, K.-H. High-Density Lipoprotein (HDL) From Elderly and Reconstituted HDL Containing Glycated Apolipoproteins A-I Share Proatherosclerotic and Prosenescent Properties With Increased Cholesterol Influx. J. Gerontol. Ser. A 2011, 66A, 511–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention (CDC); National Center for Health Statistics (NCHS). National Health and Nutrition Examination Survey Questionnaire (or Examination Protocol, or Laboratory Protocol); U.S. Department of Health and Human Services, Centers for Disease Control and Prevention: Hyattsville, MD, USA, 2019. Available online: https://wwwn.cdc.gov/nchs/nhanes/ (accessed on 21 January 2022).
- Chen, T.; Clark, J.; Riddles, M.; Mohadjer, L.; Fakhouri, T. National Health and Nutrition Examination Survey, 2015–2018: Sample Design and Estimation Procedures; National Center for Health Statistics: Washington, DC, USA, 2020.
- Chen, T.; Parker, J.; Clark, J.; Shin, H.; Rammon, J.; Burt, V. National Health and Nutrition Examination Survey: Estimation Procedures, 2011–2014; National Center for Health Statistics: Washington, DC, USA, 2018.
- Johnson, C.; Dohrmann, S.; Burt, V.; Mohadjer, L. National Health and Nutrition Examination Survey: Sample Design, 2011–2014; National Center for Health Statistics: Washington, DC, USA, 2014.
- NHANES—National Health and Nutrition Examination Survey Homepage. Available online: https://www.cdc.gov/nchs/nhanes/index.htm (accessed on 21 January 2022).
- Physical Activity Guidelines for Americans. Physical Activity Guidelines Advisory Committee Report, 2019; US Department of Health and Human Services: Washington, DC, USA, 2018.
- Fang, J.; Zhang, Z.; Ayala, C.; Thompson-Paul, A.M.; Loustalot, F. Cardiovascular Health among Non-Hispanic Asian Americans: NHANES, 2011–2016. J. Am. Heart Assoc. 2019, 8, e011324. [Google Scholar] [CrossRef] [Green Version]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.; Castro, A.F.; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T.; et al. A New Equation to Estimate Glomerular Filtration Rate. Ann. Intern. Med. 2009, 150, 604. [Google Scholar] [CrossRef] [PubMed]
- Carnuta, M.G.; Stancu, C.S.; Toma, L.; Sanda, G.M.; Niculescu, L.S.; Deleanu, M.; Popescu, A.C.; Popescu, M.R.; Vlad, A.; Dimulescu, D.R.; et al. Dysfunctional high-density lipoproteins have distinct composition, diminished anti-inflammatory potential and discriminate acute coronary syndrome from stable coronary artery disease patients. Sci. Rep. 2017, 7, 7295. [Google Scholar] [CrossRef]
- Bonizzi, A.; Piuri, G.; Corsi, F.; Cazzola, R.; Mazzucchelli, S. HDL Dysfunctionality: Clinical Relevance of Quality Rather Than Quantity. Biomedicines 2021, 9, 729. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Lin, F.-Y.; Shih, C.-M.; Au, H.-K.; Chang, Y.-J.; Nakagami, H.; Morishita, R.; Chang, N.-C.; Shyu, K.-G.; Chen, J.-W. Moderate to High Concentrations of High-Density Lipoprotein From Healthy Subjects Paradoxically Impair Human Endothelial Progenitor Cells and Related Angiogenesis by Activating Rho-Associated Kinase Pathways. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2405–2417. [Google Scholar] [CrossRef] [Green Version]
- Barter, P. Lipoprotein metabolism and CKD: Overview. Clin. Exp. Nephrol. 2014, 18, 243–246. [Google Scholar] [CrossRef]
- Reiss, A.B.; Voloshyna, I.; De Leon, J.; Miyawaki, N.; Mattana, J. Cholesterol Metabolism in CKD. Am. J. Kidney Dis. 2015, 66, 1071–1082. [Google Scholar] [CrossRef] [Green Version]
- Rysz, J.; Gluba-Brzózka, A.; Rysz-Górzyńska, M.; Franczyk, B. The Role and Function of HDL in Patients with Chronic Kidney Disease and the Risk of Cardiovascular Disease. Int. J. Mol. Sci. 2020, 21, 601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafiane, A.; Genest, J. High density lipoproteins: Measurement techniques and potential biomarkers of cardiovascular risk. BBA Clin. 2015, 3, 175–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozemek, C.; Lavie, C.J.; Rognmo, Ø. Global physical activity levels—Need for intervention. Prog. Cardiovasc. Dis. 2019, 62, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Gemming, L.; Jiang, Y.; Swinburn, B.; Utter, J.; Mhurchu, C.N. Under-reporting remains a key limitation of self-reported dietary intake: An analysis of the 2008/09 New Zealand Adult Nutrition Survey. Eur. J. Clin. Nutr. 2014, 68, 259–264. [Google Scholar] [CrossRef]
| Unweighted Total (n = 6455) | Weighted Total (n = 94,993,502) | Healthy (60%) | MetS (40%) | p-Value | |
|---|---|---|---|---|---|
| (SD) or M (Q2, Q3) | (SE) | (SE) | (SE) | ||
| Age (years) | 46.92 (17.01) | 46.17 (0.37) | 41.07 (0.44) | 53.54 (0.50) | < 0.001 |
| BMI (kg/m2) | 29.33 (7.21) | 29.33 (0.17) | 26.81 (0.19) | 32.98 (0.27) | < 0.001 |
| Waist Circumference (cm) | 99.36 (17.15) | 99.92 (0.42) | 92.63 (0.43) | 110.44 (0.55) | < 0.001 |
| WTH Ratio | 0.60 (0.10) | 0.59 (0.00) | 0.55 (0.00) | 0.66 (0.00) | < 0.001 |
| Caloric intake (Kcal/day) | 1932 (1465, 2491) * | 2103 (13) | 2136 (17) | 2054 (19) | 0.002 |
| Fasting Glucose (mg/dL) | 101 (94, 111) * | 107.92 (0.48) | 98.25 (0.38) | 121.87 (0.96) | < 0.001 |
| Triglycerides (mg/dL) | 92 (62, 138) * | 114.99 (1.67) | 84.19 (1.27) | 159.43 (3.53) | < 0.001 |
| HDL-c (mg/dL) | 53.74 (16.08) | 54.41 (0.37) | 59.07 (0.41) | 47.69 (0.46) | < 0.001 |
| LDL-c (mg/dL) | 111.40 (35.51) | 111.87 (0.72) | 110.40 (0.75) | 114.06 (1.27) | 0.012 |
| Systolic Blood Pressure (mmHg) | 123 (17.94) | 122 (0.30) | 116 (0.35) | 129 (0.50) | < 0.001 |
| Diastolic Blood Pressure (mmHg) | 70 (12.27) | 70 (0.30) | 69 (0.33) | 73 (0.40) | < 0.001 |
| MDRD eGFR (mL/min/1.73 m2) | 93.76 (25.01) | 91.19 (0.61) | 94.21 (0.74) | 86.83 (0.78) | < 0.001 |
| CKDEPI eGFR (mL/min/1.73 m2) | 97.79 (22.16) | 96.64 (0.50) | 101.07 (0.65) | 90.26 (0.69) | < 0.001 |
| hs-CRP (mg/L) | 1.9 (0.8, 4.44)) * | 3.74 (0.18) | 2.91 (0.18) | 4.86 (0.27) | < 0.001 |
| Albumin (g/L) | 42.05 (3.49) | 42.29 (0.11) | 42.70 (0.12) | 41.70 (0.13) | < 0.001 |
| HOMA-IR | 2.47 (1.48, 4.31) * | 3.83 (0.11) | 2.22 (0.06) | 6.14 (0.22) | < 0.001 |
| SCr | 0.83 (0.69, 0.98) * | 0.86 (0.00) | 0.85 (0.00) | 0.88 (0.01) | < 0.001 |
| BUN | 13 (10, 16) * | 13.88 (0.12) | 13.31 (0.13) | 14.72 (0.18) | < 0.001 |
| n (%) | % (SE) | % (SE) | % (SE) | p-value | |
| Male Sex | 3153 (48.85) | 49.72 (0.70) | 48.91 (1.07) | 50.92 (1.31) | 0.298 |
| Race/Ethnicity | |||||
| Mexican American | 1011 (15.66) | 9.44 (1.13) | 9.42 (1.22) | 9.47 (1.08) | 0.003 |
| Other Hispanic | 721 (11.17) | 6.50 (0.80) | 7.17 (0.94) | 5.52 (0.73) | |
| NH White | 2299 (35.62) | 63.47 (1.99) | 62.14 (2.29) | 65.43 (1.89) | |
| NH Black | 1345 (20.84) | 11.28 (1.11) | 12.14 (1.12) | 9.99 (1.06) | |
| NH Asian | 824 (12.77) | 5.48 (0.52) | 5.45 (0.54) | 5.52 (0.57) | |
| Other/Multi-Racial | 255 (3.95) | 3.83 (0.40) | 3.67 (0.50) | 4.06 (0.51) | |
| Low SES | 1313 (22.47) | 15.30 (1.06) | 15.31 (1.18) | 15.29 (1.36) | 0.990 |
| CKD | 934 (14.47) | 11.93 (0.52) | 7.11 (0.59) | 19.05 (0.86) | < 0.001 |
| Physically Active | 2278 (69.88) | 69.22 (1.09) | 73.40 (1.38) | 60.85 (1.78) | < 0.001 |
| Smoker | 2910 (45.08) | 46.33 (1.27) | 42.66 (1.53) | 51.76 (1.51) | < 0.001 |
| Glucose Medication | 770 (11.93) | 9.04 (0.54) | 1.65 (0.28) | 19.96 (1.16) | < 0.001 |
| Cholesterol Medication | 1166 (18.06) | 17.09 (0.68) | 4.53 (0.49) | 35.64 (1.27) | < 0.001 |
| Hypertension Medication | 1619 (25.08) | 21.77 (0.87) | 6.09 (0.54) | 44.94 (1.40) | < 0.001 |
| Coefficient | Model 1 a | Model 2 b | Model 3 c | ||||||
|---|---|---|---|---|---|---|---|---|---|
| B | SE B | p-Value | B | SE B | p-Value | B | SE B | p-Value | |
| Intercept | 111.32 | 1.61 | < 0.0001 | 70.98 | 11.03 | < 0.0001 | 80.35 | 13.70 | < 0.0001 |
| HDL-c (mg/dL) | −0.16 | 0.03 | < 0.0001 | −0.18 | 0.05 | 0.00 | −0.24 | 0.05 | < 0.0001 |
| PA | 2.53 | 1.51 | 0.10 | 2.39 | 1.52 | 0.13 | |||
| Cholesterol Med | −10.81 | 2.77 | 0.00 | −10.75 | 2.97 | 0.00 | |||
| Smoking | −1.12 | 1.68 | 0.51 | −0.49 | 1.68 | 0.77 | |||
| hsCRP | 0.13 | 0.22 | 0.56 | 0.13 | 0.24 | 0.60 | |||
| Sex | 2.18 | 1.55 | 0.17 | 2.40 | 1.76 | 0.18 | |||
| Race | 2.23 | 0.55 | 0.00 | 2.07 | 0.54 | 0.00 | |||
| Albumin | 0.87 | 0.23 | 0.00 | 0.84 | 0.24 | 0.00 | |||
| Triglycerides | −0.07 | 0.02 | 0.00 | ||||||
| WTH Ratio | −1.11 | 8.27 | 0.89 | ||||||
| Avg. Caloric Intake | 0.00 | 0.00 | 0.64 | ||||||
| R2 | 0.017 | 0.097 | 0.119 | ||||||
| Coefficient | Model 4 a | Model 5 b | Model 6 c | ||||||
|---|---|---|---|---|---|---|---|---|---|
| B | SE B | p-Value | B | SE B | p-Value | B | SE B | p-Value | |
| Intercept | 100.57 | 2.18 | < 0.0001 | 96.96 | 11.20 | < 0.0001 | 86.88 | 16.38 | < 0.0001 |
| HDL-c (mg/dL) | −0.21 | 0.04 | < 0.0001 | −0.16 | 0.04 | 0.00 | −0.17 | 0.05 | 0.00 |
| PA | 3.24 | 1.78 | 0.08 | 3.08 | 1.86 | 0.11 | |||
| Cholesterol Med | −14.42 | 1.53 | <0.0001 | −13.67 | 1.54 | < 0.0001 | |||
| Smoking | 2.18 | 1.74 | 0.22 | 2.00 | 1.83 | 0.28 | |||
| hsCRP | 0.34 | 0.20 | 0.10 | 0.29 | 0.19 | 0.13 | |||
| Sex | 3.08 | 1.31 | 0.03 | 4.13 | 1.50 | 0.01 | |||
| Race | 1.97 | 0.50 | 0.00 | 2.11 | 0.53 | 0.00 | |||
| Albumin | −0.03 | 0.22 | 0.90 | 0.03 | 0.26 | 0.90 | |||
| Triglycerides | 0.00 | 0.01 | 0.78 | ||||||
| WTH Ratio | 6.86 | 10.06 | 0.50 | ||||||
| Avg. Caloric Intake | 0.00 | 0.00 | 0.10 | ||||||
| R2 | 0.024 | 0.193 | 0.195 | ||||||
| Coefficient | Model 7 a | Model 8 b | Model 9 c | ||||||
|---|---|---|---|---|---|---|---|---|---|
| B | SE B | p-Value | B | SE B | p-Value | B | SE B | p-Value | |
| Intercept | 100.11 | 1.27 | < 0.0001 | 71.28 | 8.59 | < 0.0001 | 85.74 | 11.24 | < 0.0001 |
| HDL-c (mg/dL) | −0.05 | 0.02 | 0.0211 | −0.10 | 0.04 | 0.01 | −0.18 | 0.04 | < 0.0001 |
| PA | 3.73 | 1.26 | 0.01 | 3.40 | 1.32 | 0.02 | |||
| Cholesterol Med | −17.97 | 1.50 | < 0.0001 | −16.65 | 1.43 | < 0.0001 | |||
| Smoking | −0.25 | 1.33 | 0.85 | 0.38 | 1.40 | 0.79 | |||
| hsCRP | 0.18 | 0.16 | 0.25 | 0.23 | 0.18 | 0.20 | |||
| Sex | 1.92 | 1.25 | 0.14 | 3.14 | 1.37 | 0.03 | |||
| Race | 2.15 | 0.37 | < 0.0001 | 2.07 | 0.37 | < 0.0001 | |||
| Albumin | 0.68 | 0.19 | 0.00 | 0.60 | 0.19 | 0.00 | |||
| Triglycerides | −0.02 | 0.01 | 0.00 | ||||||
| WTH Ratio | −12.76 | 6.96 | 0.08 | ||||||
| Avg. Caloric Intake | 0.00 | 0.00 | 0.19 | ||||||
| R2 | 0.002 | 0.157 | 0.170 | ||||||
| Quantile | Total Intercept | B | p-Value | Healthy Intercept | B | p-Value | MetS Intercept | B | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| 100% | 100.11 | −0.052 | 0.0211 | 111.32 | −0.163 | < 0.0001 | 100.57 | −0.208 | <0.0001 |
| 98% | 100.25 | −0.054 | 0.0312 | 112.44 | −0.182 | < 0.0001 | 103.17 | −0.263 | <0.0001 |
| 90% | 100.38 | −0.052 | 0.0629 | 110.04 | −0.141 | 0.0002 | 105.52 | −0.316 | < 0.0001 |
| 80% | 100.13 | −0.044 | 0.2626 | 109.61 | −0.134 | 0.0039 | 108.72 | −0.384 | < 0.0001 |
| 50% | 98.31 | −0.007 | 0.9192 | 97.81 | 0.08 | 0.4763 | 108.55 | −0.376 | 0.0006 |
| Quantile | Total Group, r | p-Value | Healthy, r | p-Value | MetS, r | p-Value |
|---|---|---|---|---|---|---|
| 100% | −0.00146 | 0.9069 | −0.10475 | < 0.0001 | −0.11136 | < 0.0001 |
| 98% | 0.00306 | 0.8077 | −0.10589 | < 0.0001 | −0.12134 | < 0.0001 |
| 90% | 0.00354 | 0.7866 | −0.06336 | 0.0003 | −0.10939 | < 0.0001 |
| 80% | 0.01026 | 0.456 | −0.05131 | 0.0048 | −0.14037 | < 0.0001 |
| 50% | 0.02628 | 0.1253 | 0.0056 | 0.8091 | −0.0931 | 0.0004 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richardson, K.A.; Richardson, L.T.; Bowden, R.G. Association of High-Density Lipoprotein Cholesterol, Renal Function, and Metabolic Syndrome: An Assessment of the 2013–2018 National Health and Nutrition Examination Surveys. Kidney Dial. 2022, 2, 419-432. https://doi.org/10.3390/kidneydial2030037
Richardson KA, Richardson LT, Bowden RG. Association of High-Density Lipoprotein Cholesterol, Renal Function, and Metabolic Syndrome: An Assessment of the 2013–2018 National Health and Nutrition Examination Surveys. Kidney and Dialysis. 2022; 2(3):419-432. https://doi.org/10.3390/kidneydial2030037
Chicago/Turabian StyleRichardson, Kathleen A., Luke T. Richardson, and Rodney G. Bowden. 2022. "Association of High-Density Lipoprotein Cholesterol, Renal Function, and Metabolic Syndrome: An Assessment of the 2013–2018 National Health and Nutrition Examination Surveys" Kidney and Dialysis 2, no. 3: 419-432. https://doi.org/10.3390/kidneydial2030037