Prediction of Clinical Outcomes with Explainable Artificial Intelligence in Patients with Chronic Lymphocytic Leukemia
Abstract
1. Introduction
2. Materials and Methods
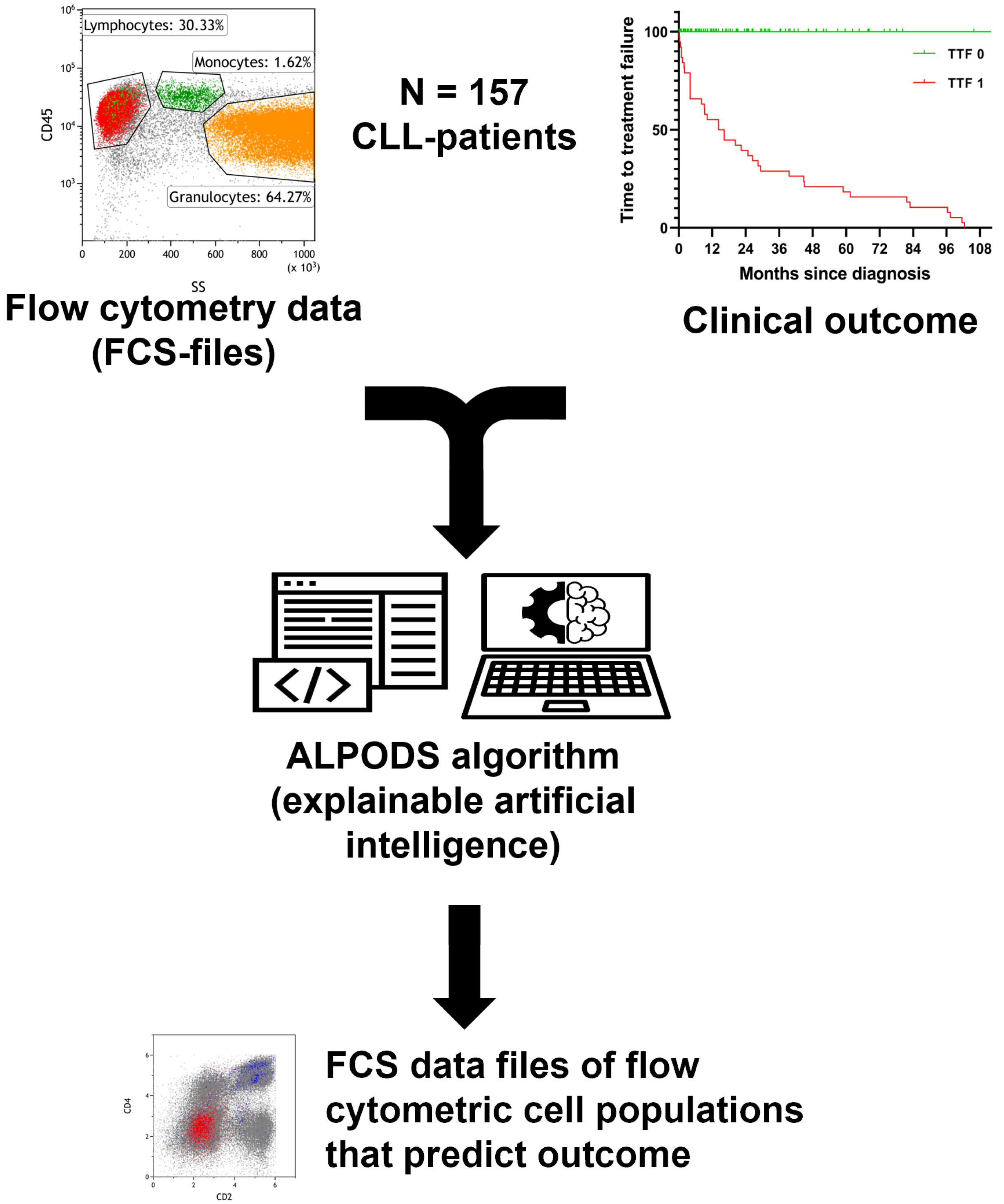
2.1. Patients, Data Acquisition, and Processing
2.2. Antigen Panel, Flow Cytometry Staining, and Analysis
2.3. Data Processing
2.4. Statistics
3. Results
3.1. Patient Characteristics
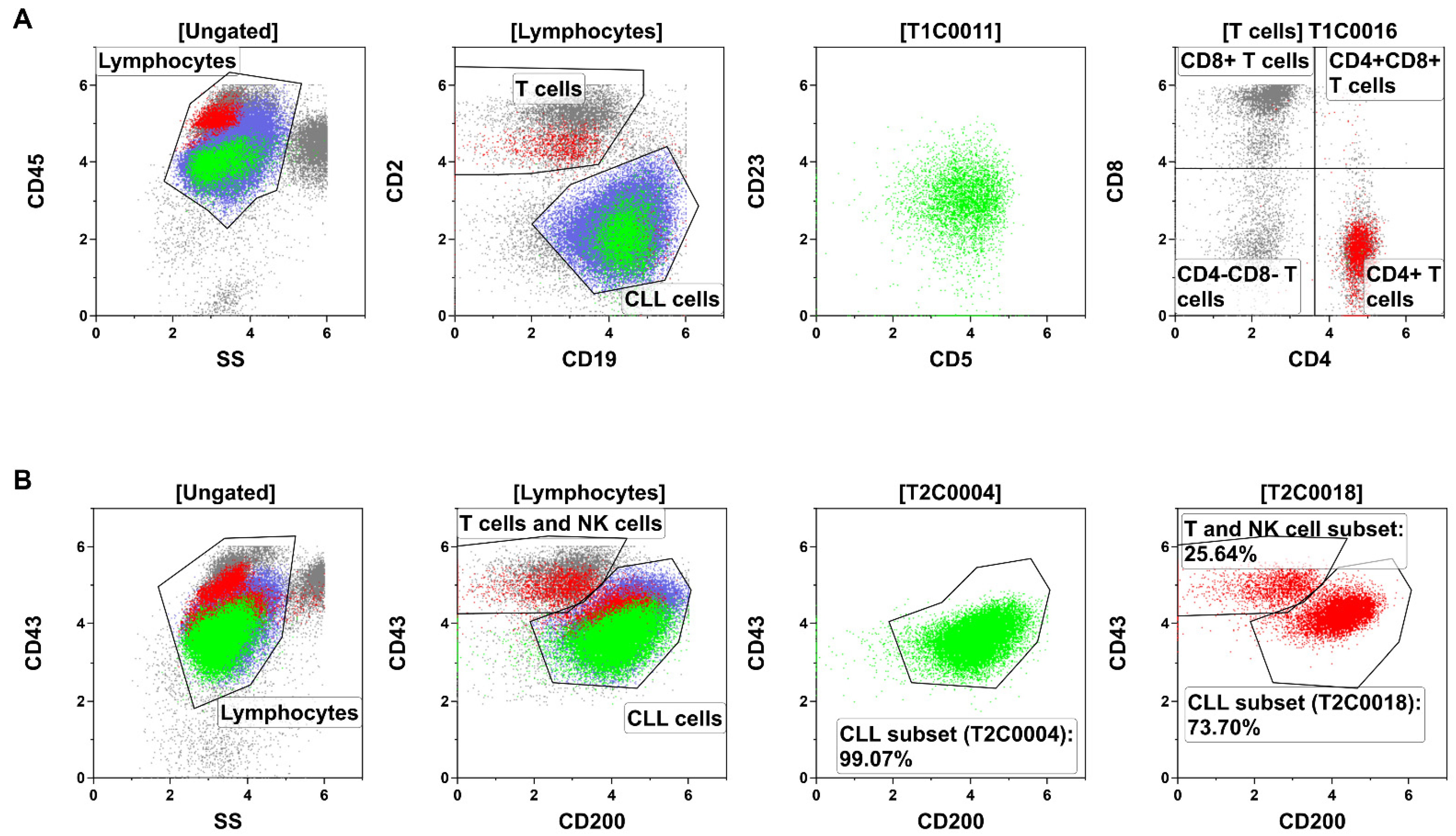
3.2. Cell Populations Identified by ALPODS
3.3. Identification of the XAI-Populations
3.4. Characterization of Predictive Subsets within CLL Cells
3.5. Clinical Significance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; de Oliveira Araujo, I.B.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Matutes, E.; Owusu-Ankomah, K.; Morilla, R.; Garcia Marco, J.; Houlihan, A.; Que, T.H.; Catovsky, D. The Immunological Profile of B-Cell Disorders and Proposal of a Scoring System for the Diagnosis of CLL. Leukemia 1994, 8, 1640–1645. [Google Scholar] [PubMed]
- Hoffmann, J.; Rother, M.; Kaiser, U.; Thrun, M.C.; Wilhelm, C.; Gruen, A.; Niebergall, U.; Meissauer, U.; Neubauer, A.; Brendel, C. Determination of CD43 and CD200 Surface Expression Improves Accuracy of B-Cell Lymphoma Immunophenotyping. Cytom. B Clin. Cytom. 2020, 98, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.R.; Sawitsky, A.; Cronkite, E.P.; Chanana, A.D.; Levy, R.N.; Pasternack, B.S. Clinical Staging of Chronic Lymphocytic Leukemia. Blood 1975, 46, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Binet, J.L.; Auquier, A.; Dighiero, G.; Chastang, C.; Piguet, H.; Goasguen, J.; Vaugier, G.; Potron, G.; Colona, P.; Oberling, F.; et al. A New Prognostic Classification of Chronic Lymphocytic Leukemia Derived from a Multivariate Survival Analysis. Cancer 1981, 48, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Kröber, A.; Bullinger, L.; Döhner, K.; Bentz, M.; Lichter, P. Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef]
- Cramer, P.; Hallek, M. Prognostic Factors in Chronic Lymphocytic Leukemia—What Do We Need to Know? Nat. Rev. Clin. Oncol. 2011, 8, 38–47. [Google Scholar] [CrossRef]
- Zenz, T.; Benner, A.; Döhner, H.; Stilgenbauer, S. Chronic Lymphocytic Leukemia and Treatment Resistance in Cancer: The Role of the P53 Pathway. Cell Cycle 2008, 7, 3810–3814. [Google Scholar] [CrossRef]
- Wang, L.; Lawrence, M.S.; Wan, Y.; Stojanov, P.; Sougnez, C.; Stevenson, K.; Werner, L.; Sivachenko, A.; DeLuca, D.S.; Zhang, L.; et al. SF3B1 and Other Novel Cancer Genes in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2011, 365, 2497–2506. [Google Scholar] [CrossRef]
- Rossi, D.; Rasi, S.; Fabbri, G.; Spina, V.; Fangazio, M.; Forconi, F.; Marasca, R.; Laurenti, L.; Bruscaggin, A.; Cerri, M.; et al. Mutations of NOTCH1 Are an Independent Predictor of Survival in Chronic Lymphocytic Leukemia. Blood 2012, 119, 521–529. [Google Scholar] [CrossRef]
- Puente, X.S.; López-Otín, C. The Evolutionary Biography of Chronic Lymphocytic Leukemia. Nat. Genet. 2013, 45, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Baliakas, P.; Hadzidimitriou, A.; Sutton, L.-A.; Rossi, D.; Minga, E.; Villamor, N.; Larrayoz, M.; Kminkova, J.; Agathangelidis, A.; Davis, Z.; et al. Recurrent Mutations Refine Prognosis in Chronic Lymphocytic Leukemia. Leukemia 2015, 29, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Damle, R.N.; Wasil, T.; Fais, F.; Ghiotto, F.; Valetto, A.; Allen, S.L.; Buchbinder, A.; Budman, D.; Dittmar, K.; Kolitz, J.; et al. Ig V Gene Mutation Status and CD38 Expression As Novel Prognostic Indicators in Chronic Lymphocytic Leukemia. Blood 1999, 94, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, T.J.; Orchard, J.A.; Ibbotson, R.E.; Davis, Z.; Thomas, P.W.; Stevenson, F.K.; Oscier, D.G. CD38 Expression and Immunoglobulin Variable Region Mutations Are Independent Prognostic Variables in Chronic Lymphocytic Leukemia, but CD38 Expression May Vary during the Course of the Disease. Blood 2002, 99, 1023–1029. [Google Scholar] [CrossRef]
- Hamblin, T.J.; Davis, Z.; Gardiner, A.; Oscier, D.G.; Stevenson, F.K. Unmutated Ig VH Genes Are Associated With a More Aggressive Form of Chronic Lymphocytic Leukemia. Blood 1999, 94, 1848–1854. [Google Scholar] [CrossRef]
- Kröber, A.; Seiler, T.; Benner, A.; Bullinger, L.; Brückle, E.; Lichter, P.; Döhner, H.; Stilgenbauer, S. VH Mutation Status, CD38 Expression Level, Genomic Aberrations, and Survival in Chronic Lymphocytic Leukemia. Blood 2002, 100, 1410–1416. [Google Scholar] [CrossRef]
- Rassenti, L.Z.; Jain, S.; Keating, M.J.; Wierda, W.G.; Grever, M.R.; Byrd, J.C.; Kay, N.E.; Brown, J.R.; Gribben, J.G.; Neuberg, D.S.; et al. Relative Value of ZAP-70, CD38, and Immunoglobulin Mutation Status in Predicting Aggressive Disease in Chronic Lymphocytic Leukemia. Blood 2008, 112, 1923–1930. [Google Scholar] [CrossRef]
- Schroers, R.; Griesinger, F.; Trümper, L.; Haase, D.; Kulle, B.; Klein-Hitpass, L.; Sellmann, L.; Dührsen, U.; Dürig, J. Combined Analysis of ZAP-70 and CD38 Expression as a Predictor of Disease Progression in B-Cell Chronic Lymphocytic Leukemia. Leukemia 2005, 19, 750–758. [Google Scholar] [CrossRef]
- Heintel, D.; Schwarzinger, I.; Chizzali-Bonfadin, C.; Thalhammer, R.; Schwarzmeier, J.; Fritzer-Szekeres, M.; Weltermann, A.; Simonitsch, I.; Lechner, K.; Jaeger, U. Association of CD38 Antigen Expression with Other Prognostic Parameters in Early Stages of Chronic Lymphocytic Leukemia. Leuk. Lymphoma 2001, 42, 1315–1321. [Google Scholar] [CrossRef]
- International CLL-IPI Working Group. An International Prognostic Index for Patients with Chronic Lymphocytic Leukaemia (CLL-IPI): A Meta-Analysis of Individual Patient Data. Lancet Oncol. 2016, 17, 779–790. [Google Scholar] [CrossRef]
- Van Gassen, S.; Callebaut, B.; Van Helden, M.J.; Lambrecht, B.N.; Demeester, P.; Dhaene, T.; Saeys, Y. FlowSOM: Using Self-Organizing Maps for Visualization and Interpretation of Cytometry Data. Cytom. A 2015, 87, 636–645. [Google Scholar] [CrossRef]
- Saeys, Y.; Van Gassen, S.; Lambrecht, B.N. Computational Flow Cytometry: Helping to Make Sense of High-Dimensional Immunology Data. Nat. Rev. Immunol. 2016, 16, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Bruggner, R.V.; Bodenmiller, B.; Dill, D.L.; Tibshirani, R.J.; Nolan, G.P. Automated Identification of Stratifying Signatures in Cellular Subpopulations. Proc. Natl. Acad. Sci. USA 2014, 111, E2770–E2777. [Google Scholar] [CrossRef] [PubMed]
- Polikowsky, H.G.; Drake, K.A. Supervised Machine Learning with CITRUS for Single Cell Biomarker Discovery. In Mass Cytometry: Methods and Protocols; McGuire, H.M., Ashhurst, T.M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 309–332. ISBN 978-1-4939-9454-0. [Google Scholar]
- Ultsch, A.; Hoffmann, J.; Röhnert, M.; Von Bonin, M.; Oelschlägel, U.; Brendel, C.; Thrun, M.C. An Explainable AI System for the Diagnosis of High Dimensional Biomedical Data. arXiv Prepr. 2021, arXiv:2107.01820. [Google Scholar]
- Hoffmann, J.; Thrun, M.C.; Röhnert, M.A.; von Bonin, M.; Oelschlägel, U.; Neubauer, A.; Ultsch, A.; Brendel, C. Identification of Critical Hemodilution by Artificial Intelligence in Bone Marrow Assessed for MRD Analysis in Acute Myeloid Leukemia: The Cinderella Method. Cytom. Part A J. Int. Soc. Anal. Cytol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Milligan, G.W.; Cooper, M.C. A Study of Standardization of Variables in Cluster Analysis. J. Classif. 1988, 5, 181–204. [Google Scholar] [CrossRef]
- Thrun, M. Projection-Based Clustering through Self-Organization and Swarm Intelligence: Combining Cluster Analysis with the Visualization of High-Dimensional Data; Springer Nature: Berlin/Heidelberg, Germany, 2018; ISBN 978-3-658-20540-9. [Google Scholar]
- Ultsch, A.; Lötsch, J. Computed ABC Analysis for Rational Selection of Most Informative Variables in Multivariate Data. PLoS ONE 2015, 10, e0129767. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Thrun, M.C.; Gehlert, T.; Ultsch, A. Analyzing the Fine Structure of Distributions. PLoS ONE 2020, 15, e0238835. [Google Scholar] [CrossRef]
- Catakovic, K.; Gassner, F.J.; Ratswohl, C.; Zaborsky, N.; Rebhandl, S.; Schubert, M.; Steiner, M.; Gutjahr, J.C.; Pleyer, L.; Egle, A.; et al. TIGIT Expressing CD4+T Cells Represent a Tumor-Supportive T Cell Subset in Chronic Lymphocytic Leukemia. OncoImmunology 2018, 7, e1371399. [Google Scholar] [CrossRef] [PubMed]
- Elston, L.; Fegan, C.; Hills, R.; Hashimdeen, S.S.; Walsby, E.; Henley, P.; Pepper, C.; Man, S. Increased Frequency of CD4+PD-1+HLA-DR+ T Cells Is Associated with Disease Progression in CLL. Br. J. Haematol. 2020, 188, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.; Gentilcore, G.; Heimersson, K.; Mozaffari, F.; Näsman-Glaser, B.; Young, E.; Rosenquist, R.; Hansson, L.; Österborg, A.; Mellstedt, H. T Cells in Chronic Lymphocytic Leukemia Display Dysregulated Expression of Immune Checkpoints and Activation Markers. Haematologica 2017, 102, 562–572. [Google Scholar] [CrossRef]
- Tötterman, T.H.; Carlsson, M.; Simonsson, B.; Bengtsson, M.; Nilsson, K. T-Cell Activation and Subset Patterns Are Altered in B-CLL and Correlate With the Stage of the Disease. Blood 1989, 74, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Peller, S.; Kaufman, S. Decreased CD45RA T Cells in B-Cell Chronic Lymphatic Leukemia Patients: Correlation With Disease Stage. Blood 1991, 78, 1569–1573. [Google Scholar] [CrossRef]
- Serrano, D.; Monteiro, J.; Allen, S.L.; Kolitz, J.; Schulman, P.; Lichtman, S.M.; Buchbinder, A.; Vinciguerra, V.P.; Chiorazzi, N.; Gregersen, P.K. Clonal Expansion within the CD4+CD57+ and CD8+CD57+ T Cell Subsets in Chronic Lymphocytic Leukemia. J. Immunol. 1997, 158, 1482–1489. [Google Scholar] [CrossRef]
- Matutes, E.; Wechsler, A.; Gomez, R.; Cherchi, M.; Catovsky, D. Unusual T-Cell Phenotype in Advanced B-Chronic Lymphocytic Leukaemia. Br. J. Haematol. 1981, 49, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.H.G.; Cawley, J.C. Suppressor t Cells in B-Cell Chronic Lymphocytic Leukaemia: Relationship to Clinical Stage. Leuk. Res. 1982, 6, 653–657. [Google Scholar] [CrossRef]
- Mittelman, A.; Denny, T.; Gebhard, D.; Cirrincione, C.; Kurland, E.; Koziner, B. Analysis of T-Cell Subsets in B-Cell Chronic Lymphocytic Leukemia: A Correlation with the Stage of Disease. Am. J. Hematol. 1984, 16, 67–73. [Google Scholar] [CrossRef]
- Platsoucas, C.D.; Galinski, M.; Kempin, S.; Reich, L.; Clarkson, B.; Good, R.A. Abnormal T Lymphocyte Subpopulations in Patients with B Cell Chronic Lymphocytic Leukemia: An Analysis by Monoclonal Antibodies. J. Immunol. 1982, 129, 2305–2312. [Google Scholar] [CrossRef]
- Witkowska, M.; Nowak, W.; Cebula-Obrzut, B.; Majchrzak, A.; Medra, A.; Robak, T.; Smolewski, P. Spontaneous in Vitro Apoptosis of de Novo Chronic Lymphocytic Leukemia Cells Correlates with Risk of the Disease Progression. Cytom. B Clin. Cytom. 2014, 86, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Jahrsdörfer, B.; Wooldridge, J.E.; Blackwell, S.E.; Taylor, C.M.; Link, B.K.; Weiner, G.J. Good Prognosis Cytogenetics in B-Cell Chronic Lymphocytic Leukemia Is Associated in Vitro with Low Susceptibility to Apoptosis and Enhanced Immunogenicity. Leukemia 2005, 19, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Oscier, D.; Else, M.; Matutes, E.; Morilla, R.; Strefford, J.C.; Catovsky, D. The Morphology of CLL Revisited: The Clinical Significance of Prolymphocytes and Correlations with Prognostic/Molecular Markers in the LRF CLL4 Trial. Br. J. Haematol. 2016, 174, 767–775. [Google Scholar] [CrossRef]
- Bulian, P.; Shanafelt, T.D.; Fegan, C.; Zucchetto, A.; Cro, L.; Nückel, H.; Baldini, L.; Kurtova, A.V.; Ferrajoli, A.; Burger, J.A.; et al. CD49d Is the Strongest Flow Cytometry–Based Predictor of Overall Survival in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2014, 32, 897–904. [Google Scholar] [CrossRef] [PubMed]

| Total (N = 157) | TTF 1 (N = 42) | TTF 0 (N = 115) | |
|---|---|---|---|
| Age (years) | |||
| Median (range) | 68 (26–91) | 70 (50–88) | 67 (26–91) |
| Sex | |||
| Female | 62 (39.5%) | 17 (40.5%) | 45 (39.1%) |
| Male | 95 (60.5%) | 25 (59.5%) | 70 (60.9%) |
| Binet | |||
| A | 83 (52.9%) | 15 (35.7%) | 68 (59.1%) |
| B | 24 (15.3%) | 7 (16.7%) | 17 (14.8%) |
| C | 12 (7.6%) | 8 (19.0%) | 4 (3.5%) |
| unknown | 31 (19.7%) | 9 (21.4%) | 22 (19.1%) |
| CLL-IPI | |||
| Low | 84 (53.5%) | 9 (21.4%) | 75 (65.2%) |
| Intermediate | 53 (33.8%) | 20 (47.6%) | 33 (28.7%) |
| High | 16 (10.2%) | 11 (26.2%) | 5 (4.3%) |
| Very High | 4 (2.5%) | 2 (4.8%) | 2 (1.7%) |
| Follow up | |||
| Median (IQR) months | 31 (9–65) | 71 (38–114) | 24 (7–46) |
| Death (N) | 16 (10.2%) | 16 (38.1%) | 0 (0%) |
| Treatment failure first line (N) | 34 (21.7%) | 34 (80.1%) | 0 (0%) |
| Richter’s syndrome (N) | 4 (2.5%) | 3 (7.1%) | 1 (0.9%) |
| Therapy (first line) | |||
| R-Bendamustin | 25 (15.9%) | 17 (40.5%) | 8 (7.0%) |
| Ibrutinib | 12 (7.6%) | 6 (14.3%) | 6 (5.2%) |
| Other | 24 (15.3%) | 15 (35.7%) | 9 (7.8%) |
| No therapy | 80 (51.0%) | 0 (0%) | 80 (70.0%) |
| Unknown | 16 (10.2%) | 4 (9.5%) | 12 (10.4%) |
| TTF 1 | TTF 0 | ROC | |||||
|---|---|---|---|---|---|---|---|
| Mean (%) | Mean (%) | SE of Difference | p-Value (MWU-Test) | AUC | 95% CI | p-Value | |
| CLL-IPI | 2.845 * | 1.322 * | 0.2712 | <0.0001 | 0.78 | 0.70–0.86 | <0.0001 |
| CD38+ | 37.05 | 22.51 | 5.414 | 0.0016 | 0.66 | 0.57–0.76 | 0.0018 |
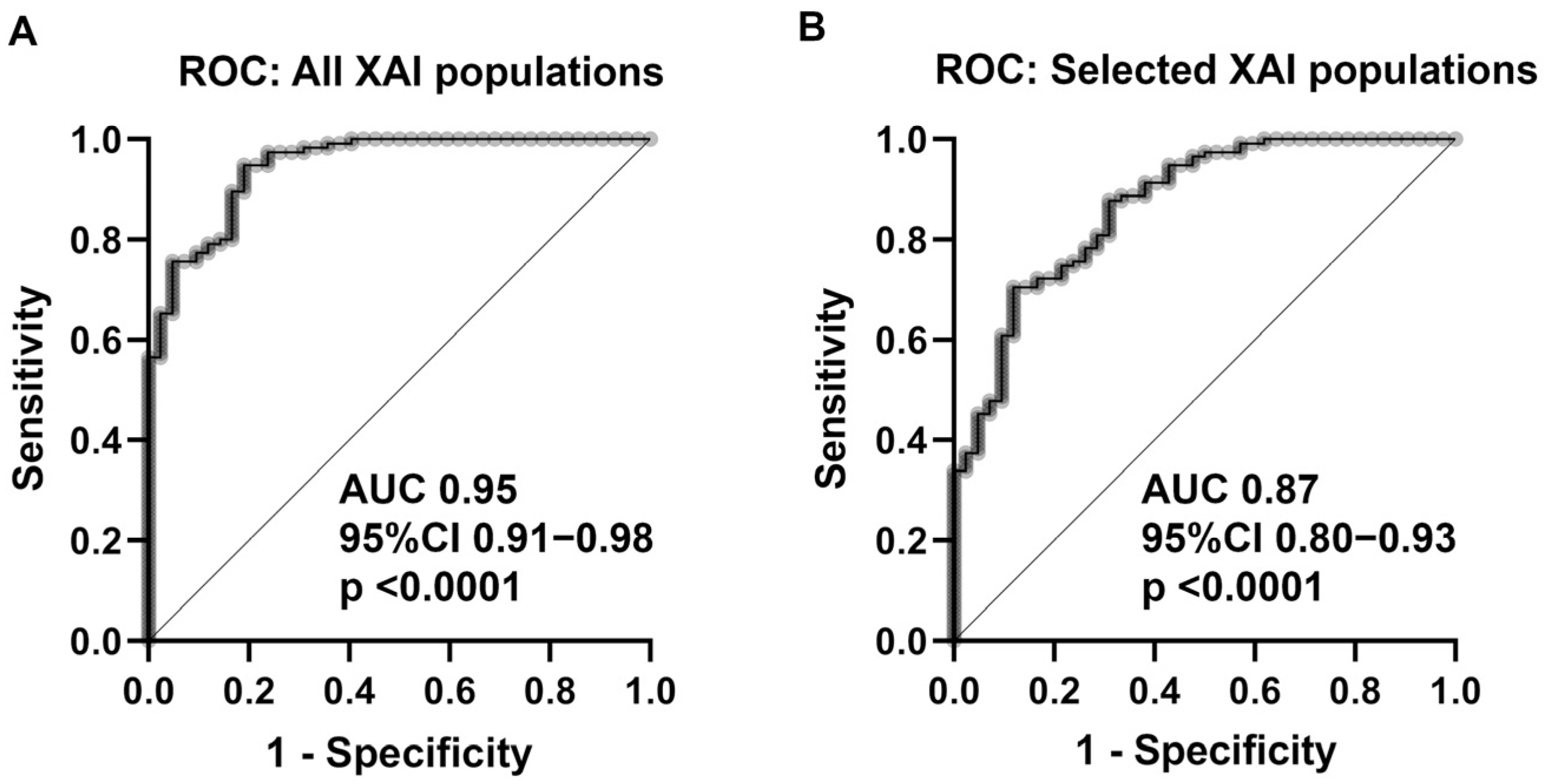
| XAI-populations | |||||||
| Total | 0.95 | 0.91–0.98 | <0.0001 | ||||
| T1C0011 | 5.36 | 1.39 | 0.81 | <0.0001 | 0.76 | 0.68–0.84 | <0.0001 |
| T1C0012 | 1.91 | 1.61 | 0.40 | 0.2650 | 0.56 | 0.45–0.66 | 0.2635 |
| T1C0016 | 4.91 | 13.51 | 1.83 | <0.0001 | 0.78 | 0.70–0.86 | <0.0001 |
| T1C0017 | 6.46 | 4.15 | 1.32 | 0.2136 | 0.57 | 0.46–0.67 | 0.2124 |
| T1C0019 | 5.19 | 6.38 | 1.01 | 0.0970 | 0.59 | 0.47–0.70 | 0.0966 |
| T1C0020 | 0.35 | 0.11 | 0.08 | 0.1514 | 0.58 | 0.47–0.68 | 0.1506 |
| T1C0023 | 2.25 | 0.57 | 0.49 | 0.5593 | 0.53 | 0.41–0.65 | 0.5573 |
| T2C0001 | 2.59 | 5.50 | 1.31 | 0.0011 | 0.67 | 0.57–0.76 | 0.0012 |
| T2C0002 | 2.85 | 0.73 | 0.61 | 0.0052 | 0.64 | 0.54–0.75 | 0.0055 |
| T2C0004 | 9.64 | 1.23 | 1.29 | 0.0002 | 0.69 | 0.58–0.80 | 0.0002 |
| T2C0009 | 1.74 | 0.84 | 0.49 | 0.6877 | 0.52 | 0.41–0.63 | 0.6859 |
| T2C0010 | 0.55 | 0.21 | 0.15 | 0.5369 | 0.53 | 0.43–0.64 | 0.5349 |
| T2C0014 | 8.19 | 12.14 | 1.97 | 0.0078 | 0.64 | 0.54–0.74 | 0.0081 |
| T2C0018 | 5.37 | 1.66 | 0.72 | 0.0001 | 0.73 | 0.63–0.82 | <0.0001 |
| T2C0020 | 4.38 | 1.75 | 0.63 | <0.0020 | 0.66 | 0.55–0.77 | 0.0022 |
| T2C0021 | 3.25 | 2.74 | 0.73 | 0.4286 | 0.04 | 0.44–0.64 | 0.4266 |
| T2C0028 | 0.85 | 0.36 | 0.14 | 0.2021 | 0.57 | 0.45–0.68 | 0.2010 |
| OR | 95% CI | p-Value | AUC | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| All XAI populations | 0.95 | 0.91–0.98 | <0.0001 | |||
| CLL-IPI | 0.78 | 0.70–0.86 | <0.0001 | |||
| Four-factor model | 0.84 | 0.77–0.91 | <0.0001 | |||
| CLL-IPI | 1.53 | 1.19–2.04 | 0.0018 | |||
| T1C0016 (CD4+ T cells) | 0.86 | 0.78–0.93 | 0.0003 | |||
| T1C0023 (CD8+ T cells) | 1.14 | 0.95–1.77 | 0.3305 | |||
| CD38 | 1.01 | 1.00–1.02 | 0.1790 | |||
| Two-factor model with IPI | 0.83 | 0.77–0.90 | <0.0001 | |||
| CLL-IPI | 1.64 | 1.29–2.16 | 0.0001 | |||
| T1C0016 (CD4+ T cells) | 0.85 | 0.78–0.92 | 0.0002 | |||
| Two-factor model w/o IPI | 0.79 | 0.71–0.87 | <0.0001 | |||
| T1C0016 (CD4+ T cells) | 0.85 | 0.78–0.91 | <0.0001 | |||
| T1C0023 (CD8+ T cells) | 1.29 | 1.05–2.14 | 0.1390 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffmann, J.; Eminovic, S.; Wilhelm, C.; Krause, S.W.; Neubauer, A.; Thrun, M.C.; Ultsch, A.; Brendel, C. Prediction of Clinical Outcomes with Explainable Artificial Intelligence in Patients with Chronic Lymphocytic Leukemia. Curr. Oncol. 2023, 30, 1903-1915. https://doi.org/10.3390/curroncol30020148
Hoffmann J, Eminovic S, Wilhelm C, Krause SW, Neubauer A, Thrun MC, Ultsch A, Brendel C. Prediction of Clinical Outcomes with Explainable Artificial Intelligence in Patients with Chronic Lymphocytic Leukemia. Current Oncology. 2023; 30(2):1903-1915. https://doi.org/10.3390/curroncol30020148
Chicago/Turabian StyleHoffmann, Joerg, Semil Eminovic, Christian Wilhelm, Stefan W. Krause, Andreas Neubauer, Michael C. Thrun, Alfred Ultsch, and Cornelia Brendel. 2023. "Prediction of Clinical Outcomes with Explainable Artificial Intelligence in Patients with Chronic Lymphocytic Leukemia" Current Oncology 30, no. 2: 1903-1915. https://doi.org/10.3390/curroncol30020148
APA StyleHoffmann, J., Eminovic, S., Wilhelm, C., Krause, S. W., Neubauer, A., Thrun, M. C., Ultsch, A., & Brendel, C. (2023). Prediction of Clinical Outcomes with Explainable Artificial Intelligence in Patients with Chronic Lymphocytic Leukemia. Current Oncology, 30(2), 1903-1915. https://doi.org/10.3390/curroncol30020148






