The Clinical Utility of FLT3 Mutation Testing in Acute Leukemia: A Canadian Consensus
Abstract
Introduction
Testing for FLT3 Mutations
Canadian Leukemia Study Group/Groupe canadien d’étude sur la leucémie
Methods
CLSG/GCEL Recommendations for FLT3 Mutation Testing in Acute Leukemia
| Q1 | Which patients should be tested for FLT3 mutations and when? |
| 1.1 | The CLSG/GCEL panel recommends that FLT3 mutation testing (ITD and TKD) be performed at the time of initial diagnosis in all untreated adult patients with:
|
| 1.2 | All adult patients meeting the criteria in Recommendation 1.1 should be tested for FLT3 mutations, regardless of their age; |
| 1.3 | AML patients with refractory disease (including primary induction failures) and relapsed disease should be retested for FLT3 mutations (ITD and TKD). |
| Q2 | Which is the preferred method for FLT3 mutation testing? |
| 2.1 | The CLSG/GCEL panel recommends that at diagnosis and following relapse/induction failure:
|
| 2.2 | Bone marrow is the preferred specimen for FLT3 mutation assessment in both newly diagnosed and R/R AML;
|
| 2.3 | DNA is the preferred analyte for upfront, PCR-based FLT3 mutation testing. |
| 2.4.1 | At this time, to conform with ELN 2017 and 2022 guidelines, FLT3-ITD and FLT3-TKD should be assessed by PCR-fragment analysis and PCR-RFLP. This requirement may change as new methodologies become available/are validated;
|
| 2.4.2 | NGS analysis may be used for multiple gene mutation assessment;
|
| 2.5 | Based on current data and ELN 2017 and 2022 recommendations, FLT3 mutation test results for all newly diagnosed and R/R AML patients should be available in electronic patient records within 2–5 calendar days. It should be noted that this timeline may become more acute, as FLT3 inhibitors may in the future be used earlier in upfront AML therapy. |
| Q3 | What is the clinical relevance of FLT3-ITD size, insertion site, and number of distinct mutations? |
| 3.1 | The available data do not support a prognostic role for FLT3-ITD size, although insertion size may be of interest in cases of relapse. It is possible that a C-terminal ITD affects the prognosis adversely. We are awaiting prospective validation. The CLSG/GCEL panel recommends that FLT3-ITD size should not play a role in clinical decision-making at this time; |
| 3.2 | An association between worse prognosis and C-terminal TKD1-ITDs has been demonstrated in the midostaurin-treated cohort in the RATIFY study *. The CLSG/GCEL panel feels that the association is plausible, and confirmation in an independent TKI-treated cohort is awaited. There is no formal evidence at this time that clinical decisions should be altered based on the insertion site; |
| 3.3 | The available evidence suggests that the number of distinct FLT3-ITDs is not correlated with prognosis. The number of FLT3-ITDs should not play a role in clinical decision-making at this time. |
| Q4 | Is there a role for FLT3-ITD analysis in MRD assessment? |
| 4.1 | MRD assessment is assuming an increasingly important role in the management of AML. Early data suggest that FLT3-ITD MRD analysis may be a stronger predictor of outcome than many other established prognostic factors defined at diagnosis and during therapy. While at present it remains to be clarified how FLT3-ITD MRD assessment will ultimately fit into an overall AML MRD testing strategy, data available at this time suggest that FLT3-ITD MRD analysis may be quite useful clinically; laboratories should plan to offer this analysis. |
| Q5 | What is the clinical relevance of the FLT3-ITD allelic burden? Should it be assessed, and if so, how? |
| 5.1 | The CLSG/GCEL panel recognizes that higher FLT3-ITD allelic burdens have long been associated with adverse outcomes. Although not mandatory for disease risk classification as per the ELN 2022 guidelines, assessment of the FLT3-ITD allelic burden remains clinically relevant from a disease management point of view. The panel feels that there will continue to be a role for FLT3-ITD allelic burden assessment to help define leukemic clonal architecture and to define treatment options (e.g., alloSCT vs. observation or maintenance therapy), in conjunction with other factors, including MRD assessment. However, there remains uncertainty regarding this measure because of the lack of standardization in assessment and due to questions regarding the impact of the FLT3-ITD allelic burden in the era of TKI therapies and MRD-based treatment decisions;
|
| 5.2 | The FLT3-ITD allelic burden should be assessed by measuring the AUC of fluorescence peaks on capillary electrophoresis (PCR-fragment analysis);
|
| 5.3 | To avoid confusion associated with the terms allelic ratio and variant allele frequency, the CLSG/GCEL panel suggests that moving forward, the FLT3-ITD allelic burden be reported as the proportion of mutant/total alleles and that neither AR nor VAF should be used;
|
| 5.4 | Until such time as there is an international standard for FLT3 testing, reporting, and interpretation, the Canadian medical community must strive to standardize national FLT3-ITD allelic burden testing and reporting to ensure that all Canadian patients receive the same care and to support the collection of consistent data on which to base future recommendations. |
| Q6 | How should the results of FLT3 mutation testing be reported? |
| 6.1 | PCR-based FLT3 mutation testing reports should include the following elements:
|
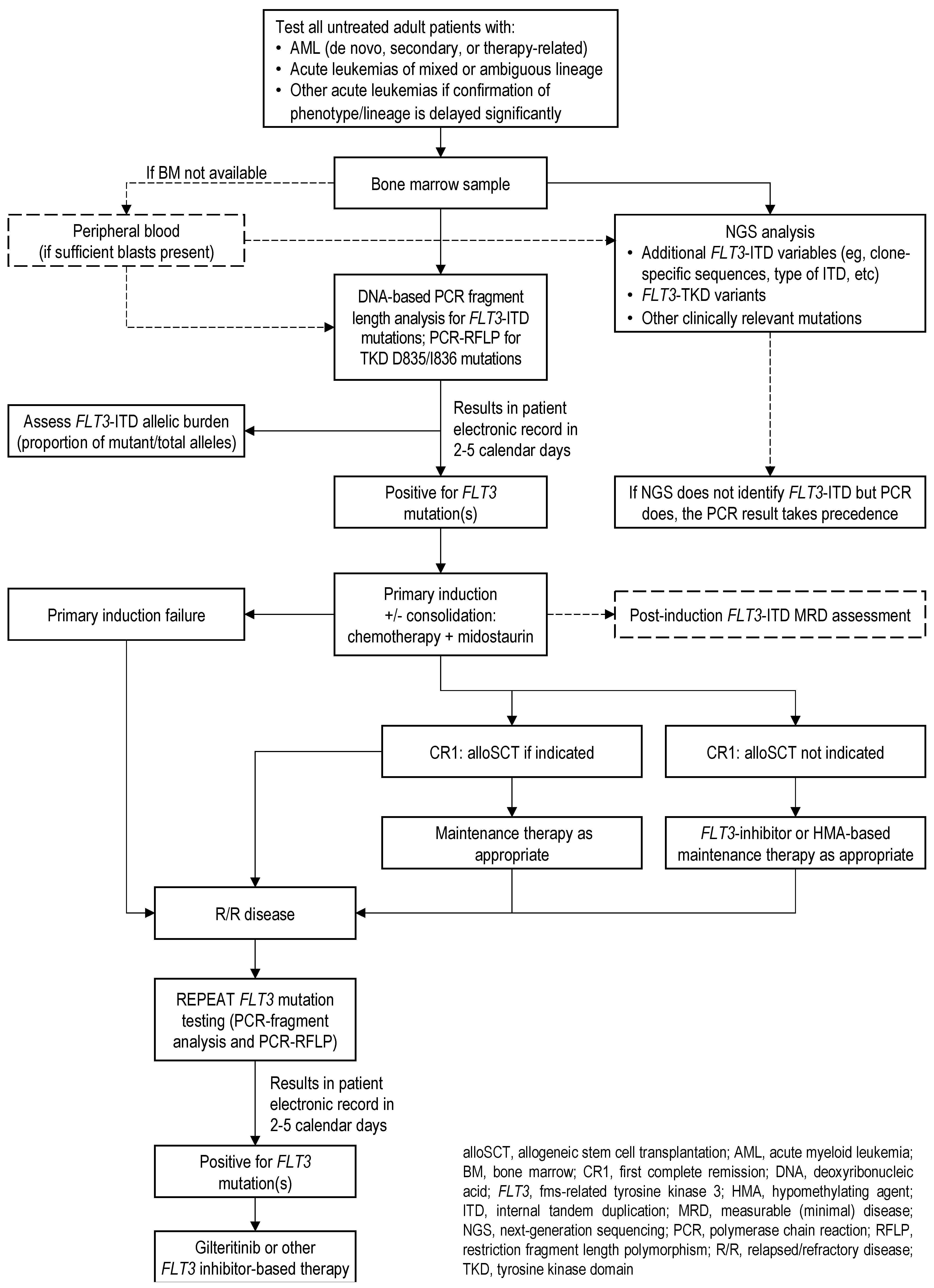
Q1. Which Patients Should Be Tested for FLT3 Mutations and When?
1.1. Which Patients Should Be Tested Upfront at the Time of Initial Diagnosis?
Recommendation 1.1
- AML (de novo, secondary, or therapy-related);
- Acute leukemias of mixed or ambiguous lineage;
- Other acute leukemias if there are significant delays in the confirmation of phenotype/lineage.
1.2. Should Patients of All Ages Be Tested?
Recommendation 1.2
1.3. Should Primary Induction Failures and Patients with R/R AML Be Retested?
Recommendation 1.3
Q2. Which Is the Preferred Method for FLT3 Mutation Testing?
Recommendation 2.1
- PCR-fragment analysis of FLT3-ITD is preferred over NGS to support the timely initiation of FLT3 inhibitor therapy;
- For FLT3-TKD detection, in the absence of an established standard, PCR-RFLP (directed to D835/I836) is preferred as it can be performed simultaneously with PCR-fragment analysis, with a relatively short combined turnaround time.
2.2. Which Is the Preferred Specimen for FLT3 Mutation Testing: Bone Marrow or Peripheral Blood?
Recommendation 2.2
- Testing on peripheral blood is acceptable if bone marrow is not available, provided that sufficient blasts are present;
- Caution is advised regarding the interpretation of results from low-blast count blood or marrow specimens.
2.3. Which Is the Preferred Analyte for FLT3 Mutation Testing: DNA or RNA?
Recommendation 2.3
2.4. Which Is the Preferred Technology for FLT3 Mutation Testing: PCR or NGS?
2.4.1. PCR-Based Testing and Analysis
2.4.2. NGS-Based Testing and Analysis
Recommendation 2.4
- PCR standardization efforts should ensure that the lower limit of detection (sensitivity) is between 1% and 5%;
- PCR primers for FLT3-ITD detection should span the juxtamembrane and part of the TKD1 domain of FLT3 encoded by exons 14 and 15;
- The technique employed should be able to detect large FLT3-ITDs (>200 base pairs, up to 400 base pairs);
- For allelic burden measurement, the PCR reaction should be performed at least in duplicate or triplicate; concordant results should be averaged for the AUC calculation.
- If PCR results are positive for FLT3-ITD but NGS fails to identify the FLT3-ITD, the PCR result takes precedence;
- When FLT3-TKD variants (particularly non-canonical variants) are detected by NGS, a short interpretation paragraph should be provided in the report;
- Non-canonical variants should be broadly classified (e.g., pathogenic, likely pathogenic, variant of uncertain significance) using existing molecular genetics somatic variant classification schemes such as AMP, ASCO, and CAP/ACP.
2.5. What Is the Recommended Turnaround Time for FLT3 Mutation Test Results?
Recommendation 2.5
3. What Is the Clinical Relevance of FLT3-ITD Size, Insertion Site, and Number of Distinct Mutations?
3.1. Does FLT3-ITD Size Matter?
Recommendation 3.1
3.2. Does the FLT3-ITD Insertion Site Matter?
Recommendation 3.2
3.3. Does the Number of Distinct FLT3-ITD Mutations Matter?
Recommendation 3.3
4. Is There a Role for FLT3-ITD Analysis in MRD Assessment?
Recommendation 4.1
5. What Is the Clinical Relevance of the FLT3-ITD Allelic Burden? Should It Be Assessed and If So, How?
5.1. What Is the Clinical Impact of the FLT3-ITD Allelic Burden?
Recommendation 5.1
- In the absence of definitive data to the contrary, the panel supports the continued use of FLT3-ITD allelic burden analysis to facilitate treatment decision-making;
- The panel agrees that all patients with either or both FLT3-ITD and -TKD at diagnosis, regardless of allelic burden, should receive midostaurin during induction and consolidation.
5.2. How Should the FLT3-ITD Allelic Burden Be Assessed?
- “PCR bias” or “template bias” (the preferential amplification of the shorter, wild-type allele during conventional PCR) can lead to a relative underestimation of the longer mutant ITD allele, leading to inaccurate estimates of the FLT3-ITD allelic burden; this problem becomes more pronounced as ITD length increases [45,73,115];
- The blast proportion in the specimen may influence the allelic burden [73];
- There is no internationally standardized method for determining the FLT3-ITD allelic burden, and even when multiple laboratories use the same methodology (in trials such as RATIFY, for example), results show considerable intra- and inter-lab variability in measurements. In the RATIFY analysis, however, the variability was significantly reduced with stringent inter-laboratory coordination and when each specimen was analyzed in triplicate [18,44].
Recommendation 5.2
- As per Recommendation 2.4, the PCR reaction should be performed at least in duplicate; concordant results should be averaged for the AUC calculation of FLT3-ITD and FLT3 wt peaks;
- If multiple FLT3-ITDs are found, the mutated fraction is determined by adding together the AUCs of all the detected FLT3-ITDs.
5.3. How Should the FLT3-ITD Allelic Burden Be Calculated and Reported?
| Measure * | Analysis † | Description | Calculation |
|---|---|---|---|
| Allelic ratio | PCR-fragment | Ratio of the AUC of mutant to wild-type alleles |  |
| Variant allele frequency | NGS | Reads of mutant alleles as a percentage of total (mutant + wt) reads | 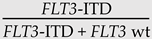 |
| Proportion of mutant alleles ‡ | PCR-fragment | AUC of mutant alleles as a proportion of total (mutant + wt) alleles | 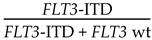 |
Recommendation 5.3
- The report should also note that ELN 2022 guidelines no longer include the FLT3-ITD allelic ratio in the risk stratification, although the FLT3-ITD allelic burden still continues to support clinical decision-making;
- This recommendation is made in the absence of an international standard for FLT3 testing, reporting, and interpretation and a lack of standardized laboratory reference values for the FLT3-ITD allelic burden. For these reasons, the report should specifically describe how the proportion of mutant/total alleles was assessed.
5.4. Can We Standardize How to Quantify and Report the FLT3-ITD Allelic Burden?
Recommendation 5.4
6. How Should Results of FLT3 Mutation Testing Be Reported?
6.1. PCR-Based FLT3 Mutation Testing Reports Should Include the following Elements
- Methodology
- ○
- Specimen type: bone marrow (preferred), whole blood, or purified blood mononuclear cells;
- ○
- Blast percentage in the specimen, if available;
- ○
- Analyte: DNA (preferred) or RNA;
- ○
- Extraction method;
- ○
- PCR method used (e.g., PCR-fragment analysis (preferred for FLT3-ITD), PCR-RLFP (preferred for FLT3-TKD D835/I836), digital PCR (dPCR), droplet digital PCR (ddPCR), and (rqPCR));
- ○
- Estimated sensitivity (limit of detection).
- Clinical information: specimen request forms should encourage prescribers to provide:
- ○
- Clinical context;
- ○
- Blast count and total white blood cell count of the submitted specimen.
- Results:
- ○
- All mutations detected;
- ○
- FLT3-ITD lower limit of detection;
- ○
- FLT3-ITD allelic burden, reported as the proportion of mutant/total alleles;
- ▪
- The method used to calculate the proportion of mutant/total alleles should be clearly defined to avoid confusion with allelic ratio and variant allele frequency;
- ○
- Insertion size, site, and distinct number of mutations (if more than one mutation is detected) play no role in clinical decision-making at this time and therefore do not need to be routinely reported.
Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALAL | acute leukemias of ambiguous lineage |
| alloSCT | allogeneic hematopoietic stem cell transplantation |
| AML | acute myeloid leukemia |
| AMLCG | (German) AML Cooperative Group |
| AMLSG | (German–Austrian) AML Study Group |
| AMP | Association for Molecular Pathology |
| AR | allelic ratio |
| ASCO | American Society of Clinical Oncology |
| AUC | area under the curve |
| CAP/ACP | Canadian Association of Pathologists/Association canadienne des pathologists |
| CLSG/GCEL | Canadian Leukemia Study Group/Groupe canadien d’étude sur la leucémie |
| CBF | core-binding factor |
| ddPCR | droplet digital PCR |
| DNA | deoxyribonucleic acid |
| dPCR | digital PCR |
| ELN | European LeukemiaNet |
| EFS | event-free survival |
| FLT3 | FMS-related tyrosine kinase 3 |
| FLT3-ITD | FLT3 internal tandem duplication |
| FLT3-TKD | FLT3 tyrosine kinase domain |
| HMA | hypomethylating agent |
| HPLC | high performance liquid chromatography |
| HTAS | high-throughput amplicon sequencing |
| ICC | International Consensus Classification |
| ITD | internal tandem duplication |
| MDS | myelodysplastic syndrome |
| MFC | multiparameter flow cytometry |
| MPAL | mixed phenotype acute leukemia |
| MRD | measurable (minimal) disease |
| NGS | next-generation sequencing |
| NPM1 | nucleophosmin 1 gene |
| OS | overall survival |
| PCR | polymerase chain reaction |
| PCR-RFLP | polymerase chain reaction-restriction fragment length polymorphism |
| RNA | ribonucleic acid |
| rqPCR | real-time quantitative PCR |
| R/R | relapsed/refractory |
| RFLP | restriction fragment length polymorphism |
| TKD | tyrosine kinase domain |
| TKI | tyrosine kinase inhibitor |
| UK MRC | United Kingdom Medical Research Council |
| VAF | variant allele frequency |
| WHO | World Health Organization |
| wt | wild-type |
References
- Fröhling, S.; Schlenk, R.F.; Breitruck, J.; Benner, A.; Kreitmeier, S.; Tobis, K.; Döhner, H.; Döhner, K. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: A study of the AML Study Group Ulm. Blood 2002, 100, 4372–4380. [Google Scholar] [CrossRef]
- Zarrinkar, P.P.; Gunawardane, R.N.; Cramer, M.D.; Gardner, M.F.; Brigham, D.; Belli, B.; Karaman, M.W.; Pratz, K.W.; Pallares, G.; Chao, Q.; et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML). Blood 2009, 114, 2984–2992. [Google Scholar] [CrossRef] [PubMed]
- Grafone, T.; Palmisano, M.; Nicci, C.; Storti, S. An overview on the role of FLT3-tyrosine kinase receptor in acute myeloid leukemia: Biology and treatment. Oncol. Rev. 2012, 6, e8. [Google Scholar] [CrossRef]
- Bacher, U.; Haferlach, C.; Kern, W.; Haferlach, T.; Schnittger, S. Prognostic relevance of FLT3-TKD mutations in AML: The combination matters—An analysis of 3082 patients. Blood 2008, 111, 2527–2537. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Schneider, F.; Hoster, E.; Schneider, S.; Dufour, A.; Benthaus, T.; Kakadia, P.M.; Bohlander, S.K.; Braess, J.; Heinecke, A.; Sauerland, M.C.; et al. Age-dependent frequencies of NPM1 mutations and FLT3-ITD in patients with normal karyotype AML (NK-AML). Ann. Hematol. 2012, 91, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kiyoi, H.; Nakano, Y.; Suzuki, R.; Kodera, Y.; Miyawaki, S.; Asou, N.; Kuriyama, K.; Yagasaki, F.; Shimazaki, C.; et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 2001, 97, 2434–2439. [Google Scholar] [CrossRef] [PubMed]
- Abu-Duhier, F.M.; Goodeve, A.C.; Wilson, G.A.; Carr, R.S.; Peake, I.R.; Reilly, J.T. FLT3 internal tandem duplication mutations are rare in agnogenic myeloid metaplasia. Blood 2002, 100, 364. [Google Scholar] [CrossRef][Green Version]
- Schlenk, R.F.; Kayser, S.; Bullinger, L.; Kobbe, G.; Casper, J.; Ringhoffer, M.; Held, G.; Brossart, P.; Lübbert, M.; Salih, H.R.; et al. Differential impact of allelic ratio and insertion site in FLT3-ITD–positive AML with respect to allogeneic transplantation. Blood 2014, 124, 3441–3449. [Google Scholar] [CrossRef]
- Dalle, I.A.; Ghorab, A.; Patel, K.; Wang, X.; Hwang, H.; Cortes, J.; Issa, G.C.; Yalniz, F.; Sasaki, K.; Chihara, D.; et al. Impact of numerical variation, allele burden, mutation length and co-occurring mutations on the efficacy of tyrosine kinase inhibitors in newly diagnosed FLT3- mutant acute myeloid leukemia. Blood Cancer J. 2020, 10, 48. [Google Scholar] [CrossRef]
- Smith, B.D.; Levis, M.; Beran, M.; Giles, F.; Kantarjian, H.; Berg, K.; Murphy, K.M.; Dauses, T.; Allebach, J.; Small, D. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood 2004, 103, 3669–3676. [Google Scholar] [CrossRef] [PubMed]
- Pratcorona, M.; Brunet, S.; Nomdedeu, J.; Ribera, J.M.; Tormo, M.; Duarte, R.; Escoda, L.; Guardia, R.; de Llano, M.P.Q.; Salamero, O.; et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: Relevance to post-remission therapy. Blood 2013, 121, 2734–2738. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Tallman, M.S.; Schiller, G.J.; Trone, D.; Gammon, G.; Goldberg, S.L.; Perl, A.E.; Marie, J.P.; Martinelli, G.; Kantarjian, H.M.; et al. Phase 2b study of 2 dosing regimens of quizartinib monotherapy in FLT3-ITD–mutated, relapsed or refractory AML. Blood 2018, 132, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Khaled, S.; Martinelli, G.; Perl, A.A.; Ganguly, S.; Russell, N.; Krämer, A.; Dombret, H.; Hogge, D.; Jonas, B.A.; et al. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): A multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2019, 20, 984–997. [Google Scholar] [CrossRef]
- Smith, C.C.; Zhang, C.; Lin, K.C.; Lasater, E.A.; Zhang, Y.; Massi, E.; Damon, L.E.; Pendleton, M.; Bashir, A.; Sebra, R.; et al. Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 “Gatekeeper” F691L Mutation with PLX3397. Cancer Discov. 2015, 5, 668–679. [Google Scholar] [CrossRef]
- Daver, N.; Schlenk, R.F.; Russell, N.H.; Levis, M.J. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia 2019, 33, 299–312. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Döhner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Perl, A.E.; Martinelli, G.; Cortes, J.E.; Neubauer, A.; Berman, E.; Paolini, S.; Montesinos, P.; Baer, M.R.; Larson, R.A.; Ustun, C.; et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3-Mutated AML. N. Engl. J. Med. 2019, 381, 1728–1740. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Tallman, M.S.; Wang, E.S.; Altman, J.K.; Appelbaum, F.R.; Bhatt, V.R.; Bixby, D.; Coutre, S.E.; De Lima, M.; Fathi, A.T.; Fiorella, M.; et al. Acute Myeloid Leukemia, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 721–749. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Schlenk, R.F.; Döhner, K.; Krauter, J.; Fröhling, S.; Corbacioglu, A.; Bullinger, L.; Habdank, M.; Späth, D.; Morgan, M.; Benner, A.; et al. Mutations and Treatment Outcome in Cytogenetically Normal Acute Myeloid Leukemia. N. Engl. J. Med. 2008, 358, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Gale, R.E.; Green, C.; Allen, C.; Mead, A.J.; Burnett, A.K.; Hills, R.K.; Linch, D.C. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood 2008, 111, 2776–2784. [Google Scholar] [CrossRef]
- Ho, A.D.; Schetelig, J.; Bochtler, T.; Schaich, M.; Schäfer-Eckart, K.; Hänel, M.; Rösler, W.; Einsele, H.; Kaufmann, M.; Serve, H.; et al. Allogeneic Stem Cell Transplantation Improves Survival in Patients with Acute Myeloid Leukemia Characterized by a High Allelic Ratio of Mutant FLT3-ITD. Biol. Blood Marrow Transplant. 2016, 22, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Linch, D.C.; Hills, R.K.; Burnett, A.K.; Khwaja, A.; Gale, R.E. Impact of FLT3ITD mutant allele level on relapse risk in intermediate-risk acute myeloid leukemia. Blood 2014, 124, 273–276. [Google Scholar] [CrossRef]
- Thiede, C.; Steudel, C.; Mohr, B.; Schaich, M.; Schäkel, U.; Platzbecker, U.; Wermke, M.; Bornhäuser, M.; Ritter, M.; Neubauer, A.; et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002, 99, 4326–4335. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Maiti, A.; Rausch, C.R.; Pemmaraju, N.; Naqvi, K.; Daver, N.G.; Kadia, T.M.; Borthakur, G.; Ohanian, M.; Alvarado, Y.; et al. 10-day decitabine with venetoclax for newly diagnosed intensive chemotherapy ineligible, and relapsed or refractory acute myeloid leukaemia: A single-centre, phase 2 trial. Lancet Haematol. 2020, 7, e724–e736. [Google Scholar] [CrossRef]
- Maiti, A.; DiNardo, C.D.; Ravandi, F.; Pemmaraju, N.; Borthakur, G.; Bose, P.; Issa, G.C.; Kadia, T.M.; Short, N.J.; Yilmaz, M.; et al. Venetoclax, FLT3 Inhibitor and Decitabine in FLT3mut Acute Myeloid Leukemia: Subgroup Analysis of a Phase II Trial. Blood 2020, 136 (Suppl. 1), 53–55. [Google Scholar] [CrossRef]
- Yilmaz, M.; Kantarjian, H.M.; Muftuoglu, M.; Kadia, T.M.; Konopleva, M.; Borthakur, G.; DiNardo, C.D.; Pemmaraju, N.; Short, N.J.; Alvarado, Y.; et al. Quizartinib with Decitabine +/− Venetoclax Is Highly Active in Patients (Pts) with FLT3-ITD Mutated (mut) Acute Myeloid Leukemia (AML): Clinical Report and Signaling Cytof Profiling from a Phase IB/II Trial. Blood 2020, 136 (Suppl. 1), 19–20. [Google Scholar] [CrossRef]
- Wang, E.S.; Montesinos, P.; Minden, M.D.; Lee, J.H.; Heuser, M.; Naoe, T.; Chou, W.C.; Liu, S.; Wu, R.; Philipose, N.; et al. Phase 3, Multicenter, Open-Label Study of Gilteritinib, Gilteritinib Plus Azacitidine, or Azacitidine Alone in Newly Diagnosed FLT3 Mutated (FLT3mut+) Acute Myeloid Leukemia (AML) Patients Ineligible for Intensive Induction Chemotherapy. Blood 2020, 136 (Suppl. 1), 27. [Google Scholar]
- Daver, N.; Venugopal, S.; Ravandi, F. FLT3 mutated acute myeloid leukemia: 2021 treatment algorithm. Blood Cancer J. 2021, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.; Perl, A.E.; Maly, J.; Levis, M.; Ritchie, E.; Litzow, M.R.; McCloskey, J.; Smith, C.C.; Schiller, G.J.; Bradley, T.; et al. Venetoclax in Combination with Gilteritinib Demonstrates Molecular Clearance of FLT3 mutation in Relapsed/Refractory FLT3-Mutated Acute Myeloid Leukemia. Blood 2021, 138, 691. [Google Scholar] [CrossRef]
- Short, N.; DiNardo, C.D.; Daver, N.; Macaron, W.; Yilmaz, M.; Borthakur, G.; Montalban-Bravo, G.; Garcia-Manero, G.; Issa, G.C.; Sasaki, K.; et al. Updated results from a Phase I/II study of the triplet combination of azacitidine, venetoclax and gilteritinib for patients with FLT3-mutated acute myeloid leukemia. Blood 2022, 140, 2007–2009. [Google Scholar] [CrossRef]
- Schuh, A.C. The Canadian AML Treatment Landscape—2019. In Proceedings of the Canadian Conference on Myelodysplastic Syndromes 2019, Vancouver, BC, Canada, 19 September 2019. [Google Scholar]
- Cucchi, D.G.J.; Denys, B.; Kaspers, G.J.L.; Janssen, J.J.W.M.; Ossenkoppele, G.J.; de Haas, V.; Zwaan, C.M.; Heuvel-Eibrink, M.M.v.D.; Philippé, J.; Csikós, T.; et al. RNA-based FLT3-ITD allelic ratio is associated with outcome and ex vivo response to FLT3 inhibitors in pediatric AML. Blood 2018, 131, 2485–2489. [Google Scholar] [CrossRef] [PubMed]
- Stirewalt, D.L.; Kopecky, K.J.; Meshinchi, S.; Engel, J.H.; Pogosova-Agadjanyan, E.L.; Linsley, J.; Slovak, M.L.; Willman, C.L.; Radich, J.P. Size of FLT3 internal tandem duplication has prognostic significance in patients with acute myeloid leukemia. Blood 2006, 107, 3724–3726. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Sun, J.; Yin, C.; Cheng, J.; Ni, J.; Jiang, L.; Wang, Q.; Yu, G.; Wei, Y.; Liu, X.; et al. Impact of FLT3-ITD allele ratio and ITD length on therapeutic outcome in cytogenetically normal AML patients without NPM1 mutation. Bone Marrow Transplant. 2020, 55, 740–748. [Google Scholar] [CrossRef]
- Ponziani, V.; Gianfaldoni, G.; Mannelli, F.; Leoni, F.; Ciolli, S.; Guglielmelli, P.; Antonioli, E.; Longo, G.; Bosi, A.; Vannucchi, A.M. The size of duplication does not add to the prognostic significance of FLT3 internal tandem duplication in acute myeloid leukemia patients. Leukemia 2006, 20, 2074–2076. [Google Scholar] [CrossRef]
- Schranz, K.; Hubmann, M.; Harin, E.; Vosberg, S.; Herold, T.; Metzeler, K.H.; Rothenberg-Thurley, M.; Janke, H.; Bräundl, K.; Ksienzyk, B.; et al. Clonal heterogeneity of FLT3-ITD detected by high-throughput amplicon sequencing correlates with adverse prognosis in acute myeloid leukemia. Oncotarget 2018, 9, 30128–30145. [Google Scholar] [CrossRef]
- Blau, O.; Berenstein, R.; Sindram, A.; Blau, I.W. Molecular analysis of different FLT3-ITD mutations in acute myeloid leukemia. Leuk. Lymphoma 2013, 54, 145–152. [Google Scholar] [CrossRef]
- Rücker, F.G.; Du, L.; Luck, T.J.; Benner, A.; Krzykalla, J.; Gathmann, I.; Voso, M.T.; Amadori, S.; Prior, T.W.; Brandwein, J.M.; et al. Molecular landscape and prognostic impact of FLT3-ITD insertion site in acute myeloid leukemia: RATIFY study results. Leukemia 2022, 36, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Meshinchi, S.; Stirewalt, D.L.; Alonzo, T.A.; Boggon, T.J.; Gerbing, R.B.; Rocnik, J.L.; Lange, B.J.; Gilliland, D.G.; Radich, J.P. Structural and numerical variation of FLT3/ITD in pediatric AML. Blood 2008, 111, 4930–4933. [Google Scholar] [CrossRef] [PubMed]
- Thiede, C.; Prior, T.W.; Lavorgna, S.; Krauter, J.; Barragán, E.; Nomdedeu, J.; Jansen, J.H.; Wei, A.H.; Zhao, W.; Li, X.; et al. FLT3mutation Assay Laboratory Cross Validation: Results from the CALGB 10603/Ratify Trial in Patients with Newly Diagnosed FLT3-Mutated Acute Myeloid Leukemia (AML). Blood 2018, 132, abs 2800. [Google Scholar] [CrossRef]
- Levis, M.; Perl, A.E. Gilteritinib: Potent targeting of FLT3 mutations in AML. Blood Adv. 2020, 4, 1178–1191. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.-M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemia: Integrating Morphological, Clinical, and Genomic Data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- National Cancer Comprehensive Network. Acute Myeloid Leukemia (Version 6.2023). Updated 24 October 2023. Available online: https://www.nccn.org/professionals/physician_gls/pdf/aml.pdf (accessed on 8 December 2023).
- Boddu, P.; Kantarjian, H.; Borthakur, G.; Kadia, T.; Daver, N.; Pierce, S.; Andreeff, M.; Ravandi, F.; Cortes, J.; Kornblau, S.M. Co-occurrence of FLT3-TKD and NPM1 mutations defines a highly favorable prognostic AML group. Blood Adv. 2017, 1, 1546–1550. [Google Scholar] [CrossRef]
- Perry, M.; Bertoli, S.; Rocher, C.; Hayette, S.; Ducastelle, S.; Barraco, F.; Labussière-Wallet, H.; Salles, G.; Recher, C.; Thomas, X.; et al. FLT3-TKD Mutations Associated With NPM1 Mutations Define a Favorable-risk Group in Patients With Acute Myeloid Leukemia. Clin. Lymphoma Myeloma Leuk. 2018, 18, e545–e550. [Google Scholar] [CrossRef]
- Oran, B.; Cortes, J.; Beitinjaneh, A.; Chen, H.-C.; de Lima, M.; Patel, K.; Ravandi, F.; Wang, X.; Brandt, M.; Andersson, B.S.; et al. Allogeneic Transplantation in First Remission Improves Outcomes Irrespective of FLT3-ITD Allelic Ratio in FLT3-ITD–Positive Acute Myelogenous Leukemia. Biol. Blood Marrow Transplant. 2016, 22, 1218–1226. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Yamaguchi, H.; Najima, Y.; Usuki, K.; Ueki, T.; Oh, I.; Mori, S.; Kawata, E.; Uoshima, N.; Kobayashi, Y.; et al. Prognostic impact of low allelic ratio FLT3-ITD and NPM1 mutation in acute myeloid leukemia. Blood Adv. 2018, 2, 2744–2754. [Google Scholar] [CrossRef] [PubMed]
- Brunet, S.; Labopin, M.; Esteve, J.; Cornelissen, J.; Socié, G.; Iori, A.P.; Verdonck, L.F.; Volin, L.; Gratwohl, A.; Sierra, J.; et al. Impact of FLT3 internal tandem duplication on the outcome of related and unrelated hematopoietic transplantation for adult acute myeloid leukemia in first remission: A retrospective analysis. J. Clin. Oncol. 2012, 30, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Wolach, O.; Stone, R.M. Optimal therapeutic strategies for mixed phenotype acute leukemia. Curr. Opin. Hematol. 2020, 27, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Hoehn, D.; Medeiros, L.J.; Chen, S.S.; Tian, T.; Jorgensen, J.L.; Ahmed, Y.; Lin, P. CD117 expression is a sensitive but nonspecific predictor of FLT3 mutation in T acute lymphoblastic leukemia and T/myeloid acute leukemia. Am. J. Clin. Pathol. 2012, 137, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Bharadwaj, M.; Levine, M.; Farnoud, N.; Pastore, F.; Getta, B.M.; Hultquist, A.; Famulare, C.; Medina, J.S.; Patel, M.A.; et al. PHF6 and DNMT3A mutations are enriched in distinct subgroups of mixed phenotype acute leukemia with T-lineage differentiation. Blood Adv. 2018, 2, 3526–3539. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, L.E.; Bendig, S.; Gu, Z.; Chen, X.; Pölönen, P.; Ma, X.; Murison, A.; Zeng, A.; Garcia-Prat, L.; Dickerson, K.; et al. Enhancer Hijacking Drives Oncogenic BCL11B Expression in Lineage-Ambiguous Stem Cell Leukemia. Cancer Discov. 2021, 11, 2846–2867. [Google Scholar] [CrossRef]
- Di Giacomo, D.; La Starza, R.; Gorello, P.; Pellanera, F.; Atak, Z.K.; De Keersmaecker, K.; Pierini, V.; Harrison, C.J.; Arniani, S.; Moretti, M.; et al. 14q32 rearrangements deregulating BCL11B mark a distinct subgroup of T-lymphoid and myeloid immature acute leukemia. Blood 2021, 138, 773–784. [Google Scholar] [CrossRef]
- Andrews, C.; Lam, W.; Sibai, H. The successful use of FLT3 inhibitors in FLT3-positive mixed phenotype acute leukemia. Leuk. Lymphoma 2020, 61, 3275–3277. [Google Scholar] [CrossRef]
- Tremblay, Z.; Wong, A.; Otis, A.S.; Pépin, M.A.; Bambace, N.; Soulières, D.; Bouchard, P.; Adam, J. Use of midostaurin in mixed phenotype acute leukemia with FLT3 mutation: A case series. Eur. J. Haematol. 2022, 108, 163–165. [Google Scholar] [CrossRef]
- Döhner, H.; Weber, D.; Krzykalla, J.; Fiedler, W.; Wulf, G.; Salih, H.; Lübbert, M.; Kühn, M.W.M.; Schroeder, T.; Salwender, H.; et al. Midostaurin plus intensive chemotherapy for younger and older patients with AML and FLT3 internal tandem duplications. Blood Adv. 2022, 6, 5345–5355. [Google Scholar] [CrossRef]
- Short, N.J.; DiNardo, C.D.; Daver, N.; Nguyen, D.; Yilmaz, M.; Kadia, T.M.; Garcia-Manero, G.; Issa, G.C.; Huang, X.; Qiao, W.; et al. A Triplet Combination of Azacitidine, Venetoclax and Gilteritinib for Patients with FLT3-Mutated Acute Myeloid Leukemia: Results from a Phase I/II Study. Blood 2021, 138, 696. [Google Scholar] [CrossRef]
- Cloos, J.; Goemans, B.F.; Hess, C.J.; van Oostveen, J.W.; Waisfisz, Q.; Corthals, S.; de Lange, D.; Boeckx, N.; Hählen, K.; Reinhardt, D.; et al. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia 2006, 20, 1217–1220. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.R.; McCormick, M.J.; Grutkoski, P.S.; Ducker, G.S.; Banerji, N.; Higgins, R.R.; Mendiola, J.R.; Reinartz, J.J. FLT3 mutations at diagnosis and relapse in acute myeloid leukemia: Cytogenetic and pathologic correlations, including cuplike blast morphology. Arch. Pathol. Lab. Med. 2010, 134, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Schmalbrock, L.K.; Dolnik, A.; Cocciardi, S.; Sträng, E.; Theis, F.; Jahn, N.; Panina, E.; Blätte, T.J.; Herzig, J.; Skambraks, S.; et al. Clonal evolution of acute myeloid leukemia with FLT3-ITD mutation under treatment with midostaurin. Blood 2021, 137, 3093–3104. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.; Luthra, R.; Yin, C.C.; Ravandi, F.; E Cortes, J.; Kantarjian, H.M.; Medeiros, L.J.; Zuo, Z. Clinical impact of change of FLT3 mutation status in acute myeloid leukemia patients. Mod. Pathol. 2012, 25, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Gilteritinib. (Xospata) Product Monograph; Astellas Pharma Inc.: Markham, ON, Canada, 2019. [Google Scholar]
- Tong, W.G.; Sandhu, V.K.; Wood, B.L.; Hendrie, P.C.; Becker, P.S.; Pagel, J.M.; Walter, R.B.; Estey, E.H. Correlation between peripheral blood and bone marrow regarding FLT3-ITD and NPM1 mutational status in patients with acute myeloid leukemia. Haematologica 2015, 100, e97–e98. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Spencer, D.H.; Abel, H.J.; Lockwood, C.M.; Payton, J.E.; Szankasi, P.; Kelley, T.W.; Kulkarni, S.; Pfeifer, J.D.; Duncavage, E.J. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J. Mol. Diagn. 2013, 15, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, S.; Lee, S.-T.; Min, Y.H.; Choi, J.R. FLT3 Internal Tandem Duplication in Patients with Acute Myeloid Leukemia Is Readily Detectable in a Single Next-Generation Sequencing Assay Using the Pindel Algorithm. Ann. Lab. Med. 2019, 39, 327–329. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Nakajima, N.; Yamaguchi, H.; Najima, Y.; Shono, K.; Marumo, A.; Omori, I.; Fujiwara, Y.; Terada, K.; Yui, S.; et al. The sensitivity of the FLT3-ITD detection method is an important consideration when diagnosing acute myeloid leukemia. Leuk. Res. Rep. 2020, 13, 100198. [Google Scholar] [CrossRef]
- Di Ciaccio, P.; Kearney, A.; Ling, S. Comparing the area under the curve and peak height methods in the calculation of FLT3-ITD allelic ratio. Int. J. Lab. Hematol. 2020, 42, e234–e236. [Google Scholar] [CrossRef]
- Kiyoi, H.; Naoe, T.; Nakano, Y.; Yokota, S.; Minami, S.; Miyawaki, S.; Asou, N.; Kuriyama, K.; Jinnai, I.; Shimazaki, C.; et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood 1999, 93, 3074–3080. [Google Scholar]
- Pratz, K.W.; Levis, M. How I treat FLT3-mutated AML. Blood 2017, 129, 565–571. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Bixby, D.; Perl, A.; Bhatt, V.R.; Altman, J.K.; Appelbaum, F.R.; de Lima, M.; Fathi, A.T.; Foran, J.M.; Gojo, I.; et al. NCCN Guidelines Insights: Acute Myeloid Leukemia, Version 2.2021. J. Natl. Compr. Cancer Netw. 2021, 19, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.M.; Levis, M.; Hafez, M.J.; Geiger, T.; Cooper, L.C.; Smith, B.; Small, D.; Berg, K.D. Detection of FLT3 Internal Tandem Duplication and D835 Mutations by a Multiplex Polymerase Chain Reaction and Capillary Electrophoresis Assay. J. Mol. Diagn. 2003, 5, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, S.; Schoch, C.; Dugas, M.; Kern, W.; Staib, P.; Wuchter, C.; Löffler, H.; Sauerland, C.M.; Serve, H.; Büchner, T.; et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: Correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002, 100, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Kayser, S.; Schlenk, R.F.; Londono, M.C.; Breitenbuecher, F.; Wittke, K.; Du, J.; Groner, S.; Späth, D.; Krauter, J.; Ganser, A.; et al. Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood 2009, 114, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Nucleic Acid Amplification Assays for Molecular Hematopathology; Approved Guideline, 2nd ed.; CLSI document MM05-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- CLSI. Molecular Methods for Clincial Genetics and Oncology Testing; Approved Guideline, 2nd ed.; CLSI document MM01-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Zwaan, C.M.; Meshinchi, S.; Radich, J.P.; Veerman, A.J.P.; Huismans, D.R.; Munske, L.; Podleschny, M.; Hählen, K.; Pieters, R.; Zimmermann, M.; et al. FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: Prognostic significance and relation to cellular drug resistance. Blood 2003, 102, 2387–2394. [Google Scholar] [CrossRef]
- Thol, F.; Kölking, B.; Damm, F.; Reinhardt, K.; Klusmann, J.-H.; Reinhardt, D.; von Neuhoff, N.; Brugman, M.H.; Schlegelberger, B.; Suerbaum, S.; et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes, Chromosom. Cancer 2012, 51, 689–695. [Google Scholar] [CrossRef]
- Tsai, H.K.; Brackett, D.G.; Szeto, D.; Frazier, R.; MacLeay, A.; Davineni, P.; Manning, D.K.; Garcia, E.; Lindeman, N.I.; Le, L.P.; et al. Targeted Informatics for Optimal Detection, Characterization, and Quantification of FLT3 Internal Tandem Duplications Across Multiple Next-Generation Sequencing Platforms. J. Mol. Diagn. 2020, 22, 1162–1178. [Google Scholar] [CrossRef]
- Tung, J.K.; Suarez, C.J.; Chiang, T.; Zehnder, J.L.; Stehr, H. Accurate Detection and Quantification of FLT3 Internal Tandem Duplications in Clinical Hybrid Capture Next-Generation Sequencing Data. J. Mol. Diagn. 2021, 23, 1404–1413. [Google Scholar] [CrossRef]
- Blätte, T.J.; Schmalbrock, L.K.; Skambraks, S.; Lux, S.; Cocciardi, S.; Dolnik, A.; Döhner, H.; Döhner, K.; Bullinger, L. getITD for FLT3-ITD-based MRD monitoring in AML. Leukemia 2019, 33, 2535–2539. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Lee, K.S.; Lee, T.G.; Lee, S.; Shin, S.; Lee, S.-T. A comparative study of next-generation sequencing and fragment analysis for the detection and allelic ratio determination of FLT3 internal tandem duplication. Diagn. Pathol. 2022, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Bibault, J.-E.; Figeac, M.; Hélevaut, N.; Rodriguez, C.; Quief, S.; Sebda, S.; Renneville, A.; Nibourel, O.; Rousselot, P.; Gruson, B.; et al. Next-generation sequencing of FLT3 internal tandem duplications for minimal residual disease monitoring in acute myeloid leukemia. Oncotarget 2015, 6, 22812–22821. [Google Scholar] [CrossRef] [PubMed]
- Ambinder, A.J.; Levis, M. Potential targeting of FLT3 acute myeloid leukemia. Haematologica 2021, 106, 671–681. [Google Scholar] [CrossRef]
- Fröhling, S.; Scholl, C.; Levine, R.L.; Loriaux, M.; Boggon, T.J.; Bernard, O.A.; Berger, R.; Döhner, H.; Döhner, K.; Ebert, B.L.; et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell 2007, 12, 501–513. [Google Scholar] [CrossRef]
- Young, D.J.; Nguyen, B.; Zhu, R.; Seo, J.; Li, L.; Levis, M.J.; Pratz, K.W.; Duffield, A.S.; Small, D. Deletions in FLT-3 juxtamembrane domain define a new class of pathogenic mutations: Case report and systematic analysis. Blood Adv. 2021, 5, 2285–2293. [Google Scholar] [CrossRef]
- Reindl, C.; Bagrintseva, K.; Vempati, S.; Schnittger, S.; Ellwart, J.W.; Wenig, K.; Hopfner, K.-P.; Hiddemann, W.; Spiekermann, K. Point mutations in the juxtamembrane domain of FLT3 define a new class of activating mutations in AML. Blood 2006, 107, 3700–3707. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Russell, N.; Wilhelm-Benartzi, C.; Othman, J.; Dillon, R.; Potter, N.; Jovanovic, J.; Gilkes, A.; Batten, L.M.; Canham, J.; Hinson, E.L.; et al. S126: Gemtuzumab-based induction chemotherapy combined with midostaurin for FLT3 mutated AML. Results from the NCRI AML19 “Midotarg” pilot trial. HemaSphere 2022, 6, 27–28. [Google Scholar] [CrossRef]
- Kottaridis, P.D.; Gale, R.E.; Langabeer, S.E.; Frew, M.E.; Bowen, D.T.; Linch, D.C. Studies of FLT3 mutations in paired presentation and relapse samples from patients with acute myeloid leukemia: Implications for the role of FLT3 mutations in leukemogenesis, minimal residual disease detection, and possible therapy with FLT3 inhibitors. Blood 2002, 100, 2393–2398. [Google Scholar] [CrossRef]
- Corces-Zimmerman, M.R.; Hong, W.-J.; Weissman, I.L.; Medeiros, B.C.; Majeti, R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA 2014, 111, 2548–2553. [Google Scholar] [CrossRef] [PubMed]
- Garg, M.; Nagata, Y.; Kanojia, D.; Mayakonda, A.; Yoshida, K.; Keloth, S.H.; Zang, Z.J.; Okuno, Y.; Shiraishi, Y.; Chiba, K.; et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood 2015, 126, 2491–2501. [Google Scholar] [CrossRef] [PubMed]
- Krönke, J.; Bullinger, L.; Teleanu, V.; Tschürtz, F.; Gaidzik, V.I.; Kühn, M.W.M.; Rücker, F.G.; Holzmann, K.; Paschka, P.; Kapp-Schwörer, S.; et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood 2013, 122, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.-C.; et al. 2021 Update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef] [PubMed]
- Grob, T.; Sanders, M.A.; Vonk, C.M.; Kavelaars, F.G.; Rijken, M.; Hanekamp, D.W.; Gradowska, P.L.; Cloos, J.; Fløisand, Y.; Kooy, M.v.M.; et al. Prognostic Value of FLT3-Internal Tandem Duplication Residual Disease in Acute Myeloid Leukemia. J. Clin. Oncol. 2023, 41, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Loo, S.; Dillon, R.; Ivey, A.; Anstee, N.S.; Othman, J.; Tiong, I.S.; Potter, N.; Jovanovic, J.; Runglall, M.; Chong, C.C.; et al. Pretransplant FLT3-ITD MRD assessed by high-sensitivity PCR-NGS determines posttransplant clinical outcome. Blood 2022, 140, 2407–2411. [Google Scholar] [CrossRef] [PubMed]
- Othman, J.; Potter, N.; Mokretar, K.; Taussig, D.; Khan, A.; Krishnamurthy, P.; Latif, A.L.; Cahalin, P.; Aries, J.; Amer, M.; et al. FLT3 inhibitors as MRD-guided salvage treatment for molecular failure in FLT3 mutated AML. Leukemia 2023, 37, 2066–2072. [Google Scholar] [CrossRef]
- Levis, M.J.; Hamadani, M.; Logan, B.; Jones, R.; Singh, A.K.; Litzow, M.; Wingard, J.R.; Papadopoulos, E.B.; Perl, A.E.; Soiffer, R.; et al. BMT-CTN 1506 (MORPHO): A randomized trial of the FLT3 inhibitor gliteritinib as post-transplant maintenance for FLT3-ITD AML. In Proceedings of the 28th Congress of the European Hematology Association, Frankfurt, Germany, 8–11 June 2023; Volume 7, p. LB2711. [Google Scholar]
- Boddu, P.C.; Kadia, T.M.; Garcia-Manero, G.; Cortes, J.; Alfayez, M.; Borthakur, G.; Konopleva, M.; Jabbour, E.J.; Daver, N.G.; DiNardo, C.D.; et al. Validation of the 2017 European LeukemiaNet classification for acute myeloid leukemia with NPM1 and FLT3-internal tandem duplication genotypes. Cancer 2019, 125, 1091–1100. [Google Scholar] [CrossRef]
- Harada, Y.; Nagata, Y.; Kihara, R.; Ishikawa, Y.; Asou, N.; Ohtake, S.; Miyawaki, S.; Sakura, T.; Ozawa, Y.; Usui, N.; et al. Prognostic analysis according to the 2017 ELN risk stratification by genetics in adult acute myeloid leukemia patients treated in the Japan Adult Leukemia Study Group (JALSG) AML201 study. Leuk. Res. 2018, 66, 20–27. [Google Scholar] [CrossRef]
- Meshinchi, S.; Alonzo, T.A.; Stirewalt, D.L.; Zwaan, M.; Zimmerman, M.; Reinhardt, D.; Kaspers, G.J.L.; Heerema, N.A.; Gerbing, R.; Lange, B.J.; et al. Clinical implications of FLT3 mutations in pediatric AML. Blood 2006, 108, 3654–3661. [Google Scholar] [CrossRef]
- Schnittger, S.; Bacher, U.; Kern, W.; Alpermann, T.; Haferlach, C.; Haferlach, T. Prognostic impact of FLT3-ITD load in NPM1 mutated acute myeloid leukemia. Leukemia 2011, 25, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Yalniz, F.; Dalle, I.A.; Kantarjian, H.; Borthakur, G.; Kadia, T.; Patel, K.; Loghavi, S.; Garcia-Manero, G.; Sasaki, K.; Daver, N.; et al. Prognostic significance of baseline FLT3-ITD mutant allele level in acute myeloid leukemia treated with intensive chemotherapy with/without sorafenib. Am. J. Hematol. 2019, 94, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.E.; Cortes, J.E.; Ganguly, S.; Khaled, S.K.; Krämer, A.; Martinelli, G.; Russell, N.H.; Chang, K.C.; Mires, D.E.; Kato, K.; et al. Effect of Co-Mutations and FLT3-ITD Variant Allele Frequency (VAF) on Response to Quizartinib or Salvage Chemotherapy (SC) in Relapsed/Refractory (R/R) Acute Myeloid Leukemia (AML). Blood 2019, 134, 737. [Google Scholar] [CrossRef]
- Engen, C.; Hellesøy, M.; Grob, T.; Al Hinai, A.; Brendehaug, A.; Wergeland, L.; Bedringaas, S.L.; Hovland, R.; Valk, P.J.M.; Gjertsen, B.T. FLT3-ITD mutations in acute myeloid leukaemia—Molecular characteristics, distribution and numerical variation. Mol. Oncol. 2021, 15, 2300–2317. [Google Scholar] [CrossRef] [PubMed]
- Straube, J.; Ling, V.Y.; Hill, G.R.; Lane, S.W. The impact of age, NPM1mut, and FLT3ITD allelic ratio in patients with acute myeloid leukemia. Blood 2018, 131, 1148–1153. [Google Scholar] [CrossRef]
- Döhner, K.; Thiede, C.; Jahn, N.; Panina, E.; Gambietz, A.; Larson, R.A.; Prior, T.W.; Marcucci, G.; Jones, D.; Krauter, J.; et al. Impact of NPM1/FLT3-ITD genotypes defined by the 2017 European LeukemiaNet in patients with acute myeloid leukemia. Blood 2020, 135, 371–380. [Google Scholar] [CrossRef]
- Bataller, A.; Oñate, G.; Diaz-Beyá, M.; Guijarro, F.; Garrido, A.; Vives, S.; Tormo, M.; Arnan, M.; Salamero, O.; Sampol, A.; et al. Acute myeloid leukemia with NPM1 mutation and favorable European LeukemiaNet category: Outcome after preemptive intervention based on measurable residual disease. Br. J. Haematol. 2020, 191, 52–61. [Google Scholar] [CrossRef]
- Balsat, M.; Renneville, A.; Thomas, X.; de Botton, S.; Caillot, D.; Marceau, A.; Lemasle, E.; Marolleau, J.-P.; Nibourel, O.; Berthon, C.; et al. Postinduction Minimal Residual Disease Predicts Outcome and Benefit from Allogeneic Stem Cell Transplantation in Acute Myeloid Leukemia with NPM1 Mutation: A Study by the Acute Leukemia French Association Group. J. Clin. Oncol. 2017, 35, 185–193. [Google Scholar] [CrossRef]
- Levis, M. FLT3 mutations in acute myeloid leukemia: What is the best approach in 2013? Hematol. Am. Soc. Hematol. Educ. Program 2013, 2013, 220–226. [Google Scholar] [CrossRef]
- Kennedy, V.E.; Smith, C.C. FLT3 Mutations in Acute Myeloid Leukemia: Key Concepts and Emerging Controversies. Front. Oncol. 2020, 10, 612880. [Google Scholar] [CrossRef]
- He, R.; Devine, D.J.; Tu, Z.J.; Mai, M.; Chen, D.; Nguyen, P.L.; Oliveira, J.L.; Hoyer, J.D.; Reichard, K.K.; Ollila, P.L.; et al. Hybridization capture-based next generation sequencing reliably detects FLT3 mutations and classifies FLT3-internal tandem duplication allelic ratio in acute myeloid leukemia: A comparative study to standard fragment analysis. Mod. Pathol. 2020, 33, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Kottaridis, P.D.; Gale, R.E.; Frew, M.E.; Harrison, G.; Langabeer, S.E.; Belton, A.A.; Walker, H.; Wheatley, K.; Bowen, D.T.; Burnett, A.K.; et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: Analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood 2001, 98, 1752–1759. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergeron, J.; Capo-Chichi, J.-M.; Tsui, H.; Mahe, E.; Berardi, P.; Minden, M.D.; Brandwein, J.M.; Schuh, A.C. The Clinical Utility of FLT3 Mutation Testing in Acute Leukemia: A Canadian Consensus. Curr. Oncol. 2023, 30, 10410-10436. https://doi.org/10.3390/curroncol30120759
Bergeron J, Capo-Chichi J-M, Tsui H, Mahe E, Berardi P, Minden MD, Brandwein JM, Schuh AC. The Clinical Utility of FLT3 Mutation Testing in Acute Leukemia: A Canadian Consensus. Current Oncology. 2023; 30(12):10410-10436. https://doi.org/10.3390/curroncol30120759
Chicago/Turabian StyleBergeron, Julie, Jose-Mario Capo-Chichi, Hubert Tsui, Etienne Mahe, Philip Berardi, Mark D. Minden, Joseph M. Brandwein, and Andre C. Schuh. 2023. "The Clinical Utility of FLT3 Mutation Testing in Acute Leukemia: A Canadian Consensus" Current Oncology 30, no. 12: 10410-10436. https://doi.org/10.3390/curroncol30120759
APA StyleBergeron, J., Capo-Chichi, J.-M., Tsui, H., Mahe, E., Berardi, P., Minden, M. D., Brandwein, J. M., & Schuh, A. C. (2023). The Clinical Utility of FLT3 Mutation Testing in Acute Leukemia: A Canadian Consensus. Current Oncology, 30(12), 10410-10436. https://doi.org/10.3390/curroncol30120759






