Abstract
Over the past few years, nutraceuticals have gained substantial attention due to the health-promoting and disease-preventing functions behind their nutritional value. The global prevalence of nutraceuticals is reflected in the increasing number of commercially available nutraceuticals and their wide range of applications. Therefore, a unique opportunity emerges for their further exploration using innovative, reliable, accurate, low cost, and high hit rate methods to design and develop next generation nutraceuticals. Towards this direction, computational techniques constitute an influential trend for academic and industrial research, providing not only the chemical tools necessary for further mechanism characterization but also the starting point for the development of novel nutraceuticals. In the present review, an overview of nutraceuticals is discussed, underscoring the crucial role of chemoinformatic platforms, chemolibraries, and in silico techniques, as well as their perspectives in the development of novel nutraceuticals. This review also aims to record the latest advances and challenges in the area of nanonutraceuticals, an innovative field that capitalizes on the assets of nanotechnology for the encapsulation of bioactive components in order to improve their release profile and therapeutic efficacy.
1. Introduction
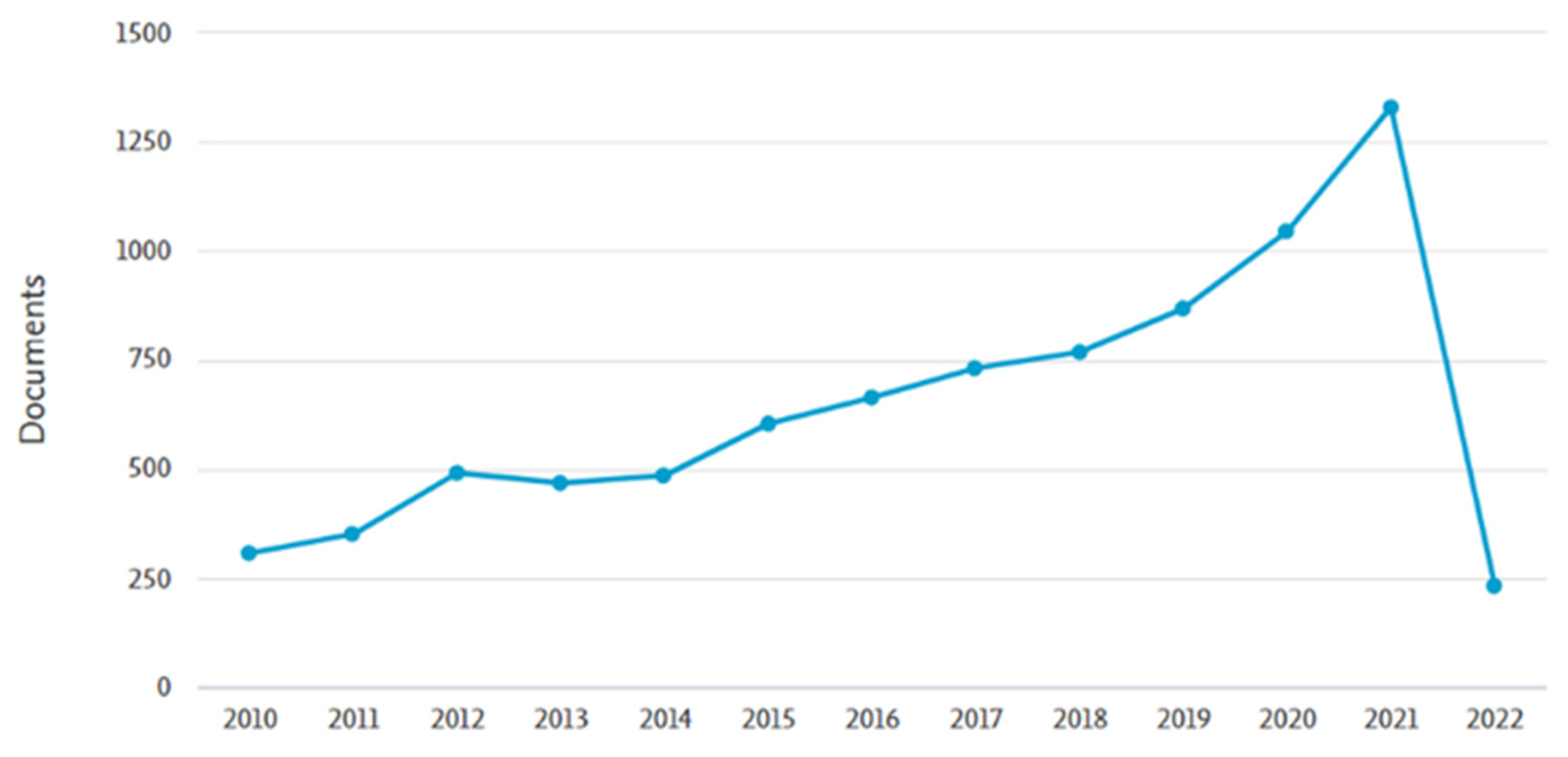
Nowadays, public awareness of health issues and concerns have created a new flourishing economy based on food-derived bioactive compounds which present health-promoting and disease-preventing functions, commonly referred to as nutraceuticals. Nutraceuticals constitute an emerging sector in the pharmaceutical and food industry, receiving considerable interest due to their functions [1]. The increased scientific community interest in the field of nutraceuticals is reflected in the fact that more than 8000 manuscripts have been published in the last decade, highlighting the unforeseen worldwide response (Figure 1).
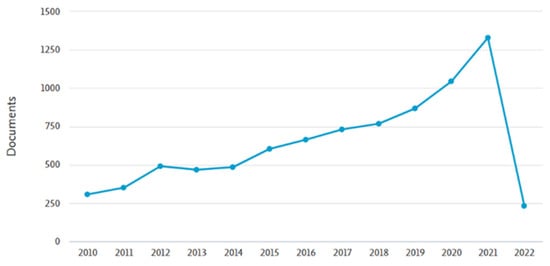
Figure 1.
Number of published manuscripts for the period 2010–2022, derived from the bibliographic database “SCOPUS” (https://pubmed.ncbi.nlm.nih.gov/?term=nutraceuticals&filter=simsearch3.fft&filter=years.2010-2022/ Last access: 11 April 2022).
Recent studies have revealed that several nutraceuticals are promising agents for the prevention and treatment of various diseases, such as allergies, Alzheimer’s disease, cardiovascular and eye disorders, cancer, obesity, diabetes, and Parkinson’s disease, as well as the regulation of immune system function and inflammation [2]. Therefore, nutraceuticals have attracted substantial interest which offers novel opportunities for the development of innovative products that will cover consumer needs for health-enhancing foods [3]. Based on the increasing number of commercially available nutraceuticals and their wide range of applications, the global nutraceutical market accounted for $289.8 billion in the year 2021 and is expected to grow to $438.9 billion by the year 2026, with a compound annual growth rate of 8.7% for the aforementioned period [4]. Furthermore, following the outbreak of the COVID-19 pandemic, the nutraceutical market is expected to increase due to the possible beneficial effects of these products on the human immune system function [5]. Additionally, the number of nutraceutical-based patents has increased, highlighting the crucial role of nutraceuticals worldwide [6].
Currently, the nutraceutical industry conforms to the practices of conventional food or pharmaceutical technology. However, current advances in the field of nanotechnology are the driving force behind the novel research strategies followed in nutraceutical development. Presently, nanosystems (structures or molecules of at least one dimension with a size from 1 to 100 nm) are incorporated in many different areas of food and health sciences. For instance, nanoengineered materials have been applied (a) for the nanopacking and improvement of the sensory attributes of foods, (b) for the smart delivery and nanofortification of functional and fortified products, and (c) for personalized treatment in nanomedicine [7]. The implementation of nano-scale materials for the encapsulation and delivery of nutraceuticals coined the concept of nanonutraceuticals [8].
Although the mechanism of action of nanoparticles is not yet fully elucidated and may differ according to the selected nanosystem, the beneficial biological properties of nanonutraceuticals are mainly attributed to the biological action of the loaded bioactive compounds [9]. For instance, nanonutraceuticals can act through the scavenging of free radicals, the improvement of antioxidant potential, and the chelation of transition metals. Various molecular pathways (including NF-κB, interleukin 6 and 1β, and TNF-α) and enzymatic functions (acetylcholinesterase, inducible nitric oxide synthase, superoxide dismutase, NADPH oxidase, etc.) are affected [10]. In addition, these nanoformulations protect the encapsulated bioactive molecules from oxidation or the action of gastrointestinal tract enzymes. Thus, through targeted delivery, they facilitate their slow and controlled release, elongate their activity, and enhance their bioefficacy [7,8].
Moreover, in recent decades, in silico virtual screening has emerged as a substantial research tool, defined as a set of computational methods that analyze large libraries of chemical compounds to identify potential hit candidates [11]. Among them, natural product (NP) libraries constitute the main tool for the discovery of novel nutraceuticals by applying virtual screening strategies. A remarkable number of academia, pharmaceutical, and food companies utilize these methods, worldwide, highlighting their contribution to the design and development process of novel compounds [12]. Computational techniques provide a wide range of possibilities to speed up the design of nutraceuticals and reduce the associated risks and costs [13].
In the field of nutraceuticals, in silico applications are still in their infancy, offering a unique opportunity for further investigation and exploitation. Recent publications have revealed new pathways toward the discovery of novel nutraceuticals by using in silico approaches [14]. The present comprehensive review is focused on the innovative concept of nutraceuticals and, in particular, on the latest advances in that field which concern the implementation of nanotechnology for the formulation of nanonutraceuticals. Furthermore, the current overview delves into the use of these state-of-the-art nanoformulations in the food and healthcare fields and also into their limitations and regulatory frameworks. In addition, it emphasizes the applications of computational methodologies and tools that facilitate the design and discovery of novel nutraceuticals. This overview illustrates the potential of computational techniques to drive the first screening steps of the nutraceutical industry in detail. Moreover, a prospective analysis of the impact of these techniques in the field of nutraceuticals is discussed.
2. Review Methodology
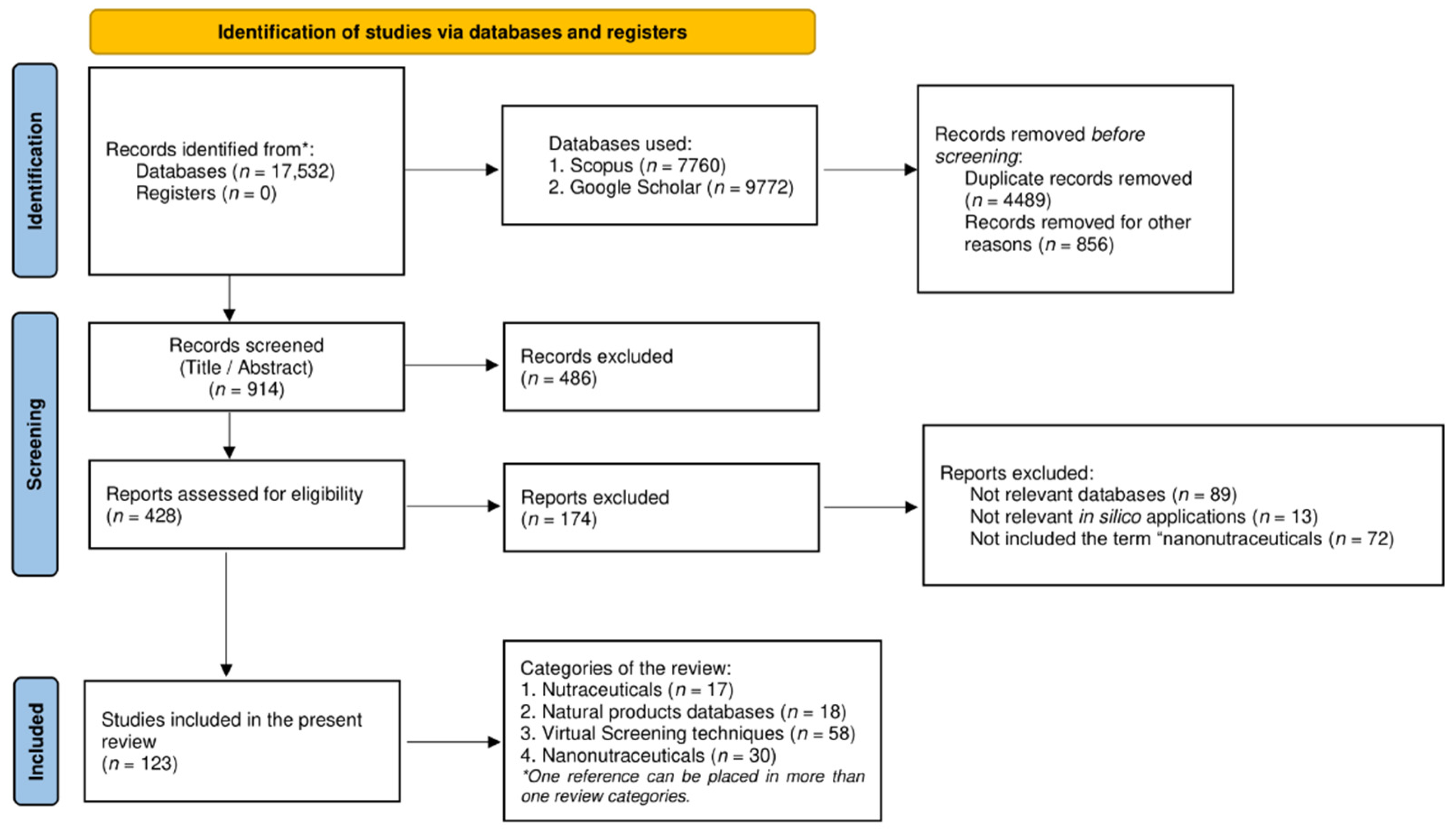
The present review focuses on collecting data regarding the current knowledge in the field of nutraceuticals in terms of in silico applications and nanotechnology advents. In order to structure this overview, a thorough bibliographic search was employed in different search engines, mainly Scopus and Google Scholar. The selected time frame was between 2015 and 2022. The period of the search was limited from 2020 to 2022, only for the terms related to nanonutraceuticals, since these concepts and their applications have been discussed in other recent review papers. The keywords used for the collection of papers were ‘Nutraceuticals AND market’, ‘Natural products databases’, ‘In silico screening’, ‘Computational techniques’, ‘Nanonutraceuticals’, ‘Nanofibers’, ‘Nanoparticles’, ‘Liposomes’, ‘Nanoemulsions’. The type of documents that were examined included original articles, reviews, and book chapters, published in the English language. The pipeline of the research methodology is illustrated in Figure 2 by a PRISMA flowchart.
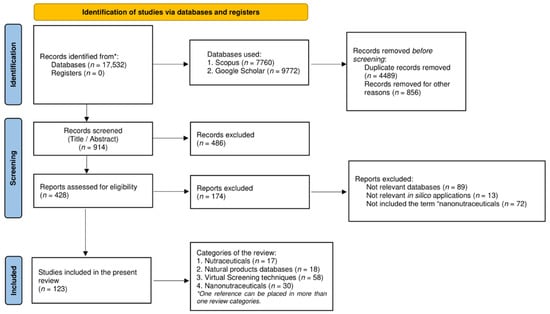
Figure 2.
PRISMA flowchart of the review process.
3. Current Knowledge in the Field of Nutraceuticals
3.1. Nutraceuticals: Definition and Introduction
The term ‘nutraceutical’ originated from the plausible combination of the words ‘nutrient’ and ‘pharmaceuticals’ and was invented in 1989 by Dr. Stephen De Felice (Chairman of the Foundation for Innovation in Medicine) [15]. According to the present definition, ‘nutraceutical’ refers to ‘a food (or part of a food) that provides medical or health benefits, including the prevention and/or treatment of a disease’ [16]. The described terminology has evolved through the years, characterizing ‘nutraceutical’ as ‘a product isolated or purified from foods that are generally sold in medicinal forms not usually associated with food’ [17]. Based on literature data, there are a plethora of definitions that describe ‘nutraceuticals’, by referring to them as ‘food, food components or nutrients providing health benefits behind their nutritional value’ [18]. To date, there is a controversy over the specific definition of ‘nutraceutical products’ based on their deliberate usage. Even though there is no clear definition of ‘nutraceuticals’ globally, the term includes health-promoting and disease-preventing functions behind the nutritional value of these products [19].
3.2. Nutraceuticals Classification
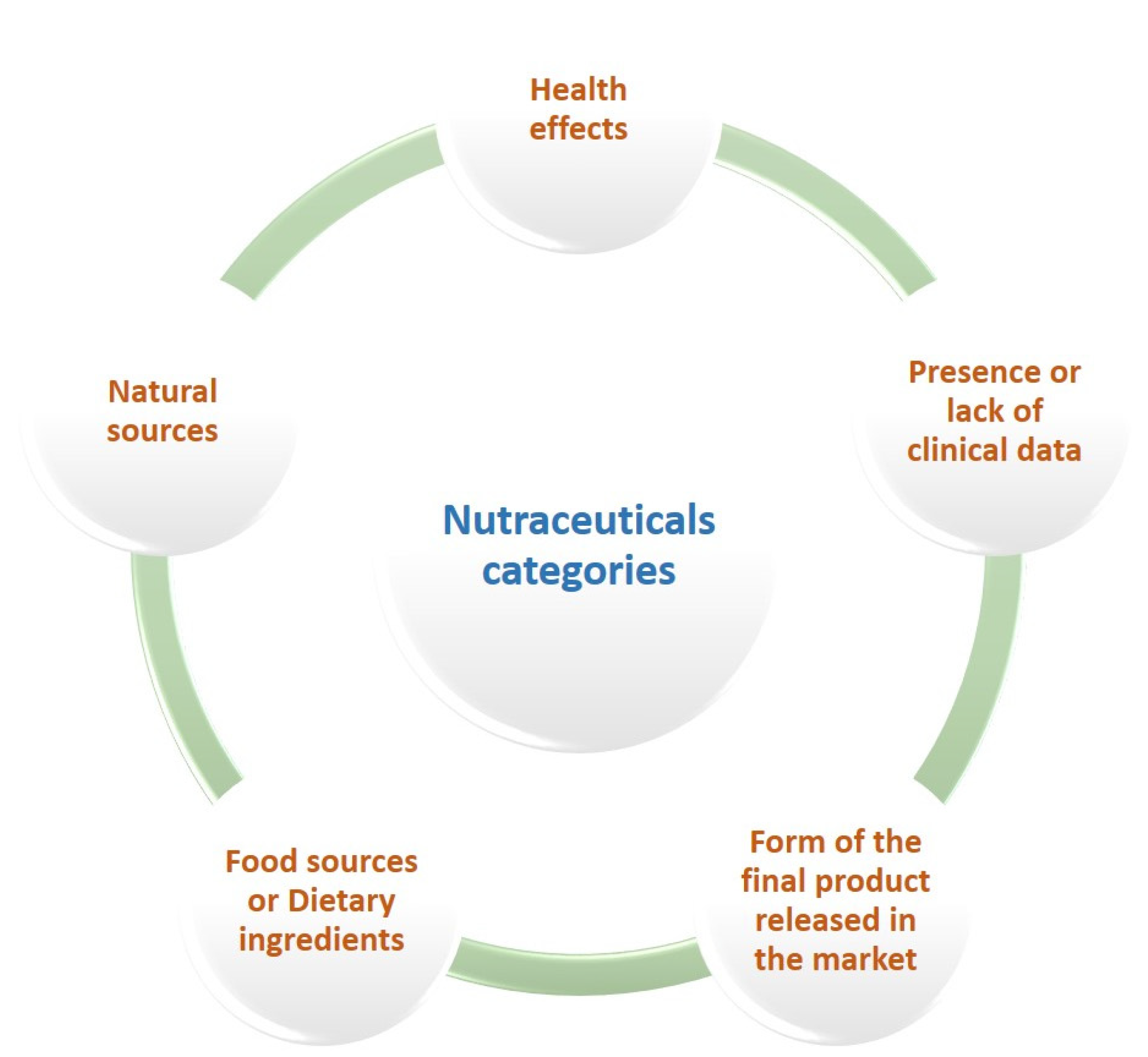
Since no scientific consensus has been reached yet over the classification of nutraceuticals, various criteria have been applied for their categorization (Figure 3). The key categories of nutraceuticals are herbal and botanical products (natural extracts or concentrates), nutrients (fatty acids, amino acids, vitamins, and minerals), functional foods, and dietary supplements, while their major natural sources are animals, plants, and microbes. If classified as nutritional ingredients, they can be divided into probiotics, prebiotics, antioxidant vitamins, polyunsaturated fatty acids, dietary fibers, polyphenols, and carotenoids [20]. Tablets, pills, creams, capsules, liquids, and powders are the most common forms touted in the global market [21]. On the basis of their health benefits, nutraceuticals serve several functions against various ailments, such as neurodegenerative and cardiovascular diseases, metabolic syndromes, congenital abnormalities, bone-related pathologies (osteoarthritis, osteoporosis), and cancer [21,22]. The term ‘established nutraceuticals’ includes the products with confirmed health-promoting effects, while the lack of validated clinical evidence describes the group of ‘potential nutraceuticals’ [20].
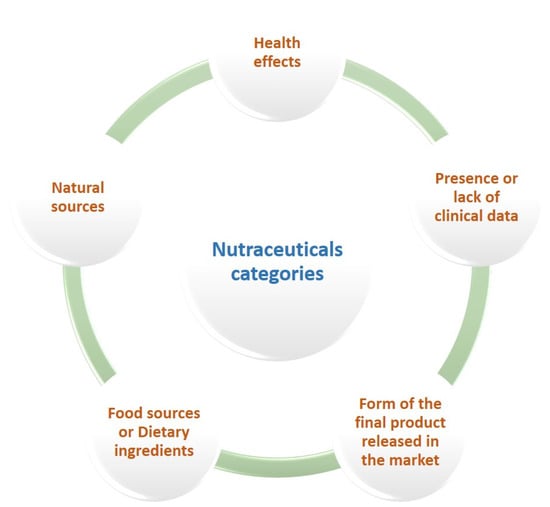
Figure 3.
Criteria used for the classification of nutraceuticals into different categories.
3.3. Regulatory Framework and Official Guidelines
Whilst nutraceuticals, as a concept, attract the interest of the current global market, there are still challenges that should be faced in order to bring nutraceuticals from bench top to bedside. Essential issues, such as the safety, toxicity, efficacy, and quality of the final products require further scrutiny to ensure consumers’ acceptance and reduce health risks. Therefore, the adoption of a shared international regulatory system, which at this point resides in the grey area between pharma and food regulations, is a key requirement.
For instance, the United States Food and Drug Administration (FDA) classifies nutraceuticals as ‘dietary supplements’ and therefore no safety or efficacy reports are demanded before approval. On the other hand, Canadian authorities have established a broader categorization of nutraceuticals, where they can be designated as drugs, foods, or natural health products. According to the Japanese regulatory bodies, which are governed by stricter legislation, nutraceuticals are termed as ‘Foods in General’, ‘Food with Health Claims’, and ‘Food for Specified Health Uses’ [3,22], while European directives remain quite equivocal since nutraceuticals can be considered as food supplements or medicinal products, according to European Food Safety Authority (EFSA) [3,23].
Focusing on the future perspectives of nutraceuticals, the official international and local authorities should set forth collective legislation that will provide a clear definition of nutraceuticals by describing a solid scientific rationale and common practices or responsibilities regarding their manufacturing, approval, and labeling [23].
4. Novel Approaches in Nutraceuticals’ Discovery
4.1. Natural Products (NPs) Databases (Chemo-Libraries)
Chemo libraries consist of the main resources for in silico applications and have emerged as a fundamental tool in the initial steps of computer-aided molecular discovery [24]. These databases are repositories of chemicals, mainly composed of synthetic and natural compounds, which provide information about their chemical scaffolds, their computable structural properties, and their vendors.
The natural compounds databases constitute the main tool for the discovery of novel nutraceuticals by applying virtual screening strategies. Natural compounds have been the center of attention of the scientific community in recent decades, rendering them in the limelight as an attractive and promising target for the discovery of novel bioactive compounds. In comparison to synthetic compounds, the key areas in which they excel are: (a) their structural diversity and complexity, often bearing numerous stereogenic centers and fused ring systems, which are poorly represented in synthetic compounds, (b) their abundance, and (c) their acceptance from consumers. It is noted that in 2017, the natural products market reached 11.5 billion USD and it is expected to rise with an annual CAGR of 19.7% between 2018 and 2026 [25].
Focusing on this knowledge-intensive scientific field, the current section attempts to present an overview of free and open access databases consisting of compounds with natural origins. These databases could be divided into two major classes, including (a) Virtual and (b) Physical Natural Product libraries. The major difference between virtual and physical libraries is that the first library contains only the chemical structures in an easily retrievable format, appropriate for computational applications, while physical libraries provide not only the chemical scaffold of NPs but also the available suppliers [26].
- Virtual Natural Product Libraries
Up-to-date, only the COlleCtion of Open Natural prodUcTs (COCONUT: https://coconut.naturalproducts.net, accessed on 11 April 2022), a free of charge and open access natural compounds database, efficiently aggregates the natural compounds chemical structures collected from various open sources (most compounds were added from the Ayurveda, Alkamid, CMNPD, and CyanoMetDB databases). Particularly, COCONUT contains 406,747 unique natural compounds in a readable format (.SMILES file) which are easily and quickly downloaded. Apart from chemical structures, COCONUT provides information about the stereochemical forms, organisms, natural geographical presence, and diverse pre-computed molecular properties of natural compounds [27].
The Natural Products Atlas (NPAtlas: https://www.npatlas.org/, accessed on 11 April 2022) constitutes another recently created open-access database, incorporating 24,594 natural compounds. It is a well-annotated database, including detailed information (structure, name, organisms source, isolation references, total syntheses, and cases of structural reassignment) about natural compounds, but unfortunately, it involves only microbial natural compounds [28].
The FooDB (https://foodb.ca/, accessed on 11 April 2022), a food-related chemical database, obtains >23,000 food chemicals in a searchable and downloadable format. Up to today, it is the most informative public resource of food ingredients, offering a unique opportunity for the identification of dietary components by performing virtual screening methods [29].
Based on the proven beneficial effects of functionally useful plants, as food and medicine, the Collective Molecular Activities of Useful Plants database (CMAUP: http://bidd.group/CMAUP/index.html, accessed on 11 April 2022) collected and classified in a downloadable format 47,645 plant ingredients derived from 5645 plants. The novelty of the aforementioned freely available database is that it possesses information not only for the chemical structure, name, and predicted physicochemical properties of the ingredients but also reports the ZINC code pointing out potential suppliers [30].
Marine natural products (MNPs) are considered important sources of biologically active agents that regulate a variety of biological functions, offering a major impact on human health [31,32,33,34]. The Comprehensive Marine Natural Products database (CMNPD: https://www.cmnpd.org/, accessed on 11 April 2022) is a freely available database that provides abundant information. The complete dataset could be downloaded via https://docs.cmnpd.org/downloads (accessed on 11 April 2022) in a ready-to-use format for virtual screening [35].
Another database conveniently downloadable in a readable format is the South African Natural Compounds Database (SANCDB: https://sancdb.rubi.ru.ac.za/, accessed on 11 April 2022), comprising more than 1000 compounds isolated from the plant and marine life in South Africa. Compared to other natural databases, SANCDB incorporates available analogues from MolPort (https://www.molport.com/, accessed on 11 April 2022) and Mcule (https://mcule.com/, accessed on 11 April 2022), two commercially available vendors, overcoming the major problem of the commercial availability of the compounds. Additionally, it facilitates virtual screening, including chemical scaffolds in a ready-to-dock format [36]. Table 1 enlists the most common free access and downloadable virtual natural compounds databases.

Table 1.
List of the most significant open-access virtual natural compounds databases.
- Physical Natural Product Libraries
The ZINC 15 database (http://zinc15.docking.org, accessed on 11 April 2022) constitutes the most comprehensive resource, which includes readily purchasable compounds (over 230 million compounds in a ready-to-dock format), overcoming the limitations of the compounds’ commercial availability. Particularly, the field of natural compounds consists of over 80,000 ready-to-use compounds, derived from a plethora of vendors. In addition, it categorizes natural compounds according to the following vendors: Analyticon Discovery (www.ac-discovery.com, accessed on 11 April 2022), AfroDB [41], Compound cloud (https://compoundcloud.bioascent.com/, accessed on 11 April 2022), Indofine (www.indofinechemical.com, accessed on 11 April 2022), MolPort (www.molport.com, accessed on 11 April 2022), MicroSource Natural Products (www.msdicovery.com, accessed on 11 April 2022), Nubbe (www.nubbe.iq.unesp.br, accessed on 11 April 2022), Specs (www.specs.net, accessed on 11 April 2022), TimTec (www.timtec.net, accessed on 11 April 2022) and UEFS (www.uefs.br, accessed on 11 April 2022), offering a direct assessment of purchasability and price of compounds. Therefore, ZINC 15 combines the information about the structure collection and potential suppliers, rendering it an ideal tool for virtual screening applications. Another advantage of the ZINC 15 database of particular interest is that, apart from chemical structures and vendors, it provides physicochemical properties and analogs of the compounds that can also be examined as potential hits [42].
Analyticon Discovery (https://ac-discovery.com/, accessed on 11 April 2022) is a free access database, that provides a continuously growing collection of purified natural compounds. In particular, the library could be divided into the following subsets: (a) MEGx which offers about 5000 purified natural compounds originating from plants and microorganisms, (b) MACROx comprises over of 1800 macrocycle compounds, and (c) FRGx with over 200 fragments. Additionally, Analyticon Discovery includes a semisynthetic NP-derived compound subset (NATx) with over 26,000 compounds. Finally, polyphenols and flavonoids collections are also available, offering a unique opportunity for further exploration in the fields of the development of novel taste-modulating or health-promoting ingredients for the food industry [26].
Ambinter (https://www.ambinter.com/, accessed on 11 April 2022) and Greenpharma (www.greenpharma.com, accessed on 11 April 2022) constitute two collaborative companies, offering a set of ~8000 natural compounds (alkaloids, phenols, phenolic acids, terpenoids, and others) in .SDF format ready to use for virtual screening. Additionally, the above-mentioned companies propose more than 11,000 semi-synthetic derivatives of natural compounds [26].
One of the largest natural compound libraries is InterBioScreen (https://www.ibscreen.com/, accessed on 11 April 2022), listing over 68,000 well-annotated natural compounds derived from a variety of sources, such as plants and microorganisms. The presented library is easily and quickly downloaded in a readable format (.SMILES and .SDF) [26].
The MolPort (https://www.molport.com/, accessed on 11 April 2022) database is another natural compound vendor of paramount importance since it stores in downloadable files over 10,000 unique natural and over 100,000 natural-like products from a variety of suppliers (.SMILES and .SDF). Therefore, its usage facilitates in silico screening applications since it possesses available-to-purchase natural products.
A collection of more than 3000 natural compounds and 396 food additive-related compounds are supplied from MedChemExpress (https://www.medchemexpress.com/, accessed on 11 April 2022). For data accessibility, a query is required, and purchasable compounds in .SDF format is received. The main advantage of the present database is that all compounds have indicated bioactivity and safety.
INDOFINE Chemical Company (https://indofinechemical.com/, accessed on 11 April 2022) includes around 1900 NPs and semisynthetic compounds, in a ready-to-screen format (.SDF), focused on flavonoids. The library consists of flavonoids, flavones, isoflavones, flavanones, coumarins, chromones, chalcones, and lipids especially. The chemical scaffolds of Indofine are offered and are classified according to compound types.
The most common free access and downloadable physical natural compounds databases are presented in Table 2.

Table 2.
List of the most significant open-access physical natural compounds databases.
4.2. Virtual Screening (VS) Techniques
As it is generally known, the identification of bioactive molecules constitutes an expensive, time-consuming, and laborious inter-disciplinary process. As a result, innovative approaches are continuously developed, aiming to optimize and simplify this procedure. Among them, Virtual Screening (VS) is one of the most important and widespread strategies that has been applied for the determination of potentially bioactive molecules. In recent years, a variety of tools and software that can be performed in VS were utilized to reduce the selection of promising compounds that will be tested experimentally. Particularly, VS objectives are to accelerate the discovery process, increase the number of compounds to be tested experimentally, and rationalize their choice [13,43]. Additionally, the classification of NPs into libraries contributes effectively to VS, facing and tackling issues related to the extraction, purification, and purchasability of NPs [26].
The most commonly used methods for VS of NP libraries include Molecular Docking, Quantitative Structure-Activity Models (QSAR), Molecular Docking, Pharmacophore Modeling, and Molecular Dynamics (MD) Simulations. The main advantage of these methods is that they lead to reducing the selection of compounds that will be tested experimentally [44].
In the field of nutraceuticals, in silico approaches such as QSAR, molecular docking, and molecular dynamic simulations have been utilized, aiming to unravel bioactive food components with health-promoting and disease-preventing properties [45]. The present section provides a brief description of the fundamental idea of the above techniques as well as their state-of-the-art applications in the field of nutraceuticals.
- Quantitative Structure-Activity Relationship (QSAR)
In general, Quantitative Structure-Activity Relationship (QSAR) analysis is a ligand-based computational technique that attempts to correlate the structural properties (chemical structures) and the biological activity of a compounds’ dataset [46]. The underlying principle of QSAR models is based on the hypothesis that structurally similar compounds may exhibit similar biological activities [47]. The creation of QSAR models is causally linked with equations that relate a dependent variable (i.e., an observed activity) with a number of calculated descriptors, including physicochemical, constitutional, and topological properties [48,49]. For this purpose, various multivariate statistical regression (Multiple Linear Regression—MLR, Principal Component Analysis—PCA, and Partial Least Square Analysis—PLS) and Machine Learning (ML) tools are applied in an effort to generate appropriate algorithms [50]. Table 3 presents a list of available software for molecular descriptor calculations. The building of the model is followed by the validation process in which the accuracy of the method is verified. The produced model can be used as a prediction tool to prioritize compounds that have the potential to display biological activity and to reduce the number of the compounds that will be tested experimentally [51]. Therefore, it is a widely used process with a broad spectrum of applications in the pharmaceutical landscape [52]. Regarding nutraceuticals, the relationship between food ingredients and a variety of properties has already been studied on several occasions [53].

Table 3.
List of several available softwares to calculate molecular descriptors.
- Molecular Docking
Molecular Docking is the most commonly used in silico technique, which predicts the interaction between a small molecule (ligand) and a protein (receptor) at the atomic level. This approach enables the characterization of the behavior of small molecules in the binding site of a target protein as well as the elucidation of the fundamental biochemical process behind this interaction [54]. It is a structure-based approach which requires a high-resolution 3D illustration of the examined target derived from (a) X-ray crystallography [55], (b) Nuclear Magnetic Resonance Spectroscopy [56], and (c) Cryo-Electron Microscopy [57]. Until now, numerous computational tools and algorithms have been developed, including commercial or free-of-charge software (Table 4). Molecular Docking finds a plethora of applications mainly in the field of drug discovery and design [58]. It should be noted that during recent years, a constantly increasing interest has been observed concerning the applications of molecular docking in food science [59].

Table 4.
The most commonly used ligand-receptor Molecular Docking software.
In the nutraceutical landscape, molecular docking studies have been employed to provide information about the initial steps of nutraceutical research that precede the in vitro studies [45]. Herein, the most relevant applications of molecular docking in the assessment of the potential health-promoting benefits of nutraceuticals are reviewed.
- Pharmacophore Modeling
A pharmacophore model illustrates in a 3D arrangement, the chemical features, which are crucial for the molecular recognition of a ligand by a macromolecule, offering a putative explanation for the binding affinity of structurally diverse ligands to a common target [72]. It can be generated either in a structure-based way, by predicting the potential interactions between the target and the ligand, or in a ligand-based way, by overlaying a group of active molecules and creating common chemical features that may be responsible for their bioactivity [73]. Currently, a variety of 3D pharmacophore modeling generators have been constructed, containing commercially available software and academic programs (Table 5) [74].

Table 5.
List of several 3D pharmacophore modeling software.
Once a pharmacophore model is created, it can be exploited as a query to screen a chemical library. Compounds that satisfy the query pharmacophore features are retrieved and expected to exhibit bioactivity. The described process, commonly known as Pharmacophore-based Virtual Screening, represents a mainstream tool of VS with a plethora of applications in the drug discovery process [75,76].
- Molecular Dynamics Simulations (MD simulation)
Molecular Dynamics Simulation is another powerful computational tool that captures the behavior of proteins, ligand-protein complexes, and other biomolecules in full atomic detail and at very fine temporal resolution [84]. It is a well-established technique which provides a molecular perspective to observe the behavior of atoms, molecules, and particulates [85]. Based on Newton’s equation of motion, MD predicts the physical movements of atoms and molecules using interatomic potentials or molecular mechanics force fields, offering the opportunity to comprehend the overall behavior of molecular systems during the motion of individual atoms [84].
Up to now, several force fields [86] and tools have been developed and are available for MD simulations. GROMACS [87], AMBER [88,89], Nanoscale MD (NAMD) [90], CHARMM-GUI [91], and DESMOND are the most commonly used tools for MD simulations.
In recent years, the impact of MD simulations in molecular biology and drug discovery has expanded drastically [92]. MD simulations have gained ground in deciphering functional mechanisms of proteins and other biomolecules, unraveling the structural basis of disease, and designing and optimizing the production of small molecules [84].
- Applications of in Silico Screening Techniques in the Field of Nutraceuticals
Although the application of in silico screening for the discovery of novel nutraceuticals is still in its first steps, studies proving the significant role of these methodologies have been carried out. Therefore, in this section, relevant applications of in silico techniques in the evaluation of the potential health benefits of nutraceuticals are described.
Nowadays, the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a life-threatening disease causing thousands of deaths daily, is responsible for a current global health crisis. Therefore, the scientific community has the made treatment and prevention of SARS-CoV-2 infection its first priority [93]. It has been proven that nutraceuticals contribute effectively to reducing the chances of SARS-CoV-2 infection, but also in alleviating COVID-19 symptoms [94].
Towards this direction, Gyebi et al. (2021) performed a structure-based virtual screening to suggest inhibitors of 3-Chymotrypsin-Like Protease (3CLpro) of SARS-CoV-2 from Vernonia amygdalina and Occinum gratissimum. In particular, they applied docking studies in the active site of 3CLpro, aiming to predict the binding affinity of an in-house library, which includes 173 phytochemicals from Vernonia amygdalina and Occinum gratissimum. Docking results defined a hit list of 10 phytochemicals with strong binding affinities in the catalytic center of 3CLpro from three related strains of coronavirus (SARS-CoV, MERS-CoV, and HKU4). Subsequently, drug-likeness prediction revealed two terpenoids, neoandrographolide and vernolide, as the most promising inhibitors of SARS-CoV-2 3CLpro. The selected compounds were subjected to Molecular Dynamics simulations and the results showed that the examined terpenoid-enzyme complexes exhibited strong interactions and structural stability, which could be adapted in experimental models for the development of preventive nutraceuticals against coronavirus diseases [95]. Furthermore, Kodchakorn et al. (2020) employed a combination of in silico screening techniques to determine natural compounds with high calculated binding affinity on the homology structure of coronavirus protease (SARS-CoV-2 PR). Molecular Docking and Molecular Dynamics simulations were applied to eight natural compounds (Andrographolide, anthocyanin-β-d-glucoside, capsaicin, curcumin, cyanidin, cyanidin-3-O-glucoside, sesamin, and hesperidin). Result analysis indicated that all natural compounds presented favorable binding affinities, providing preliminary data for the development of novel nutraceuticals [96]. On the other hand, Kumar et al. (2019) utilized Molecular Docking to screen a library, consisting of 106 well-known nutraceuticals, against four SARS-CoV-2 targets (S protein (Receptor Binding Domain)-ACE2 complex, Mpro, PLpro, and Nsp15). The results suggested that among the tested nutraceuticals, folic acid and its derivatives, such as tetrahydrofolic acid and 5-methyl tetrahydrofolic acid, were the most promising and could serve as a starting material for further in vitro and in vivo experiments [14]. Recently, Baig et al. (2022) studied the in silico inhibitory activity of 58 compounds, derived from the miraculous herb Nigella sativa, against the SARS-CoV-2 target in an effort to propose potential compounds as SARS-CoV-2 inhibitors. Three compounds, α-hederin, rutin, and nigellamine A2 were identified as the most promising molecules and further investigation is necessary to prove the ability of Nigella sativa to inhibit SARS-CoV-2 targets [97].
5. Nanotechnology: A Powerful Toolbox in the Field of Nutraceuticals
5.1. Health Effects and Limitations of Nanonutraceuticals
Nanonutraceuticals outweigh traditional nutraceutical formulations since they can (a) enhance the solubility and stability of the encapsulated natural bioactive compounds and (b) increase their absorption and biological efficacy by diminishing the off-target release and minimizing their side effects [8]. The up-to-date reported nanosized delivery systems include polymeric nanonutraceuticals (nanocapsules and nanospheres), carbon-based nanomaterials (i.e., fullerene and graphene particles), lipid-based formulations (solid lipid nanoparticles (SLNs), lipid nanocapsules, micelles, nanosuspensions, lipid–polymer hybrid nanoparticles, nanostructured lipid carriers, and liposomes), metal-based nanoparticles (silver and gold nanoparticles), dendrimers, nanoemulsions, exosomes, niosomes, quantom dots, nanoshells, nanofilms, and nanofibers. In the majority of cases, the therapeutic cargo of these nanocarriers is attributed to biologically active constituents, such as minerals, vitamins, polyphenols (i.e., resveratrol, rutin, tannins, anthocyanins, catechins and flavonoids, curcuminoids, berberine, etc.), carotenoids (lycopene, β-carotene, astaxanthin, etc.), ω-3 fatty acids, phytosterols, and probiotics (Lactobacillus and Bifidobacterium bacteria). When these nanovehicles are loaded with phytochemicals (i.e., curcumin, resveratrol, vitamin E, etc.), they are specified as nano-phytomedicines or nano-phytoceuticals. The first insights concerning these nanoformulations showed that they act as more efficient delivery systems of phytoconstituents [98,99].
Based on the latest scientific evidence, the role of nutraceuticals in the prevention and treatment of several pathologies is multifarious. The scope of health-related applications of nanonutraceuticals is extended from the display of antioxidant, antimicrobial, anti-inflammatory, wound healing, pain relief, and immunomodulatory properties to the management of age-related neurogenerative conditions (i.e., Alzheimer’s and Parkinson’s disease), cancer, diabetes, skin diseases and recently, of pre- and post-COVID-19 infections [99,100]. Recent examples of nanonutraceuticals in therapeutics and healthcare are presented in Table 6.

Table 6.
Potential therapeutical effects of recent nanonutraceuticals.
Special focus should be paid to the tuning of probiotics and prebiotics to their nanosized products, known as nanoprobiotics and nanoprebiotics, respectively. According to the International Scientific Association for Probiotics and Prebiotics (ISAPP) definition, live microorganisms confer beneficial effects on human health by modulating the immune system, producing antimicrobial compounds, interacting with the gut microbiota of the host, and improving gut barrier integrity are characterized as probiotics. The substrates or natural compounds that the host microbiota use to improve the health of the host are acknowledged as prebiotics [105]. The encapsulation of probiotics and prebiotics to nanoplatforms reduces any possible side effects, enhances their stability, absorption, and bioactivity, increases their fermentability and indigestibility in the GI tract, and triggers their selective stimulation and targeted activity [106,107].
Nonetheless, the research community should address some issues regarding the toxicity and safety implications as well as the manufacturing challenges of nanonutraceuticals. At first, these nanoformulations must be fully characterized on the basis of their physicochemical properties, especially their size and shape, which may induce tissue damage or inadvertent permeation of non-targeted cell membranes. In addition, further clinical data from in vivo animal models should be collected and evaluated to decipher the mechanisms of action of these nanoproducts, improve their absorption and metabolism by the gastroinstestinal (GI) tract, and eliminate any possible immunotoxicity. Furthermore, the commercialization of nanoproducts is strongly related to the establishment of scaled-up cost-effective processes, which ensure the reproducibility, reliability, and high quality of the final product. The outcomes of these trials will lead to the establishment of guidelines and standardized protocols for the safe monitoring of nanonutraceuticals, which, eventually, will curb the concerns of the consumers regarding their use [8,108].
5.2. The Latest Updates Regarding Nanonutraceuticals Applications
Based on the most recent projections regarding the demands of the current nutraceuticals market [4], nanonutraceuticals will be at the forefront of research and industry strategies in the upcoming years. Indicative examples of the newest applications of nanoformulations, recorded in the last two years (2021–2022) are exhibited in Table 7.

Table 7.
Examples of nanonutraceuticals reported in the last two years (2021–2022).
The entrapment of curcumin in monoglycerides oleogels, which formed oil nanoemulsions of high stability, resulted in higher encapsulation efficiency and a more controlled release of this bioactive molecule [109]. Nanoparticles increased the solubility and bioavailability of thymoquinone and have been used in a cancer mice model as a complementary therapy to prevent cases of nephrotoxicity caused by the cisplatin chemotherapy [110]. Thymoquinone nanoemulsions also exhibited strong anti-ulcer properties [111]. Costunolide is another natural anticancer agent of the sesquiterpene group whose anti-tumor properties were enhanced when it was loaded in an α-cyclodextrin nanoemulsion [112]. Oil-in-water nanoemulsions of resveratrol demonstrated higher cytotoxic activity and improved significantly the solubility, bioavailability, and in vivo efficacy of this polyphenol [113].
Lipid nanocarriers, such as solid lipid nanoparticles (SLNs), enhanced the therapeutic effect of phytoconstituents, such as berberine, due to their lipid nature that facilitates the absorption and the targeted delivery of the bioactive compounds [114]. The combination of two unsaturated fatty acids, oleic and linoleic acid, formed liposomes, known as ufasomes, which achieved delivery of less or non-polar molecules, such as oleuropein, and increased their antioxidant activity [115]. Liposomes containing phosphatidylcholine and the liquid lipid Plurol Oleique also acted as carriers for the ocular delivery of thymoquinone and decreased its possible adverse effects (i.e., the toxicity of high doses, low cell absorption, and permeability) [116]. Liposomal formulations (cationic liposomal formulation) containing thymoquinone were also investigated for their anticancer properties [117]. Furthermore, phospholipid liposomes loaded with quercetin and mint oil were used against oral cavities [118].
Corn starch and sodium alginate-based nanofibers were applied as coatings to protect the probiotic activity of Bifidobacteria and lactic acid bacteria in yogurts and under gastrointestinal conditions in a simulated system [119,120]. Moreover, a nanostructured hydrogel formed by a lupin-derived peptide proved to have significant antioxidant properties, paving the way for the implementation of food-derived peptides in nanotechnology [122]. A year later, the same research group used synthetic analogues of lupin β-conglutin and soybean glycinin bioactive peptides in gel nanoformulation as DPP-4 and ACE inhibitors [121]. Based on the results of Faruk et al. (2022), a soybean nano-isoflavone presented potential therapeutic activity against the degenerative effect of D-galactose [123].
To sum up, there is mounting evidence that the engineering of natural products into nanoformulations is emerging as a straightforward approach, able to improve the low solubility, reduced bioavailability, low stability, non-site specific targeting, and possible degradation of conventional nutraceuticals by gastrointestinal fluid.
6. Conclusions
In the last few years, a new perception has been shaped in the general public regarding the incorporation of natural products and functional foods into everyday life. Thus, the market of nutraceuticals greatly expanded due to their acknowledged health benefits against several pathologies and their increased therapeutic efficacy compared to known conventional formulations. Rapid progress in the field of natural compound databases and chemoinformatics tools facilitates the design and development of novel nutraceuticals with enhanced bioactivities by applying in silico screening methodologies. Furthermore, the combination of in silico techniques with modern nanonization strategies is the key driver in all the innovations related to nutraceuticals. Therefore, nanonutraceuticals are considered the next generation nutraceuticals since they present improved properties, such as enhanced stability and solubility and improved absorption and bioavailability, and thus, more targeted delivery and upgraded therapeutic efficacy. Nonetheless, further investigation and clinical data are required to draw safe conclusions regarding the toxicity and safety of these nanoformulations. Finally, it is important to stress that both the research community as well as international and local authorities should establish shared legislation and common protocols to ensure the safety of consumers.
Author Contributions
Conceptualization, T.T., E.K. and P.Z.; methodology, T.T. and E.K.; investigation, K.T., P.C. and V.J.S.; data curation, T.T., K.T., P.C. and E.K.; writing—original draft preparation, T.T., K.T., P.C. and E.K.; writing—review and editing, T.T., V.J.S., E.K. and P.Z.; visualization, K.T. and P.C.; supervision, E.K. and P.Z.; project administration, V.J.S. and P.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dikmen, B.Y.; Filazi, A. Chapter 69—Nutraceuticals: Turkish Perspective. In Nutraceuticals; Gupta, R.C., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 971–981. [Google Scholar]
- Nasri, H.; Baradaran, A.; Shirzad, H.; Rafieian-Kopaei, M. New Concepts in Nutraceuticals as Alternative for Pharmaceuticals. Int. J. Prev. Med. 2014, 5, 1487–1499. [Google Scholar] [PubMed]
- Chopra, A.S.; Lordan, R.; Horbańczuk, O.K.; Atanasov, A.G.; Chopra, I.; Horbańczuk, J.O.; Jóźwik, A.; Huang, L.; Pirgozliev, V.; Banach, M.; et al. The Current Use and Evolving Landscape of Nutraceuticals. Pharmacol. Res. 2022, 175, 106001. [Google Scholar] [CrossRef] [PubMed]
- BCC Research. Nutraceuticals Market Size, Share & Growth Analysis Report. Available online: https://www.bccresearch.com/market-research/food-and-beverage/nutraceuticals-global-markets.html (accessed on 13 March 2022).
- Mordor Intelligence. Global Nutraceuticals Market Size, Share, Trends, Growth (2022–27). Available online: https://www.mordorintelligence.com/industry-reports/global-nutraceuticals-market-industry (accessed on 13 March 2022).
- Daliu, P.; Santini, A.; Novellino, E. A Decade of Nutraceutical Patents: Where Are We Now in 2018? Expert Opin. Ther. Patents 2018, 28, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Pérez, K.M.; Ruiz-Pulido, G.; Medina, D.I.; Parra-Saldivar, R.; Iqbal, H.M. Insight of Nanotechnological Processing for Nano-Fortified Functional Foods and Nutraceutical—Opportunities, Challenges, and Future Scope in Food for Better Health. Crit. Rev. Food Sci. Nutr. 2021, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Paolino, D.; Mancuso, A.; Cristiano, M.C.; Froiio, F.; Lammari, N.; Celia, C.; Fresta, M. Nanonutraceuticals: The New Frontier of Supplementary Food. Nanomaterials 2021, 11, 792. [Google Scholar] [CrossRef] [PubMed]
- Daliu, P.; Santini, A.; Novellino, E. From Pharmaceuticals to Nutraceuticals: Bridging Disease Prevention and Management. Expert Rev. Clin. Pharmacol. 2019, 12, 1–7. [Google Scholar] [CrossRef]
- Hesari, M.; Mohammadi, P.; Khademi, F.; Shackebaei, D.; Momtaz, S.; Moasefi, N.; Farzaei, M.H.; Abdollahi, M. Current Advances in the Use of Nanophytomedicine Therapies for Human Cardiovascular Diseases. Int. J. Nanomed. 2021, 16, 3293–3315. [Google Scholar] [CrossRef]
- Murugan, N.A.; Podobas, A.; Gadioli, D.; Vitali, E.; Palermo, G.; Markidis, S. A Review on Parallel Virtual Screening Softwares for High-Performance Computers. Pharmaceuticals 2022, 15, 63. [Google Scholar] [CrossRef]
- Suay-García, B.; Bueso-Bordils, J.I.; Falcó, A.; Antón-Fos, G.M.; Alemán-López, P.A. Virtual Combinatorial Chemistry and Pharmacological Screening: A Short Guide to Drug Design. Int. J. Mol. Sci. 2022, 23, 1620. [Google Scholar] [CrossRef]
- Haga, J.H.; Ichikawa, K.; Date, S. Virtual Screening Techniques and Current Computational Infrastructures. Curr. Pharm. Des. 2016, 22, 3576–3584. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, A.K.; Gupta, A.; Pandey, A.K. In-Silico Approaches to Study Therapeutic Efficacy of Nutraceuticals. In Phytochemistry: An In-Silico and In-Vitro Update: Advances in Phytochemical Research; Kumar, S., Egbuna, C., Eds.; Springer: Singapore, 2019; pp. 479–490. [Google Scholar]
- Andlauer, W.; Fürst, P. Nutraceuticals: A Piece of History, Present Status and Outlook. Food Res. Int. 2002, 35, 171–176. [Google Scholar] [CrossRef]
- De Felice, S.L. The Nutraceutical Revolution: Its Impact on Food Industry R&D. Trends Food Sci. Technol. 1995, 6, 59–61. [Google Scholar] [CrossRef]
- Pandey, M.; Verma, R.; Saraf, S. Nutraceuticals: New Era of Medicine and Health. Asian J. Pharm. Clin. Res. 2010, 3, 2010. [Google Scholar]
- Andrew, R.; Izzo, A.A. Principles of Pharmacological Research of Nutraceuticals. Br. J. Pharmacol. 2017, 174, 1177–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aronson, J.K. Defining “Nutraceuticals”: Neither Nutritious nor Pharmaceutical. Br. J. Clin. Pharmacol. 2017, 83, 8–19. [Google Scholar] [CrossRef]
- Ansari, S.; Chauhan, B.; Kalam, N.; Kumar, G. Current Concepts and Prospects of Herbal Nutraceutical: A Review. J. Adv. Pharm. Technol. Res. 2013, 4, 4–8. [Google Scholar] [CrossRef]
- De, S.; Gopikrishna, A.; Keerthana, V.; Girigoswami, A.; Girigoswami, K. An Overview of Nanoformulated Nutraceuticals and Their Therapeutic Approaches. Curr. Nutr. Food Sci. 2021, 17, 392–407. [Google Scholar] [CrossRef]
- Blaze, J. A Comparison of Current Regulatory Frameworks for Nutraceuticals in Australia, Canada, Japan, and the United States. Innov. Pharm. 2021, 12, 8. [Google Scholar] [CrossRef]
- Santini, A.; Cammarata, S.M.; Capone, G.; Ianaro, A.; Tenore, G.C.; Pani, L.; Novellino, E. Nutraceuticals: Opening the Debate for a Regulatory Framework: Nutraceutical Regulatory Framework. Br. J. Clin. Pharmacol. 2018, 84, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Lagunin, A.A.; Goel, R.K.; Gawande, D.Y.; Pahwa, P.; Gloriozova, T.A.; Dmitriev, A.V.; Ivanov, S.M.; Rudik, A.V.; Konova, V.I.; Pogodin, P.V.; et al. Chemo- and Bioinformatics Resources for in Silico Drug Discovery from Medicinal Plants beyond Their Traditional Use: A Critical Review. Nat. Prod. Rep. 2014, 31, 1585–1611. [Google Scholar] [CrossRef]
- Global Natural & Organic Personal Care Market 2018–2026: Growth Trends, Key Players and Competitive Strategies. Available online: https://www.prnewswire.com/news-releases/global-natural--organic-personal-care-market-2018-2026-growth-trends-key-players-and-competitive-strategies-300675255.html (accessed on 13 March 2022).
- Chen, Y.; de Bruyn Kops, C.; Kirchmair, J. Data Resources for the Computer-Guided Discovery of Bioactive Natural Products. J. Chem. Inf. Model. 2017, 57, 2099–2111. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT Online: Collection of Open Natural Products Database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naveja, J.J.; Rico-Hidalgo, M.P.; Medina-Franco, J.L. Analysis of a Large Food Chemical Database: Chemical Space, Diversity, and Complexity. F1000Research 2018, 7, 993. [Google Scholar] [CrossRef]
- Zeng, X.; Zhang, P.; Wang, Y.; Qin, C.; Chen, S.; He, W.; Tao, L.; Tan, Y.; Gao, D.; Wang, B.; et al. CMAUP: A Database of Collective Molecular Activities of Useful Plants. Nucleic Acids Res. 2019, 47, D1118–D1127. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, A.; Naughton, L.M.; Montánchez, I.; Dobson, A.D.W.; Rai, D.K. Current Status and Future Prospects of Marine Natural Products (MNPs) as Antimicrobials. Mar. Drugs 2017, 15, 272. [Google Scholar] [CrossRef]
- Pradhan, B.; Nayak, R.; Patra, S.; Jit, B.P.; Ragusa, A.; Jena, M. Bioactive Metabolites from Marine Algae as Potent Pharmacophores against Oxidative Stress-Associated Human Diseases: A Comprehensive Review. Molecules 2021, 26, 37. [Google Scholar] [CrossRef]
- Ghosh, S.; Sarkar, T.; Pati, S.; Kari, Z.A.; Edinur, H.A.; Chakraborty, R. Novel Bioactive Compounds From Marine Sources as a Tool for Functional Food Development. Front. Mar. Sci. 2022, 9, 832957. [Google Scholar] [CrossRef]
- Khanam, S.; Prakash, A. Biomedical Applications and Therapeutic Potential of Marine Natural Products and Marine Algae. IP J. Nutr. Metab. Health Sci. 2021, 4, 76–82. [Google Scholar] [CrossRef]
- Lyu, C.; Chen, T.; Qiang, B.; Liu, N.; Wang, H.; Zhang, L.; Liu, Z. CMNPD: A Comprehensive Marine Natural Products Database towards Facilitating Drug Discovery from the Ocean. Nucleic Acids Res. 2021, 49, D509–D515. [Google Scholar] [CrossRef]
- Diallo, B.N.; Glenister, M.; Musyoka, T.M.; Lobb, K.; Tastan Bishop, Ö. SANCDB: An Update on South African Natural Compounds and Their Readily Available Analogs. J. Cheminform. 2021, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Pilon, A.C.; Valli, M.; Dametto, A.C.; Pinto, M.E.F.; Freire, R.T.; Castro-Gamboa, I.; Andricopulo, A.D.; Bolzani, V.S. NuBBEDB: An Updated Database to Uncover Chemical and Biological Information from Brazilian Biodiversity. Sci. Rep. 2017, 7, 7215. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.W.; Lin, Y.C.; Chang, H.S.; Wang, C.C.; Chen, I.S.; Jheng, J.L.; Li, J.H. TIPdb-3D: The Three-Dimensional Structure Database of Phytochemicals from Taiwan Indigenous Plants. Database 2014, 2014, bau055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.Y.C. TCM Database@Taiwan: The World’s Largest Traditional Chinese Medicine Database for Drug Screening In Silico. PLoS ONE 2011, 6, e15939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degtyarenko, K.; de Matos, P.; Ennis, M.; Hastings, J.; Zbinden, M.; McNaught, A.; Alcántara, R.; Darsow, M.; Guedj, M.; Ashburner, M. ChEBI: A Database and Ontology for Chemical Entities of Biological Interest. Nucleic Acids Res. 2008, 36, D344–D350. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Zofou, D.; Babiaka, S.B.; Meudom, R.; Scharfe, M.; Lifongo, L.L.; Mbah, J.A.; Mbaze, L.M.; Sippl, W.; Efange, S.M.N. AfroDb: A Select Highly Potent and Diverse Natural Product Library from African Medicinal Plants. PLoS ONE 2013, 8, e78085. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- da Silva Rocha, S.F.L.; Olanda, C.G.; Fokoue, H.H.; Sant’Anna, C.M.R. Virtual Screening Techniques in Drug Discovery: Review and Recent Applications. Curr. Top. Med. Chem. 2019, 19, 1751–1767. [Google Scholar] [CrossRef]
- Lin, X.; Li, X.; Lin, X. A Review on Applications of Computational Methods in Drug Screening and Design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef] [Green Version]
- Carpio, L.E.; Sanz, Y.; Gozalbes, R.; Barigye, S.J. Computational Strategies for the Discovery of Biological Functions of Health Foods, Nutraceuticals and Cosmeceuticals: A Review. Mol. Divers. 2021, 25, 1425–1438. [Google Scholar] [CrossRef]
- Neves, B.J.; Braga, R.C.; Melo-Filho, C.C.; Moreira-Filho, J.T.; Muratov, E.N.; Andrade, C.H. QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery. Front. Pharmacol. 2018, 9, 1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akamatsu, M. Current State and Perspectives of 3D-QSAR. Curr. Top. Med. Chem. 2002, 2, 1381–1394. [Google Scholar] [CrossRef] [PubMed]
- Rybińska-Fryca, A.; Sosnowska, A.; Puzyn, T. Representation of the Structure—A Key Point of Building QSAR/QSPR Models for Ionic Liquids. Materials 2020, 13, 2500. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Kar, S.; Das, R.N. Chemical Information and Descriptors. In Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment; Elsevier: Amsterdam, The Netherlands, 2015; pp. 47–80. [Google Scholar]
- Dudek, A.Z.; Arodz, T.; Gálvez, J. Computational Methods in Developing Quantitative Structure-Activity Relationships (QSAR): A Review. Comb. Chem. High Throughput Screen 2006, 9, 213–228. [Google Scholar] [CrossRef]
- Cherkasov, A.; Muratov, E.; Fourches, D.; Varnek, A.; Baskin, I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.; Todeschini, R.; et al. QSAR Modeling: Where Have You Been? Where Are You Going To? J. Med. Chem. 2013, 57, 4977–5010. [Google Scholar] [CrossRef] [Green Version]
- Achary, P.G.R. Applications of Quantitative Structure-Activity Relationships (QSAR) Based Virtual Screening in Drug Design: A Review. Mini Rev. Med. Chem. 2020, 20, 1375–1388. [Google Scholar] [CrossRef]
- Pripp, A.H.; Isaksson, T.; Stepaniak, L.; Sørhaug, T.; Ardö, Y. Quantitative Structure Activity Relationship Modelling of Peptides and Proteins as a Tool in Food Science. Trends Food Sci. Technol. 2005, 16, 484–494. [Google Scholar] [CrossRef]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Smyth, M.S.; Martin, J.H. X Ray Crystallography. Mol. Pathol. 2000, 53, 8–14. [Google Scholar] [CrossRef]
- Sugiki, T.; Kobayashi, N.; Fujiwara, T. Modern Technologies of Solution Nuclear Magnetic Resonance Spectroscopy for Three-Dimensional Structure Determination of Proteins Open Avenues for Life Scientists. Comput. Struct. Biotechnol. J. 2017, 15, 328–339. [Google Scholar] [CrossRef]
- Nakane, T.; Kotecha, A.; Sente, A.; McMullan, G.; Masiulis, S.; Brown, P.M.G.E.; Grigoras, I.T.; Malinauskaite, L.; Malinauskas, T.; Miehling, J.; et al. Single-Particle Cryo-EM at Atomic Resolution. Nature 2020, 587, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, X.; Huang, Y.; Wang, C.; Chen, F.; Yang, L.; Ling, L.; Che, Z.; Chen, X. Recent Developments in Molecular Docking Technology Applied in Food Science: A Review. Int. J. Food Sci. Technol. 2020, 55, 33–45. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, W.J.; Balius, T.E.; Mukherjee, S.; Brozell, S.R.; Moustakas, D.T.; Lang, P.T.; Case, D.A.; Kuntz, I.D.; Rizzo, R.C. DOCK 6: Impact of New Features and Current Docking Performance. J. Comput. Chem. 2015, 36, 1132–1156. [Google Scholar] [CrossRef] [Green Version]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking11Edited by F. E. Cohen. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [Green Version]
- Corbeil, C.R.; Williams, C.I.; Labute, P. Variability in Docking Success Rates Due to Dataset Preparation. J. Comput. Aided Mol. Des. 2012, 26, 775–786. [Google Scholar] [CrossRef] [Green Version]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- McGann, M. FRED and HYBRID Docking Performance on Standardized Datasets. J. Comput. Aided Mol. Des. 2012, 26, 897–906. [Google Scholar] [CrossRef] [PubMed]
- McGann, M. FRED Pose Prediction and Virtual Screening Accuracy. J. Chem. Inf. Model. 2011, 51, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Kelley, B.P.; Brown, S.P.; Warren, G.L.; Muchmore, S.W. POSIT: Flexible Shape-Guided Docking for Pose Prediction. J. Chem. Inf. Model. 2015, 55, 1771–1780. [Google Scholar] [CrossRef] [PubMed]
- van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bitencourt-Ferreira, G.; de Azevedo, W.F. Docking with SwissDock. Methods Mol. Biol. 2019, 2053, 189–202. [Google Scholar] [CrossRef]
- Seidel, T.; Bryant, S.D.; Ibis, G.; Poli, G.; Langer, T. 3D Pharmacophore Modeling Techniques in Computer-Aided Molecular Design Using LigandScout. In Tutorials in Chemoinformatics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 279–309. [Google Scholar]
- Yang, S.Y. Pharmacophore Modeling and Applications in Drug Discovery: Challenges and Recent Advances. Drug Discov. Today 2010, 15, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Schaller, D.; Šribar, D.; Noonan, T.; Deng, L.; Nguyen, T.N.; Pach, S.; Machalz, D.; Bermudez, M.; Wolber, G. Next Generation 3D Pharmacophore Modeling. WIREs Comput. Mol. Sci. 2020, 10, e1468. [Google Scholar] [CrossRef] [Green Version]
- Grabowski, M.; Murgueitio, M.S.; Bermudez, M.; Rademann, J.; Wolber, G.; Weindl, G. Identification of a Pyrogallol Derivative as a Potent and Selective Human TLR2 Antagonist by Structure-Based Virtual Screening. Biochem. Pharmacol. 2018, 154, 148–160. [Google Scholar] [CrossRef]
- Grabowski, M.; Murgueitio, M.S.; Bermudez, M.; Wolber, G.; Weindl, G. The Novel Small-Molecule Antagonist MMG-11 Preferentially Inhibits TLR2/1 Signaling. Biochem. Pharmacol. 2020, 171, 113687. [Google Scholar] [CrossRef]
- Barnum, D.; Greene, J.; Smellie, A.; Sprague, P. Identification of Common Functional Configurations Among Molecules. J. Chem. Inf. Comput. Sci. 1996, 36, 563–571. [Google Scholar] [CrossRef]
- Baroni, M.; Cruciani, G.; Sciabola, S.; Perruccio, F.; Mason, J.S. A Common Reference Framework for Analyzing/Comparing Proteins and Ligands. Fingerprints for Ligands and Proteins (FLAP): Theory and Application. J. Chem. Inf. Model. 2007, 47, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Koes, D.R.; Camacho, C.J. Pharmer: Efficient and Exact Pharmacophore Search. J. Chem. Inf. Model. 2011, 51, 1307–1314. [Google Scholar] [CrossRef] [Green Version]
- Dixon, S.L.; Smondyrev, A.M.; Knoll, E.H.; Rao, S.N.; Shaw, D.E.; Friesner, R.A. PHASE: A New Engine for Pharmacophore Perception, 3D QSAR Model Development, and 3D Database Screening: 1. Methodology and Preliminary Results. J. Comput. Aided Mol. Des. 2006, 20, 647–671. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Krieger, J.M.; Li, H.; Bahar, I. Pharmmaker: Pharmacophore Modeling and Hit Identification Based on Druggability Simulations. Protein Sci. 2020, 29, 76–86. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Dror, O.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PharmaGist: A Webserver for Ligand-Based Pharmacophore Detection. Nucleic Acids Res. 2008, 36, W223–W228. [Google Scholar] [CrossRef] [Green Version]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Huang, K.; Miao, M.; Feng, B.; Campanella, O.H. Molecular Dynamics Simulation for Mechanism Elucidation of Food Processing and Safety: State of the Art. Compr. Rev. Food Sci. Food Saf. 2019, 18, 243–263. [Google Scholar] [CrossRef] [Green Version]
- González, M.A. Force Fields and Molecular Dynamics Simulations. École Thématique Société Française Neutron 2011, 12, 169–200. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Tripathi, T. Molecular Dynamics Simulation of Protein and Protein–Ligand Complexes. In Computer-Aided Drug Design; Singh, D.B., Ed.; Springer: Singapore, 2020; pp. 133–161. [Google Scholar]
- Tagde, P.; Tagde, S.; Tagde, P.; Bhattacharya, T.; Monzur, S.M.; Rahman, M.H.; Otrisal, P.; Behl, T.; ul Hassan, S.S.; Abdel-Daim, M.M.; et al. Nutraceuticals and Herbs in Reducing the Risk and Improving the Treatment of COVID-19 by Targeting SARS-CoV-2. Biomedicines 2021, 9, 1266. [Google Scholar] [CrossRef] [PubMed]
- Savant, S.; Srinivasan, S.; Kruthiventi, A.K. Potential Nutraceuticals for COVID-19. Nutr. Diet. Suppl. 2021, 13, 25–51. [Google Scholar] [CrossRef]
- Gyebi, G.A.; Elfiky, A.A.; Ogunyemi, O.M.; Ibrahim, I.M.; Adegunloye, A.P.; Adebayo, J.O.; Olaiya, C.O.; Ocheje, J.O.; Fabusiwa, M.M. Structure-Based Virtual Screening Suggests Inhibitors of 3-Chymotrypsin-Like Protease of SARS-CoV-2 from Vernonia Amygdalina and Occinum Gratissimum. Comput. Biol. Med. 2021, 136, 104671. [Google Scholar] [CrossRef]
- Kodchakorn, K.; Poovorawan, Y.; Suwannakarn, K.; Kongtawelert, P. Molecular Modelling Investigation for Drugs and Nutraceuticals against Protease of SARS-CoV-2. J. Mol. Graph. Model. 2020, 101, 107717. [Google Scholar] [CrossRef]
- Baig, A.; Srinivasan, H. SARS-CoV-2 Inhibitors from Nigella Sativa. Appl. Biochem. Biotechnol. 2022, 194, 1051–1090. [Google Scholar] [CrossRef]
- Gupta, S.; Tejavath, K.K. Nano Phytoceuticals: A Step Forward in Tracking Down Paths for Therapy Against Pancreatic Ductal Adenocarcinoma. J. Clust. Sci. 2022, 1–21. [Google Scholar] [CrossRef]
- Dubey, A.K.; Chaudhry, S.K.; Singh, H.B.; Gupta, V.K.; Kaushik, A. Perspectives on Nano-Nutraceuticals to Manage Pre and Post COVID-19 Infections. Biotechnol. Rep. 2022, 33, e00712. [Google Scholar] [CrossRef] [PubMed]
- Shende, P.; Mallick, C. Nanonutraceuticals: A Way towards Modern Therapeutics in Healthcare. J. Drug Deliv. Sci. Technol. 2020, 58, 101838. [Google Scholar] [CrossRef]
- Ferrado, J.B.; Perez, A.A.; Ruiz, M.C.; León, I.E.; Santiago, L.G. Chrysin-Loaded Bovine Serum Albumin Particles as Bioactive Nanosupplements. Food Funct. 2020, 11, 6007–6019. [Google Scholar] [CrossRef] [PubMed]
- El-Far, A.H.; Godugu, K.; Salaheldin, T.A.; Darwish, N.H.E.; Saddiq, A.A.; Mousa, S.A. Nanonutraceuticals: Anti-Cancer Activity and Improved Safety of Chemotherapy by Costunolide and Its Nanoformulation against Colon and Breast Cancer. Biomedicines 2021, 9, 990. [Google Scholar] [CrossRef] [PubMed]
- Sikk, A.M. Review on Defensive Roles of Thymoquinone Nanobiosensoring Prospective in Opposition to Cancer. J. Cancer Clin. Res. 2021, 4, 297–299. [Google Scholar]
- Rahat, I.; Rizwanullah, M.; Gilani, S.J.; Bin-Jummah, M.N.; Imam, S.S.; Kala, C.; Asif, M.; Alshehri, S.; Sharma, S.K. Thymoquinone Loaded Chitosan—Solid Lipid Nanoparticles: Formulation Optimization to Oral Bioavailability Study. J. Drug Deliv. Sci. Technol. 2021, 64, 102565. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and Prebiotics in Intestinal Health and Disease: From Biology to the Clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Ashaolu, T.J. Emerging Applications of Nanotechnologies to Probiotics and Prebiotics. Int. J. Food Sci. Technol. 2021, 56, 3719–3725. [Google Scholar] [CrossRef]
- Durazzo, A.; Nazhand, A.; Lucarini, M.; Atanasov, A.G.; Souto, E.B.; Novellino, E.; Capasso, R.; Santini, A. An Updated Overview on Nanonutraceuticals: Focus on Nanoprebiotics and Nanoprobiotics. Int. J. Mol. Sci. 2020, 21, 2285. [Google Scholar] [CrossRef] [Green Version]
- Rambaran, T.F. A Patent Review of Polyphenol Nano-Formulations and Their Commercialization. Trends Food Sci. Technol. 2022, 120, 111–122. [Google Scholar] [CrossRef]
- Palla, C.A.; Aguilera-Garrido, A.; Carrín, M.E.; Galisteo-González, F.; Gálvez-Ruiz, M.J. Preparation of Highly Stable Oleogel-Based Nanoemulsions for Encapsulation and Controlled Release of Curcumin. Food Chem. 2022, 378, 132132. [Google Scholar] [CrossRef] [PubMed]
- Harakeh, S.; Qari, Y.; Tashkandi, H.; Almuhayawi, M.; Saber, S.H.; aljahdali, E.; El-Shitany, N.; Shaker, S.; Lucas, F.; Alamri, T.; et al. Thymoquinone Nanoparticles Protect against Cisplatin-Induced Nephrotoxicity in Ehrlich Carcinoma Model without Compromising Cisplatin Anti-Cancer Efficacy. J. King Saud Univ. Sci. 2022, 34, 101675. [Google Scholar] [CrossRef]
- Radwan, M.F.; El-Moselhy, M.A.; Alarif, W.M.; Orif, M.; Alruwaili, N.K.; Alhakamy, N.A. Optimization of Thymoquinone-Loaded Self-Nanoemulsion for Management of Indomethacin-Induced Ulcer. Dose-Response 2021, 19, 155932582110136. [Google Scholar] [CrossRef]
- Alhakamy, N.A.; Badr-Eldin, S.M.; Ahmed, O.A.A.; Aldawsari, H.M.; Okbazghi, S.Z.; Alfaleh, M.A.; Abdulaal, W.H.; Neamatallah, T.; Al-hejaili, O.D.; Fahmy, U.A. Green Nanoemulsion Stabilized by In Situ Self-Assembled Natural Oil/Native Cyclodextrin Complexes: An Eco-Friendly Approach for Enhancing Anticancer Activity of Costunolide against Lung Cancer Cells. Pharmaceutics 2022, 14, 227. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, F.; Maurizi, L.; Forte, J.; Marazzato, M.; Hanieh, P.; Conte, A.; Ammendolia, M.; Marianecci, C.; Carafa, M.; Longhi, C. Resveratrol-Loaded Nanoemulsions: In Vitro Activity on Human T24 Bladder Cancer Cells. Nanomaterials 2021, 11, 1569. [Google Scholar] [CrossRef] [PubMed]
- Javed Iqbal, M.; Quispe, C.; Javed, Z.; Sadia, H.; Qadri, Q.R.; Raza, S.; Salehi, B.; Cruz-Martins, N.; Abdulwanis Mohamed, Z.; Sani Jaafaru, M.; et al. Nanotechnology-Based Strategies for Berberine Delivery System in Cancer Treatment: Pulling Strings to Keep Berberine in Power. Front. Mol. Biosci. 2021, 7, 624494. [Google Scholar] [CrossRef]
- Cristiano, M.C.; Froiio, F.; Mancuso, A.; Cosco, D.; Dini, L.; Di Marzio, L.; Fresta, M.; Paolino, D. Oleuropein-Laded Ufasomes Improve the Nutraceutical Efficacy. Nanomaterials 2021, 11, 105. [Google Scholar] [CrossRef]
- Landucci, E.; Bonomolo, F.; De Stefani, C.; Mazzantini, C.; Pellegrini-Giampietro, D.E.; Bilia, A.R.; Bergonzi, M.C. Preparation of Liposomal Formulations for Ocular Delivery of Thymoquinone: In Vitro Evaluation in HCEC-2 e HConEC Cells. Pharmaceutics 2021, 13, 2093. [Google Scholar] [CrossRef]
- Rachamalla, H.K.; Bhattacharya, S.; Ahmad, A.; Sridharan, K.; Madamsetty, V.S.; Mondal, S.K.; Wang, E.; Dutta, S.K.; Jan, B.L.; Jinka, S.; et al. Enriched Pharmacokinetic Behavior and Antitumor Efficacy of Thymoquinone by Liposomal Delivery. Nanomedicine 2021, 16, 641–656. [Google Scholar] [CrossRef]
- Castangia, I.; Manconi, M.; Allaw, M.; Perra, M.; Orrù, G.; Fais, S.; Scano, A.; Escribano-Ferrer, E.; Ghavam, M.; Rezvani, M.; et al. Mouthwash Formulation Co-Delivering Quercetin and Mint Oil in Liposomes Improved with Glycol and Ethanol and Tailored for Protecting and Tackling Oral Cavity. Antioxidants 2022, 11, 367. [Google Scholar] [CrossRef]
- Ghorbani, S.; Maryam, A. Encapsulation of Lactic Acid Bacteria and Bifidobacteria Using Starch-sodium Alginate Nanofibers to Enhance Viability in Food Model. J. Food Process. Preserv. 2021, 45, e16048. [Google Scholar] [CrossRef]
- Atraki, R.; Azizkhani, M. Survival of Probiotic Bacteria Nanoencapsulated within Biopolymers in a Simulated Gastrointestinal Model. Innov. Food Sci. Emerg. Technol. 2021, 72, 102750. [Google Scholar] [CrossRef]
- Pugliese, R.; Bartolomei, M.; Bollati, C.; Boschin, G.; Arnoldi, A.; Lammi, C. Gel-Forming of Self-Assembling Peptides Functionalized with Food Bioactive Motifs Modulate DPP-IV and ACE Inhibitory Activity in Human Intestinal Caco-2 Cells. Biomedicines 2022, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- Pugliese, R.; Arnoldi, A.; Lammi, C. Nanostructure, Self-Assembly, Mechanical Properties, and Antioxidant Activity of a Lupin-Derived Peptide Hydrogel. Biomedicines 2021, 9, 294. [Google Scholar] [CrossRef]
- Faruk, E.M.; Fouad, H.; Hasan, R.A.A.; Taha, N.M.; El-Shazly, A.M. Inhibition of Gene Expression and Production of INOS and TNF-α in Experimental Model of Neurodegenerative Disorders Stimulated Microglia by Soy Nano-Isoflavone/Stem Cell-Exosomes. Tissue Cell 2022, 76, 101758. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).